Developing an Echocardiography-Based, Automatic Deep Learning Framework for the Differentiation of Increased Left Ventricular Wall Thickness Etiologies
Abstract
1. Introduction
2. Methods
2.1. Population Selection
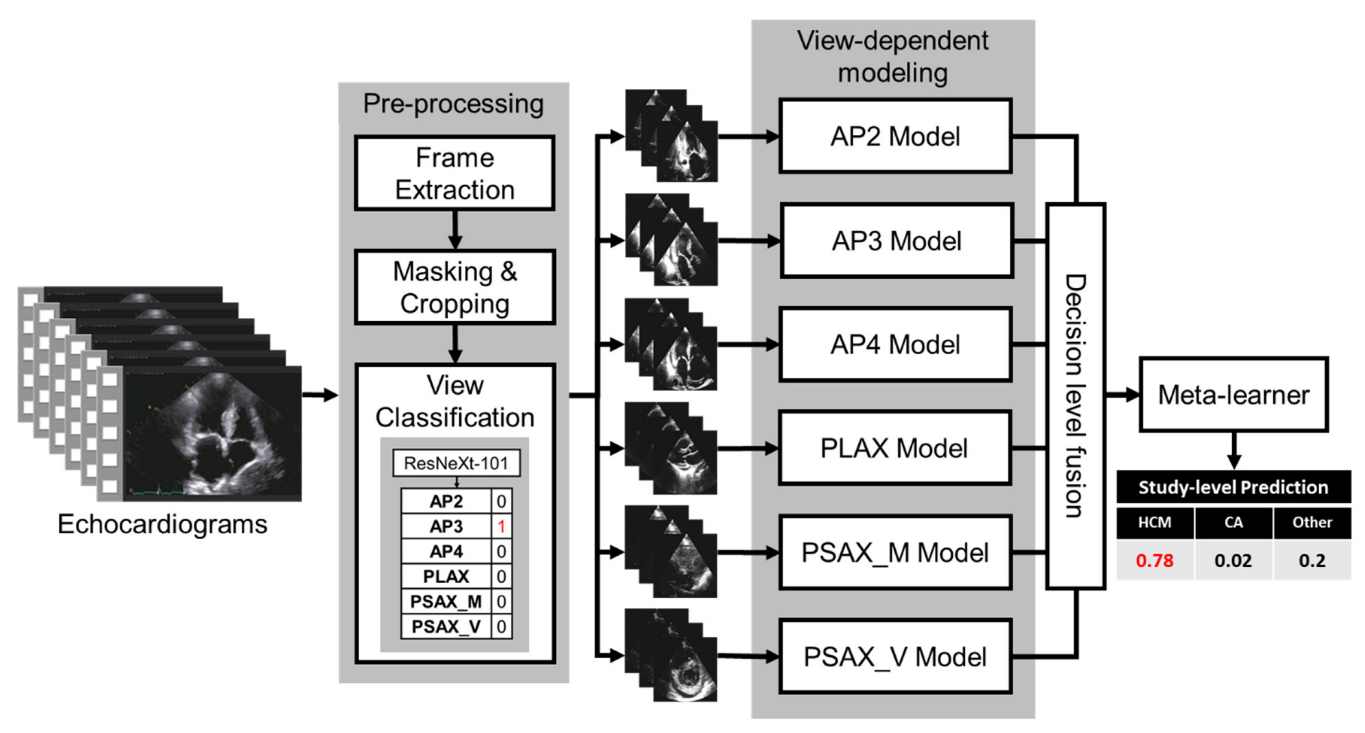
2.2. Proposed Fusion Model Architecture
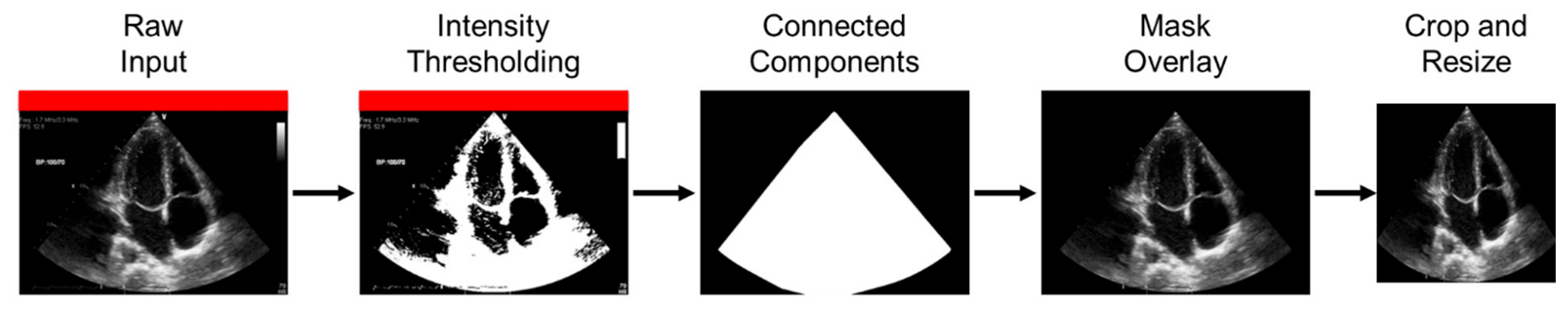
2.3. Preprocessing
2.4. View Classifier
2.5. View-Dependent Modeling Paradigm
2.6. Fusion Model
2.7. Model Performance
Model Interpretability: GRADCAM and the Ablation Study
3. Results
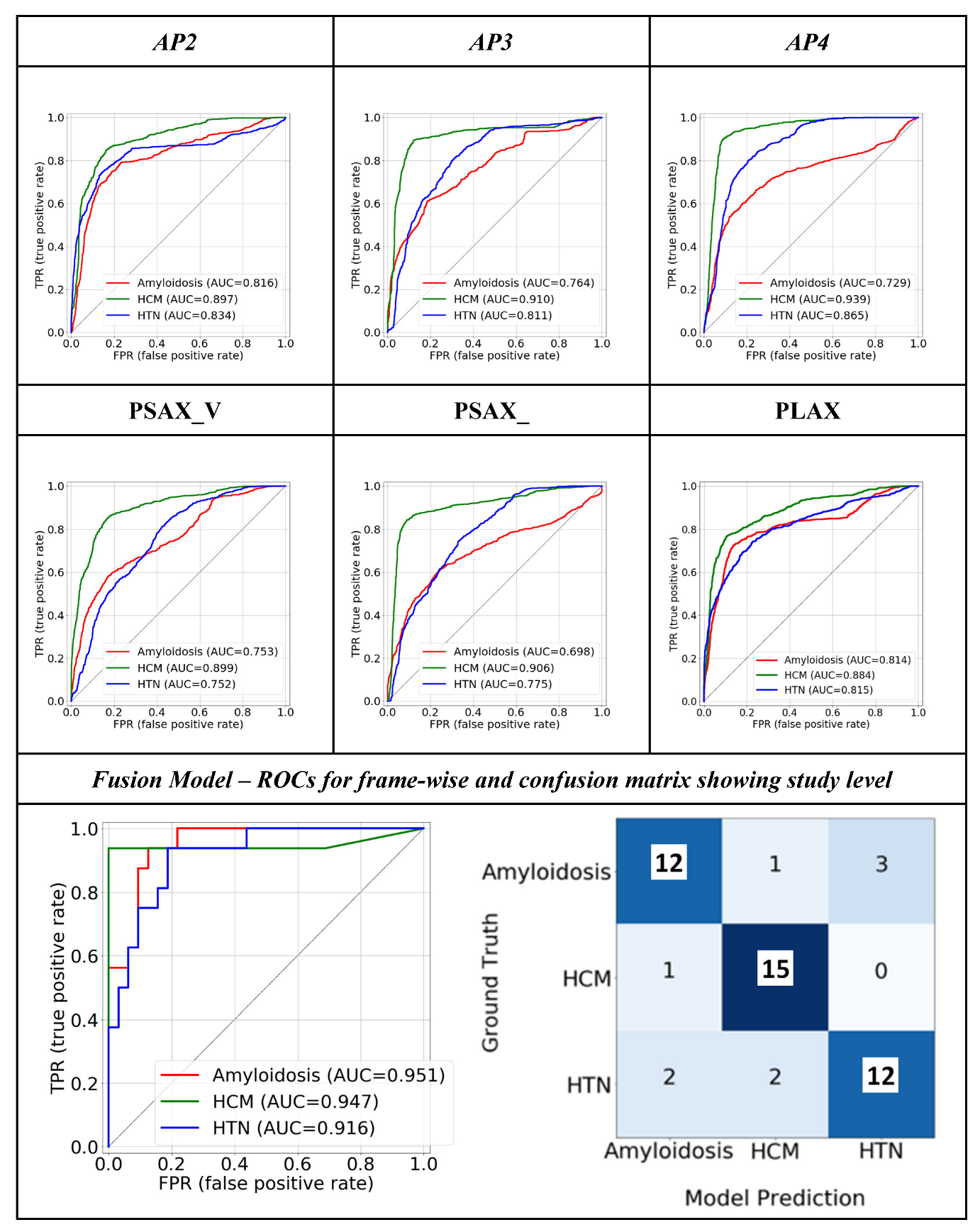
3.1. Quantitative Performance
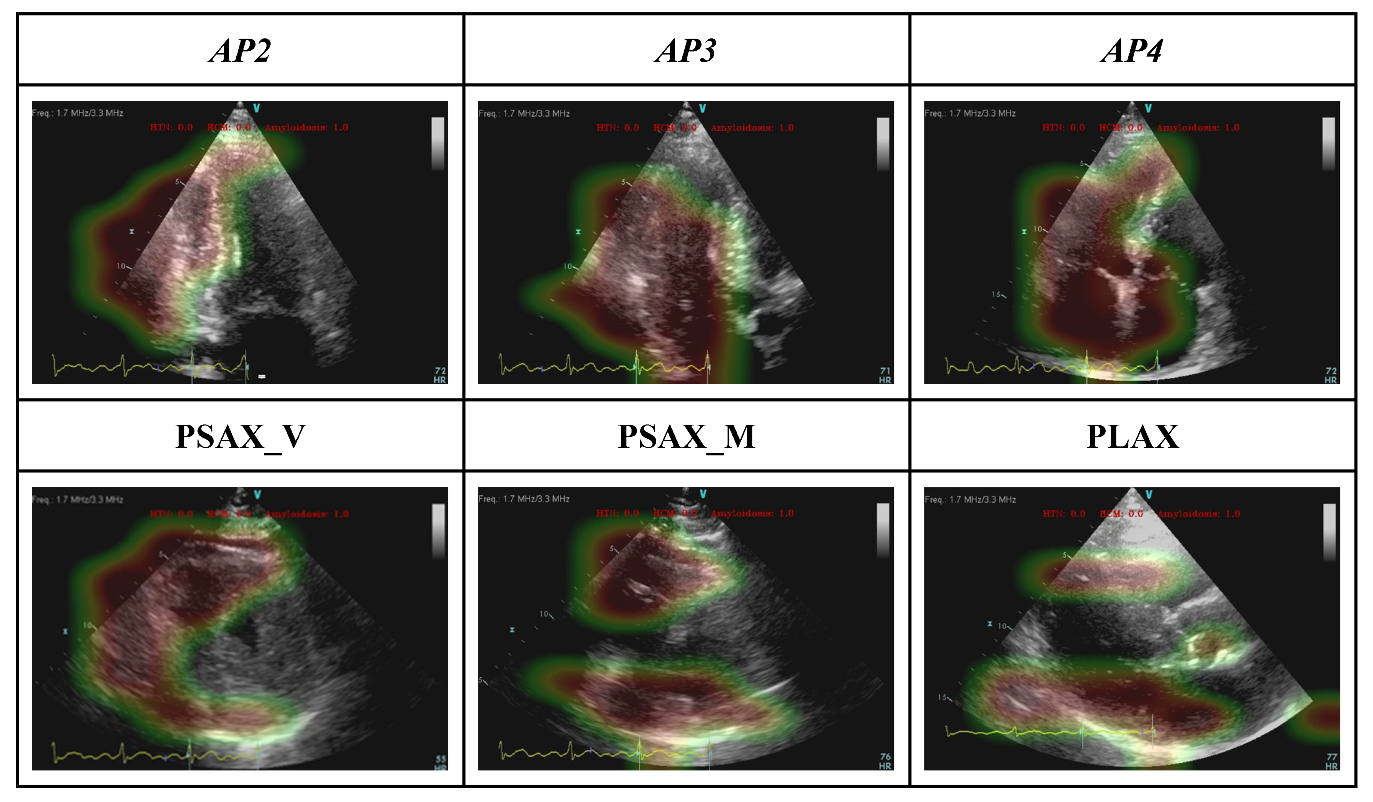
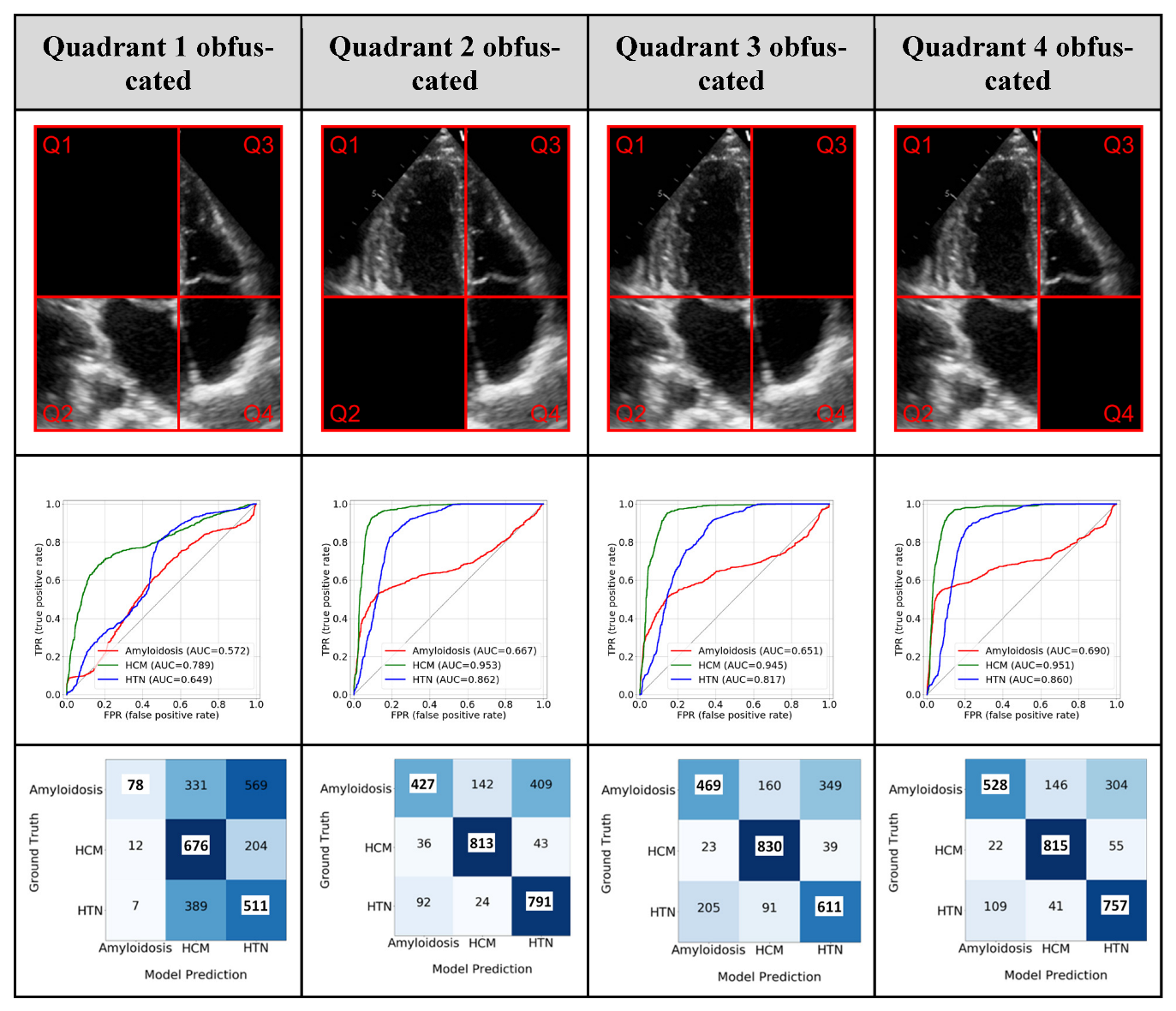
3.2. Model Interpretability: GRADCAM
3.3. Model Interpretability: Ablation Study
3.4. Weights of the Individual Model Outputs Learnt by the Meta Learner
4. Discussion
4.1. An Echo-Based, End-to-End Deep Learning Model with A Fusion Architecture
4.2. Fusion: Using Information from all the Views
4.3. Clinical Application of the Model
5. Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | artificial intelligence |
| AP2 | apical 2-chamber view |
| AP3 | apical 3-chamber view |
| AP4 | apical 4-chamber view |
| AUROC | area under the operating characteristic curve |
| CA | cardiac amyloidosis |
| HCM | hypertrophic cardiomyopathy |
| HTN | hypertensive heart disease |
| LV | left ventricular |
| PLAX | parasternal long-axis view |
| PSAX_V | parasternal short-axis view, at the valve level |
| PSAX_M | parasternal short-axis view, at the mid-ventricular level |
| TTE | transthoracic echocardiography |
References
