Abstract
The paper explored the problem of automatic diagnosis based on immunohistochemical image analysis. The issue of automated diagnosis is a preliminary and advisory statement for a diagnostician. The authors studied breast cancer histological and immunohistochemical images using the following biomarkers progesterone, estrogen, oncoprotein, and a cell proliferation biomarker. The authors developed a breast cancer diagnosis method based on immunohistochemical image analysis. The proposed method consists of algorithms for image preprocessing, segmentation, and the determination of informative indicators (relative area and intensity of cells) and an algorithm for determining the molecular genetic breast cancer subtype. An adaptive algorithm for image preprocessing was developed to improve the quality of the images. It includes median filtering and image brightness equalization techniques. In addition, the authors developed a software module part of the HIAMS software package based on the Java programming language and the OpenCV computer vision library. Four molecular genetic breast cancer subtypes could be identified using this solution: subtype Luminal A, subtype Luminal B, subtype HER2/neu amplified, and basalt-like subtype. The developed algorithm for the quantitative characteristics of the immunohistochemical images showed sufficient accuracy in determining the cancer subtype “Luminal A”. It was experimentally established that the relative area of the nuclei of cells covered with biomarkers of progesterone, estrogen, and oncoprotein was more than 85%. The given approach allows for automating and accelerating the process of diagnosis. Developed algorithms for calculating the quantitative characteristics of cells on immunohistochemical images can increase the accuracy of diagnosis.
1. Introduction
According to the American Statistical Register, in 2020, there were approximately 2.3 million new breast cancer cases and 685,000 breast cancer death cases worldwide. Breast cancer incidence and mortality varied among countries, with age-standardized incidence ranging from the highest of 112.3 per 100,000 population in Belgium to the lowest of 80 per 100,000 population in Iran, and the age-standardized mortality was from the highest of 41.0 per 100,000 population on the island of Fiji to the lowest of 6.4 per 100,000 population in South Korea.
The disease analysis that has been conducted since 2000 revealed that the peak age of breast cancer in some countries in Asia and Africa was ten years earlier than in Europe or America. Regarding breast cancer tendencies, the age-standardized incidence rates increased significantly in China and South Korea and decreased in the United States between 2000 and 2012. Meanwhile, the age-standardized mortality rates increased substantially in China and South Korea but decreased in the United Kingdom, the United States, and Australia between 2000 and 2015. Analyzing the data, we can conclude that regardless of the region of residence, the problem of cancer is relevant and requires tools for diagnosis.
Immunohistochemistry is one of the effective tools for breast cancer preoperative diagnosis.
Immunohistochemistry (IHC) is used to find specific protein products, observe the unique characteristics of breast cancer, differentiate breast cancer from cancer in other locations, obtain information about genetic changes, study prognostic factors, and provide their effective treatment. Technical developments allow for the use of IHC as a comfortable diagnostic tool and its application in advanced protocols in combination with other histochemical methods. Standardization and quality control are critical issues for the acceptable use of IHC in breast cancer practice.
The American Society of Clinical Oncology and the College of American Pathologists convened an international expert group that conducted a systematic review and evaluation of the literature in partnership with Cancer Care Ontario and developed recommendations for optimal ER/PgR test efficiency. The Commission recommends determining the status of ER and PgR for all invasive breast cancers and recurrent breast cancers. It is recommended that ER and PgR tests be considered positive if the sample contains at least 1% positive tumor nuclei during testing in the typical reactivity of internal (usual epithelial elements) and external controls.
Biomedical images cannot be archived. Therefore, the image is uploaded into memory in its original format. The next stage is preliminary processing including input parameter identification, adaptive filtering, and brightness/contrast adjusting.
Images obtained from a microscope are characterized by noise and the absence of precise contours of cell nuclei. Therefore, it is necessary to develop an algorithm for image preprocessing. The ultimate goal of the preprocessing stage is to remove impulse noise and align the histogram.
Thus, the results of the immunohistochemical study are immunohistochemical images. Immunohistochemical images are processed using artificial intelligence for diagnosis.
Artificial intelligence is widely used in medicine. These can be both software (stationary and mobile) and hardware modules and devices, which allows for the speeding up and automation of diagnostics. The relevance of breast cancer research and the use of AI for these tasks are explored in the research study [].