- Schirmer, H.; Lunde, P.; Rasmussen, K. Prevalence of left ventricular hypertrophy in a general population; The Tromsø Study. Eur. Heart J. 1999, 20, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Cuspidi, C.; Sala, C.; Negri, F.; Mancia, G.; Morganti, A. Prevalence of left-ventricular hypertrophy in hypertension: An updated review of echocardiographic studies. J. Hum. Hypertens. 2012, 26, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Ommen, S.R.; Mital, S.; Burke, M.A.; Day, S.M.; Deswal, A.; Elliott, P.; Evanovich, L.L.; Hung, J.; Joglar, J.A.; Kantor, P.; et al. 2020 AHA/ACC Guideline for the Diagnosis and Treatment of Patients With Hypertrophic Cardiomyopathy. Circulation 2020, 142, e558–e631. [Google Scholar] [PubMed]
- Garcia-Pavia, P.; Rapezzi, C.; Adler, Y.; Arad, M.; Basso, C.; Brucato, A.; Burazor, I.; Caforio, A.L.P.; Damy, T.; Eriksson, U.; et al. Diagnosis and treatment of cardiac amyloidosis: A position statement of the ESC Working Group on Myocardial and Pericardial Diseases. Eur. Heart J. 2021, 42, 1554–1568. [Google Scholar] [CrossRef]
- Kittleson, M.M.; Maurer, M.S.; Ambardekar, A.V.; Bullock-Palmer, R.P.; Chang, P.P.; Eisen, H.J.; Nair, A.P.; Nativi-Nicolau, J.; Ruberg, F.L.; Cardiology, O.; et al. Cardiac Amyloidosis: Evolving Diagnosis and Management: A Scientific Statement From the American Heart Association. Circulation 2020, 142, e7–e22. [Google Scholar] [CrossRef]
- Litjens, G.; Ciompi, F.; Wolterink, J.M.; Vos BD de Leiner, T.; Teuwen, J.; Išgum, I. State-of-the-Art Deep Learning in Cardiovascular Image Analysis. JACC Cardiovasc. Imaging 2019, 12, 1549–1565. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Laak, J.A.W.M.; van der Ginneken, B.; van Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Madani, A.; Ong, J.R.; Tibrewal, A.; Mofrad, M.R.K. Deep echocardiography: Data-efficient supervised and semi-supervised deep learning towards automated diagnosis of cardiac disease. Npj Digit. Med. 2018, 1, 59. [Google Scholar] [CrossRef]
- Kwon, J.; Kim, K.; Jeon, K.; Park, J. Deep learning for predicting in-hospital mortality among heart disease patients based on echocardiography. Echocardiogr 2019, 36, 213–218. [Google Scholar] [CrossRef]
- Østvik, A.; Salte, I.M.; Smistad, E.; Nguyen, T.M.; Melichova, D.; Brunvand, H.; Haugaa, K.; Edvardsen, T.; Grenne, B.; Lovstakken, L. Myocardial Function Imaging in Echocardiography Using Deep Learning. IEEE Trans. Med. Imaging 2021, 40, 1340–1351. [Google Scholar] [CrossRef]
- Wahlang, I.; Maji, A.K.; Saha, G.; Chakrabarti, P.; Jasinski, M.; Leonowicz, Z.; Jasinska, E. Deep Learning Methods for Classification of Certain Abnormalities in Echocardiography. Electronics 2021, 10, 495. [Google Scholar] [CrossRef]
- Ghorbani, A.; Ouyang, D.; Abid, A.; He, B.; Chen, J.H.; Harrington, R.A.; Liang, D.H.; Ashley, E.A.; Zou, J.Y. Deep learning interpretation of echocardiograms. Npj Digit. Med. 2020, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Duffy, G.; Cheng, P.P.; Yuan, N.; He, B.; Kwan, A.C.; Shun-Shin, M.J.; Alexander, K.M.; Ebinger, J.; Lungren, M.P.; Rader, F.; et al. High-Throughput Precision Phenotyping of Left Ventricular Hypertrophy with Cardiovascular Deep Learning. JAMA Cardiol. 2022, 7, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Yao, X.; Wu, B.; Zhou, H.; Xia, S.; Su, W.; Wu, Y.; Zheng, X. Using deep learning method to identify left ventricular hypertrophy on echocardiography. Int J. Cardiovasc Imaging 2021, 38, 759–769. [Google Scholar] [CrossRef]