With hardware development, new software tools appeared that use immunohistochemical images to make a diagnosis. Scientists have also paid great attention to the development of software that combines algorithms for image processing, segmentation, object detection, etc. Analysis of the latest publications in this area is provided in Section 2.
Modern software complexes such as ImageJ are characterized by a large number of tools for the manual or semi-automatic image processing of experimental samples. The disadvantages of this system are the need for computer vision knowledge and time for complex and monotonous work. In addition, some programs have functionality that allows highlighting only areas with cell nuclei. Therefore, diagnosticians need to independently calculate the cell nuclei parameters. We offer a software package that allows for automatic calculation of the micro-objects’ quantitative characteristics in immunohistochemical images. To do this, the program uses a knowledge base for preprocessing and segmentation algorithms. The result of the program is the identification of the disease subtype. Taking into account the current trends in machine learning, in future research, it is planned to use neural networks with the U-net architecture for automatic image segmentation, which will allow for more accurate identification of micro-objects in the image.
The main contribution of this paper can be summarized as follows:
- We developed an algorithm for image preprocessing that was based on adaptive median filtering with experimental determination of the image noise level, and identification of the filter window size, which allowed for a reduction in the impulse noise level on the input image;
- We proposed a combined segmentation algorithm based on the watershed and threshold segmentation algorithms to calculate the area and identify the cell staining intensity. It will allow for the determination of informative indicators for breast cancer subtype identification;
- We developed a method of the automatic statement of specified diagnosis based on the preliminary processing algorithms and histological and immunohistochemical image segmentation using brightness indicators and relative area. This made it possible to determine the breast cancer subtype automatically;
- We developed a software module within the HIAMS software system, implemented in the Java programming language using the OpenCV computer vision library.
The article consists of the following structural parts: (1) Introduction: the relevance of the immunohistochemical study of breast cancer is shown. (2) Literature analysis: publications on automatic diagnosis are analyzed. (3) Materials and methods: the method of diagnosis based on the image immunohistochemical analysis is described. (4) Results, comparisons, discussions: the developed module of automatic diagnosis and network data is described, and the developed system’s comparative analysis with known ones is carried out. (5) Conclusions: our conclusions are presented.
2. Literature Review
Let us overview the artificial intelligence tools to analyze immunohistochemical images and automated diagnosis.
In [], the authors investigated the possibility of automated breast cancer diagnosis. Immunohistochemical (IHC) images, image segmentation algorithms, and neural network methods were analyzed in [,]. An automatic breast cancer diagnosis was performed using textural features, entropy, and classifiers SVM, CNN, and DTree. These methods of automation and data analysis also require time and hardware. However, their efficiency and speed are better than manual diagnostics based on biomarkers [,,,,]. There are several problems with the accuracy and reliability of the dataset, data gaps, noise, anomalies, etc. The use of biomarkers in digital image analysis was considered in [].
The significant contribution of scientists to the development of algorithms for the automatic selection and calculation of cell nuclei parameters emphasizes the importance of this problem. The use of biomarkers allows for the highlighting of necessary nuclei in the image. However, the absence of clear contours, touching and crossing of nuclei in the immunohistochemical image causes the development of preprocessing and segmentation algorithms.
Existing means of artificial intelligence do not provide sufficient prediction accuracy in diagnosis. The use of deep learning methods to classify many classes of pathologies based on image analysis was studied in [,,]. CNN architecture optimization using bio-inspired algorithms for breast cancer detection was considered in [].
In [], the authors investigated the segmentation and classification of the IHC image nuclei using biomarkers. The authors used two semi-automatic software: NuclearQuant v. 1.13 and Pannoramic Viewer v. 1.13. 1.14. Each of these programs determines the status of biomarkers on the analyzed micro-object.
Software tools for breast cancer diagnosis were discussed in []. The authors defined a system of diagnosis and prognosis of the disease, Diaprog, which used the data of the classified and archived care records. In this study, a significant emphasis was placed on data processing, however, the algorithm for calculating the quantitative cell nuclei characteristics and making a diagnosis based on them is not given.