- Zhang, J.; Gajjala, S.; Agrawal, P.; Tison, G.H.; Hallock, L.A.; Beussink, N.L.; Lassen, M.H.; Fan, E.; Aras, M.A.; Jordan, C.; et al. Fully Automated Echocardiogram Interpretation in Clinical Practice. Circulation 2018, 138, 1623–1635. [Google Scholar] [CrossRef]
- Luckie, M.; Khattar, R.S. Systolic anterior motion of the mitral valve—Beyond hypertrophic cardiomyopathy. Heart 2008, 94, 1383. [Google Scholar] [CrossRef]
- Huang, S.-C.; Pareek, A.; Zamanian, R.; Banerjee, I.; Lungren, M.P. Multimodal fusion with deep neural networks for leveraging CT imaging and electronic health record: A case-study in pulmonary embolism detection. Sci. Rep. 2020, 10, 22147. [Google Scholar] [CrossRef]
- Huang, S.-C.; Pareek, A.; Seyyedi, S.; Banerjee, I.; Lungren, M.P. Fusion of medical imaging and electronic health records using deep learning: A systematic review and implementation guidelines. Npj Digit. Med. 2020, 3, 136. [Google Scholar] [CrossRef]
- Sengupta, P.P.; Shrestha, S.; Berthon, B.; Messas, E.; Donal, E.; Tison, G.H.; Min, J.K.; D’hooge, J.; Voigt, J.-U.; Dudley, J.; et al. Proposed Requirements for Cardiovascular Imaging-Related Machine Learning Evaluation (PRIME): A Checklist Reviewed by the American College of Cardiology Healthcare Innovation Council. JACC Cardiovasc. Imaging 2020, 13, 2017–2035. [Google Scholar] [CrossRef]
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated Residual Transformations for Deep Neural Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 5987–5995. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. Arxiv 2016. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks Via Gradient-Based Localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Lang, R.M.; Badano, L.P.; Mor-Avi, V.; Afilalo, J.; Armstrong, A.; Ernande, L.; Flachskampf, F.A.; Foster, E.; Goldstein, S.A.; Kuznetsova, T.; et al. Recommendations for Cardiac Chamber Quantification by Echocardiography in Adults: An Update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. Eur. Heart J. Cardiovasc. Imaging 2015, 16, 233–271. [Google Scholar] [CrossRef]
- Nagueh, S.F.; Bierig, S.M.; Budoff, M.J.; Desai, M.; Dilsizian, V.; Eidem, B.; Goldstein, S.A.; Hung, J.; Maron, M.S.; Ommen, S.R.; et al. American Society of Echocardiography Clinical Recommendations for Multimodality Cardiovascular Imaging of Patients with Hypertrophic Cardiomyopathy Endorsed by the American Society of Nuclear Cardiology, Society for Cardiovascular Magnetic Resonance, and Society of Cardiovascular Computed Tomography. J. Am. Soc. Echocardiog 2011, 24, 473–498. [Google Scholar]
- Chen, C.; Qin, C.; Qiu, H.; Tarroni, G.; Duan, J.; Bai, W.; Rueckert, D. Deep Learning for Cardiac Image Segmentation: A Review. Front. Cardiovasc. Med. 2020, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Pagourelias, E.D.; Mirea, O.; Duchenne, J.; Cleemput, J.V.; Delforge, M.; Bogaert, J.; Kuznetsova, T.; Voigt, J.-U. Echo Parameters for Differential Diagnosis in Cardiac Amyloidosis. Circ. Cardiovasc. Imaging 2017, 10, e005588. [Google Scholar] [CrossRef] [PubMed]