The article in [] was devoted to the software tool for analyzing IHC images for the quantitative assessment of tissue pathology. The authors developed an automated IHC_Tool procedure with TIFF images at a magnification of _200 to quantify the cell traits. Automatic classification of cancer cells using machine learning was analyzed in []. The method of automated machine learning for differentiation of the invasion method was formed in the article.
In [], the authors described the use of a Faster R-CNN object detector with four function extractors: Resnet-50, VGG-16, Inception-V2, and Resnet-101 for automatic lymphocyte detection and counting. In [], the authors presented an automatic computer-aided diagnosis system based on the Multimodal fusion of Breast Cancer (MF-CAD).
In [], the researchers investigated new methods of segmentation and calculation of the IHC image cancer cell nuclei. These methods segment nuclei based on modified superpixel segmentation. In [], IHC methods for the use of immunotherapy in oncology were explored. The authors described various highly multiplexed methods that allow for the simultaneous detection of multiple markers on a single tissue section. The authors in [] discussed the methods of analysis of IHC images based on deep learning using CNN and U-Net. However, only a few studies have focused on automatic diagnosis using many biomarkers.
Thus, these articles provided the impetus to develop algorithms and software systems to speed up and improve the quality of the diagnosing process. However, these programs do not provide a diagnosis based on the immunohistochemical images of different types of biomarkers. In addition, there is a need to develop universal segmentation algorithms for different types of images.
The authors in [] analyzed classical and new approaches for automatic diagnosis in oncology using machine learning. A broad analysis of classical and in-depth teaching methods used in the histological image analysis of images was presented. In articles [,], the technique of image analysis based on the pathologist-tree network was investigated, and an automatic system of analysis of the IHC images was developed. In addition, an automated rapid visualization system using a synchronized 12-LED illuminator was developed.
Feasibility in the quantification of Ki-67, ER, PR, and HER2 biomarkers was proven in []. However, the article did not provide algorithms and software tools for diagnosis based on the analyzed biomarkers.
In [,], the approach to automatic biomedical image segmentation using U-Net convolutional neural network technology is presented. However, in this case, taking into account the specifics of immunohistochemical images, a different approach to segmentation was chosen. An adaptive method of biomedical image segmentation based on metrics was developed in []. This approach uses the rules based on segmentation algorithms. In [,,,,,], strategies to analyze biomedical images based on data processing algorithms are presented. The research findings demonstrate approaches to the development of adaptive methods of cytological and histological image preprocessing and segmentation based on fuzzy logic. However, there is a need to develop alternative methods of immunohistochemical processing and the automatic identification of the cancer subtype based on the obtained indicators.
3. Materials and Methods
We used the following symbols to describe the method of the diagnostic statement:
- —
- Pr is progesterone;
- —
- Er is estrogen;
- —
- HER2/neu is the oncoprotein;
- —
- Ki-67 is the cell proliferation biomarker;
- —
- Sw is the area of a field of view window;
- —
- Sp is the area of positive cells in the field of view;
- —
- δs is the ratio of the area of positive cells in the field of view to the area of the field of view window;
- —
- KI is the color intensity coefficient;
- —
- is the degree of tumor differentiation based on the histological image analysis;
- —
- is the subtype Luminal A of breast cancer (BC);
- —
- is the BC subtype Luminal B;
- —
- is the BC amplified subtype;
- —
- is the BC subtype basaltic;
- —
- is the relative area of the cell nuclei (estrogen biomarker);
- —
- is the relative area of the cell nuclei (progesterone biomarker);
- —
- is the relative area of the cell nuclei (biomarker oncoprotein);
- —
- is the relative area of the cell nuclei (a biomarker of cell proliferation);
- —
- is the level of color intensity of the cell nuclei (biomarker estrogen);
- —
- is the level of color intensity of the cell nuclei (a biomarker of cell proliferation);
- —
- TL is the lower segmentation threshold (thresholding);
- —
- TH is the upper segmentation threshold (thresholding).
3.1. Method of Diagnostic Statement Based on Immunohistochemical Image Analysis
To make a diagnosis, we used the histological images IG. As a result of the action of biomarkers such as progesterone, estrogen, oncoprotein, and the cell proliferation biomarker, for each histological image, we obtained four immunohistochemical images: . Thus, the input is the following set of images:
To make an accurate specified diagnosis, we analyzed the immunohistochemical images in two leading indicators, and .