- Nagata, Y.; Takeuchi, M.; Mizukoshi, K.; Wu, V.C.-C.; Lin, F.-C.; Negishi, K.; Nakatani, S.; Otsuji, Y. Intervendor Variability of Two-Dimensional Strain Using Vendor-Specific and Vendor-Independent Software. J. Am. Soc. Echocardiog 2015, 28, 630–641. [Google Scholar] [CrossRef] [PubMed]
- Risum, N.; Ali, S.; Olsen, N.T.; Jons, C.; Khouri, M.G.; Lauridsen, T.K.; Samad, Z.; Velazquez, E.J.; Sogaard, P.; Kisslo, J. Variability of Global Left Ventricular Deformation Analysis Using Vendor Dependent and Independent Two-Dimensional Speckle-Tracking Software in Adults. J. Am. Soc. Echocardiog 2012, 25, 1195–1203. [Google Scholar] [CrossRef] [PubMed]


| Characteristics | Subtypes | HCM (305) | Amyloidosis (244) | HTN/Others (254) |
|---|---|---|---|---|
| Age | 58.44 (+/−15.05) | 69.25 (+/−11.15) | 64.25 (+/−14.31) | |
| Gender | Male | 175 (57.37%) | 196 (80.33%) | 178 (70.08%) |
| Female | 130 (42.62%) | 48 (19.67%) | 76 (29.92%) | |
| Race | White | 256 (83.93%) | 214 (87.7%) | 218 (85.83%) |
| Black or African American | 17 (5.57%) | 12 (4.92%) | 16 (6.3%) | |
| American Indian | 4 (1.31%) | 2 (0.81%) | 2 (0.79%) | |
| Asian | 9 (2.95%) | 2 (0.81%) | 9 (3.54%) | |
| Other/Unknown | 19 (6.23%) | 14 (5.73%) | 9 (3.54%) | |
| Ethnicity | Hispanic or Latino | 10 (3.28%) | 12 (4.91%) | 17 (6.69%) |
| Not Hispanic or Latino | 276 (90.49%) | 224 (91.8%) | 227 (89.37%) | |
| Unknown | 19 (6.23%) | 8 (3.27%) | 10 (3.94%) | |
| Comorbidities at the time of TTE | Hypertension | 153 (38.73%) | 82 (33.6%) | 116 (45.67%) |
| Coronary Artery Disease | 71 (23.27%) | 70 (28.68%) | 71 (27.95%) | |
| Diabetics (Type I and Type II) | 37 (9.37%) | 21 (8.6%) | 41 (16.14%) | |
| Chronic Kidney Disease | 45 (11.4%) | 51 (20.9%) | 35 (13.78%) | |
| Congestive Heart failure | 4 (1.01%) | 2 (0.81%) | 3 (1.18%) | |
| Single View Models | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| AP2 | AP3 | AP4 | |||||||
| Precision | Recall | F1-Score | Precision | Recall | F1-Score | Precision | Recall | F1-Score | |
| CA | 0.68 [±0.0067] | 0.72 [±0.0047] | 0.70 [±0.0046] | 0.64 [±0.0054] | 0.49 [±0.0045] | 0.56 [±0.0041] | 0.63 [±0.0076] | 0.58 [±0.0083] | 0.61 [±0.0067] |
| HCM | 0.75 [±0.0048] | 0.81 [±0.0044] | 0.78 [±0.0036] | 0.82 [± 0.0045] | 0.85 [±0.0046] | 0.83 [±0.0036] | 0.77 [±0.0067] | 0.93 [±0.0047] | 0.84 [±0.0056] |
| HTN/others | 0.76 [±0.0015] | 0.64 [±0.0016] | 0.70 [±0.0013] | 0.59 [± 0.0017] | 0.71 [±0.0014] | 0.65 [±0.0015] | 0.73 [±0.0016] | 0.64 [±0.0017] | 0.68 [±0.0013] |
| PLAX | PSAX_V | PSAX_M | |||||||
| Precision | Recall | F1-Score | Precision | Recall | F1-Score | Precision | Recall | F1-Score | |
| CA | 0.70 [±0.0069] | 0.70 [±0.0051] | 0.72 [±0.0034] | 0.57 [±0.0069] | 0.62 [±0.0053] | 0.59 [±0.0054] | 0.57 [±0.0057] | 0.58 [±0.0050] | 0.57 [±0.0042] |
| HCM | 0.85 [±0.0044] | 0.66 [±0.0041] | 0.74 [±0.0033] | 0.76 [±0.0046] | 0.78 [±0.0042] | 0.77 [±0.0036] | 0.87 [±0.0039] | 0.78 [±0.0049] | 0.82 [±0.0029] |
| HTN/others | 0.62 [±0.0017] | 0.73 [±0.0017] | 0.67 [±0.0014] | 0.58 [±0.0019] | 0.52 [±0.0018] | 0.55 [±0.0016] | 0.56 [±0.0018] | 0.60 [±0.0017] | 0.58 [±0.0015] |
| Fusion Model | |||||||||
| Precision | Recall | F1-Score | |||||||
| CA | 0.80 [±0.0167] | 0.75 [±0.0165] | 0.77 [±0.0134] | ||||||
| HCM | 0.83 [±0.0143] | 0.94 [±0.009] | 0.88 [±0.0100] | ||||||