The method of the specified diagnosis consists of the following steps:
Image preprocessing.
Each of these images is a microscopic image with pulsed noise. Therefore, it is necessary to preprocess the images. To do this, we calculated the peak signal-to-noise ratio (PSNR).
Let I be the input image (histological or immunohistochemical image). As a result of median filtering over the input image, we obtain:
where I is the input image and is the result of the median filtering.
The next step is to quantify the noise level of the image. We used the value of the peak signal-to-noise ratio (PSNR) [].
We calculated the standard deviation (MSE) between the two images to calculate this value.
where and are the filtered original images, respectively, size m × n. The value of PSNR is determined as follows:
where is the maximum value accepted by the pixel of the image. Experimental studies have established the following parameters of the median filter window:
where mw is the size of the median filter window.
To reduce the level of impulse noise, we used a median filter with a window size of mw. We present the image filtering as follows:
where is the input image; mw is the filter window; is the image after filtering.
To calculate the average brightness of the image, we used the following transformation:
where n is the total number of pixels in the image; Ri, Gi, Bi are the values of red, green, and blue channels; and the th pixel of the image, respectively [].
The following parameters α were selected experimentally depending on Y’s average brightness level. Using the α parameter, we adjusted the image brightness. Therefore, this image was better processed during the segmentation. Selection of the parameter was carried out by experimental selection of the value of α with step 2 and further analysis of the obtained image after segmentation.
Based on the defined parameter α, we performed the following image transformation
3.2. Segmentation and Calculation of Cell Staining Intensity Area
At this stage, we segmented the obtained images to determine the cell staining intensity area. We chose a combination of a watershed algorithm based on markers and a threshold. As a result of the algorithm use, we obtained a mask with a segmented image, where the pixels of one segment were marked with the same label and formed a connected area. The main disadvantage of this algorithm is the use of a pre-processing procedure for images with a large number of local minima. The standard watershed algorithm of the OpenCV library was selected as the basis of the algorithm, which includes elements of threshold segmentation, erode and dilate operations, and the generation of markers. The main emphasis was placed on the choice of the lower and upper thresholds.
Each image type is unique. Therefore, the lower (TL) and upper (TH) segmentation thresholds for a specific image type were experimentally selected:
We present the segmentation by the watershed method as follows:
The next step is to calculate the ratio of the positive cell area in the field of view to the area of the field of the view window.
To calculate the intensity of cells, we imposed the segmented image on the input image and calculated the intensity of the selected areas.
The image intensity Ki was calculated according to the scale from 1 to 3. In usual practice, diagnosticians describe the intensity with the words “high”, “medium”, and “low” based on their own subjective experience. To develop an automatic system, we converted qualitative characteristics into quantitative ones. The sources of information were the image samples and descriptions made by diagnosticians.
3.3. Determination of Breast Cancer Molecular Genetic Subtype
The degree of tumor differentiation is divided into three types:
- —a highly differentiated tumor;
- —a moderately differentiated tumor;
- —a low differentiated tumor.
The molecular genetic subtype of breast cancer is determined based on four biomarkers.
Luminal subtype A is characterized for , and is defined by the following system of features:
Luminal subtype B is characterized for , and is defined by the following system of features:
The amplified subtype HER2/neu is characterized for and defined by the following system of features:
The following system of features defines the basal-like subtype:
4. Results, Comparison, and Discussion
4.1. Dataset Description
For the computer experiments, the immunohistochemical image dataset of breast cancer was used []. Table 1 shows the parameters of the selected dataset.

Table 1.
Characteristics of an immunohistochemical image dataset.
The sample for automatic segmentation was divided into training and tests in a 60 to 40 percent ratio.
4.2. Software Module Structure
The software module was implemented using the Java programming language and the openCV library. Figure 1 shows a generalized structure.
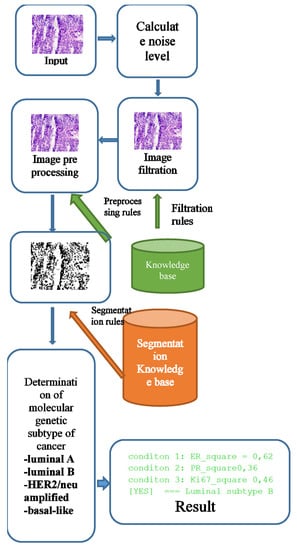
Figure 1.