| HTN/others | 0.80 [±0.0151] | 0.75 [±0.01473] | 0.77 [±0.01394] | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Chao, C.-J.; Jeong, J.J.; Farina, J.M.; Seri, A.R.; Barry, T.; Newman, H.; Campany, M.; Abdou, M.; O’Shea, M.; et al. Developing an Echocardiography-Based, Automatic Deep Learning Framework for the Differentiation of Increased Left Ventricular Wall Thickness Etiologies. J. Imaging 2023, 9, 48. https://doi.org/10.3390/jimaging9020048
Li J, Chao C-J, Jeong JJ, Farina JM, Seri AR, Barry T, Newman H, Campany M, Abdou M, O’Shea M, et al. Developing an Echocardiography-Based, Automatic Deep Learning Framework for the Differentiation of Increased Left Ventricular Wall Thickness Etiologies. Journal of Imaging. 2023; 9(2):48. https://doi.org/10.3390/jimaging9020048
Chicago/Turabian StyleLi, James, Chieh-Ju Chao, Jiwoong Jason Jeong, Juan Maria Farina, Amith R. Seri, Timothy Barry, Hana Newman, Megan Campany, Merna Abdou, Michael O’Shea, and et al. 2023. "Developing an Echocardiography-Based, Automatic Deep Learning Framework for the Differentiation of Increased Left Ventricular Wall Thickness Etiologies" Journal of Imaging 9, no. 2: 48. https://doi.org/10.3390/jimaging9020048
APA StyleLi, J., Chao, C.-J., Jeong, J. J., Farina, J. M., Seri, A. R., Barry, T., Newman, H., Campany, M., Abdou, M., O’Shea, M., Smith, S., Abraham, B., Hosseini, S. M., Wang, Y., Lester, S., Alsidawi, S., Wilansky, S., Steidley, E., Rosenthal, J., ... Banerjee, I. (2023). Developing an Echocardiography-Based, Automatic Deep Learning Framework for the Differentiation of Increased Left Ventricular Wall Thickness Etiologies. Journal of Imaging, 9(2), 48. https://doi.org/10.3390/jimaging9020048