Software module of the generalized structure.
The knowledge base for the selection of filtering and segmentation algorithms was implemented in the form of “IF–THEN” type rules. An example of the rules for selecting the parameters of the filtering algorithms and the brightness level adjustment parameters is given in Section 3.1. The filter algorithm was selected according to the following rules:
- IF the peak signal-to-noise ratio <= 20, THEN the median filter window = 5 × 5;
- IF the peak signal-to-noise ratio >20, THEN the median filter window = 3 × 3;
The rules for choosing the parameters of the segmentation algorithms are given in Section 3.2. The rules in “IF_THEN” format are as follows:
- IF Image Type = progesterone THEN thresholds lower = 160 AND thresholds upper = 180;
- IF Image Type = estrogen THEN thresholds lower = 180 AND thresholds upper = 210;
- IF Image Type = oncoprotein THEN thresholds lower = 40 AND thresholds upper = 230;
- IF Image Type = cell proliferation biomarker THEN thresholds lower = 160 AND thresholds upper = 180.
The input parameters were the following image characteristics: noise level, peak signal-to-noise ratio, and image type (Her2/neu, Ki-67, Er, Pr). A filtering or segmentation algorithm and its parameters were provided for each range of input values. Training was carried out by testing images with predefined parameters. This stage is computationally complex and requires parallelization. The best results were recorded into the knowledge base. When a new image is received, the parameters of the algorithms are automatically selected depending on the selected image input values.
A stack of immunohistochemical and histological images enters the software module. First, we calculated the noise level. A median filtering was performed based on the noise level with a window size corresponding to the parameter selection rules.
Images obtained as a result of microscopic examination were characterized by uneven illumination, obscuration areas, or, conversely, excessive illumination. An image preprocessing unit was used to adjust the brightness level. Brightness adjustment was based on the rules stored in the knowledge base.
It is necessary to determine the relative area of the cell nuclei in the image and the average level of brightness of the cell nuclei to make a diagnosis. To do this, it is necessary to segment the images and superimpose the segmented image on the original to calculate the brightness level.
After the segmentation stage, the brightness level is calculated. The next step is to calculate the cancer subtype’s conditions directly.
Class “Main” is an entry point to the software module. This class provides methods for determining the image path and storing intermediate versions of the image after performing certain operations such as filtering. ArrayList is used to store image parameters that are used for further processing. The fields “ER_”, “PR_”, “HERN2_”, “KI67” are designed to store information about the parameters that are used to determine a particular cancer subtype.
|
An example of a code for determining the relative area and level of brightness is as follows:
|
This class was designed for automatic immunohistochemical and cytological image segmentation. The class consisted of a constructor, additional methods, and fields for storing an image object in the OpenCv Mat format.
4.3. Computer Experiments
A software module was developed for computer experiments, which automatically defines a preliminary diagnosis. The software module is a part of the HIAMS software package []. Examples of immunohistochemical images are shown in Figure 2.
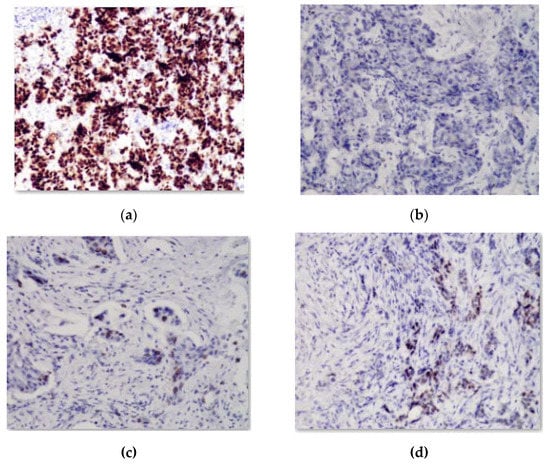
Figure 2.
Image with reaction to (a) estrogen; (b) oncoprotein, (c) cell proliferation biomarker; (d) progesterone.
These images were obtained from the database of a private immunohistochemical laboratory. The image database was closed.
Figure 3 shows the original image and the generated mask, reflecting the areas of interest. The image is the mask generated automatically without human intervention based on the developed adaptive algorithm of image preprocessing. As can be seen from Figure 3, the studied images were not of the same type. With automatic diagnosing, many calculations were performed on several images. Relative area and cell intensity were used to assess the accuracy of the developed method of preliminary diagnosing. Figure 4 shows the ratio of the correctly defined parameters (area, intensity) to the total number of images in the study. To determine the subtype “Luminal A”, one of the conditions was to calculate the area of cells in the image processed with the biomarker “progesterone”.
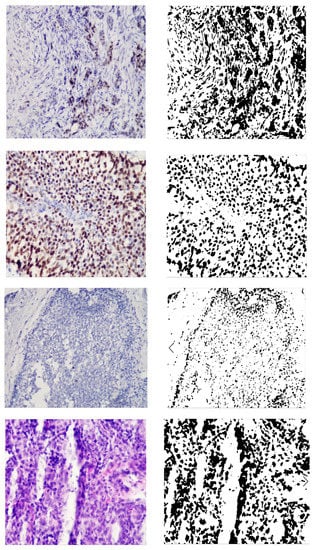
Figure 3.
Original and generated images.
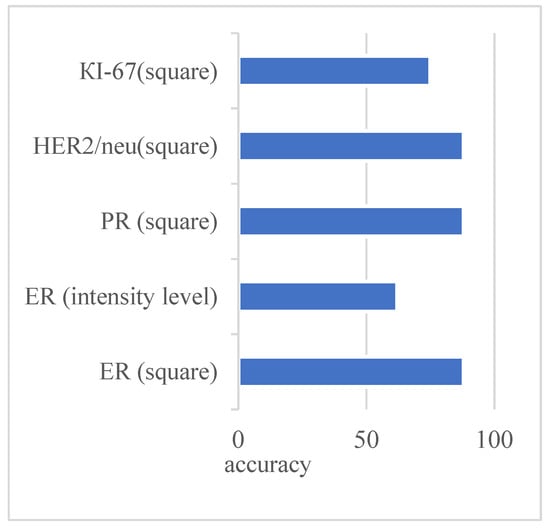
Figure 4.
Indicators of the correctly detected parameters for Luminal A subtype.
Several parameters and conditions characterize each molecular genetic subtype of cancer. Figure 4 shows the number of correctly defined parameters for the subtype Luminal A based on the experimental studies.
The figure shows that the parameters were determined at almost the same level. Only the intensity parameter for the ER image had a low result—62%.
4.4. Comparison of Results of Automated Microscopy Systems
Table 2 shows a comparative analysis of some of the automated microscopy systems and developed systems. The indicators for comparison are the following: segmentation algorithms, automatic calculation of quantitative characteristics, automatic calculation of brightness and area, storage in a database, and diagnosis according to the Nottingham scale.

Table 2.
Comparative analysis of automated microscopy systems (“+”—criterion is present, “−”—criterion is absent, “+/−”—criterion is implemented in automated mode).
Thus, most automated microscopy systems have a set of algorithms that allow for the calculation of the characteristics of the cell nuclei. However, unlike the developed system, analogs do not have the functionality for automatic diagnosis. Diagnosing in the known automated microscopy systems is only possible in manual or automated mode.
5. Conclusions
Advances in artificial intelligence have greatly influenced the development of modern medicine. The visualization of processes in diagnosing various organs makes it possible to identify pathological processes in the early stages. Processing of images obtained during visualization is an urgent and complex problem. A cancer diagnosis is based on cytological, histological, and immunohistochemical image analysis. The use of immunohistochemical images, obtained under the influence of biomarkers, allows for accurate diagnoses. We present a new method of the specified diagnosis of breast cancer subtypes. Pre-processing improved the quality of the input immunohistochemical images. The segmentation was performed based on the watershed and threshold segmentation algorithms. The authors developed the algorithm for determining the molecular genetic subtype of breast cancer: “Luminal A”, “Luminal B”, HER2/neu amplified, and basal-like. Experiments to determine the subtype of breast cancer “Luminal A” based on the calculation of the area and intensity of cells in the image showed high accuracy (more than 80%). Only the intensity parameter for ER showed a result of 62%.
In further studies, the initial histological images should be automatically classified. In addition, in limited initial samples, it is necessary to artificially generate immunohistological images to test the developed algorithms for automatic diagnosis.
Author Contributions
Conceptualization and methodology, O.B.; Software and validation, O.P.; Formal analysis, B.D.; Data curation, T.D.; Writing—original draft preparation, G.M.; Project administration, I.I. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available in this paper. The data presented in this study are available on request from the corresponding author. The data are not publicly available due to restrictions of privacy.
Acknowledgments
The authors would like to thank the Armed Forces of Ukraine because this research was only possible because of the resilience and courage of the Ukrainian Army.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ibrahim, A.; Gamble, P.; Jaroensri, R.; Abdelsamea, M.; Mermel, C.; Chen, P.-H.; Rakha, E. Artificial intelligence in digital breast pathology: Techniques and applications. Breast 2020, 49, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Punitha, S.; Al-Turjman, F.; Thompson, S. An automated breast cancer diagnosis using feature selection and parameter optimization in ANN. Comput. Electr. Eng. 2021, 90, 106958. [Google Scholar] [CrossRef]
- Chattoraj, S.; Vishwakarma, K. Classification of histopathological breast cancer images using iterative VMD aided Zernike moments & textural signatures. arXiv 2018, arXiv:1801.04880. Available online: http://arxiv.org/abs/1801.04880 (accessed on 10 May 2022).
- Cordeiro, C.; Ioshii, S.; Alves, J.; Oliveira, L. An Automatic Patch-based Approach for HER-2 Scoring in Immunohistochemical Breast Cancer Images Using Color Features. arXiv 2018, arXiv:1805.05392. Available online: http://arxiv.org/abs/1805.05392 (accessed on 10 May 2022).
- An, Y.; Kim, S.; Kang, B.; Park, C.; Jung, N.; Kim, J. Breast cancer in very young women (<30 years): Correlation of imaging features with clinicopathological features and immunohistochemical subtypes. Eur. J. Radiol. 2015, 84, 1894–1902. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wan, J.; Xu, G.; Xiang, L.-H.; Fang, Y.; Ding, S.-S.; Jiang, X.; Sun, L.-P.; Zhang, Y.-F. Conventional US and 2-D Shear Wave Elastography of Virtual Touch Tissue Imaging Quantification: Correlation with Immunohistochemical Subtypes of Breast Cancer. Ultrasound Med. Biol. 2019, 45, 2612–2622. [Google Scholar] [CrossRef]
- Usman, H.; Abidin, F. Digital image analysis of immunohistochemistry KI-67 using QuPath software in breast cancer. JKKI 2021, 12, 34–43. [Google Scholar] [CrossRef]
- Chen, J.-M.; Li, Y.; Xu, J.; Gong, L.; Wang, L.-W.; Liu, W.-L.; Liu, J. Computer-aided prognosis on breast cancer with hematoxylin and eosin histopathology images: A review. Tumor Biol. 2017, 39, 101042831769455. [Google Scholar] [CrossRef]
- Holten-Rossing, H.; Klingberg, H. AI deep learning tumour detection directly on ER, PR and KI-67 IHC slides yields a single slide automated workflow with high concordance to manual scoring. In Proceedings of the 6th Digital Pathology & AI Congress, London, UK, 5–6 December 2019. [Google Scholar] [CrossRef]
- Jakobsen, M.; Teerapakpinyo, C.; Shuangshoti, S.; Keelawat, S. Comparison between digital image analysis and visual assessment of immunohistochemical HER2 expression in breast cancer. Pathol.-Res. Pract. 2018, 214, 2087–2092. [Google Scholar] [CrossRef]
- Mi, W.; Li, J.; Guo, Y.; Ren, X.; Liang, Z.; Zhang, T.; Zou, H. Deep Learning-Based Multi-Class Classification of Breast Digital Pathology Images. CMAR 2021, 13, 4605–4617. [Google Scholar] [CrossRef]
- Robertson, S.; Azizpour, H.; Smith, K.; Hartman, J. Digital image analysis in breast pathology-from image processing techniques to artificial intelligence. Transl. Res. 2017, 194, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Guo, X.; Tian, F. Application of Artificial Intelligence Technology in Pathological Image Analysis of Breast Tissue. J. Phys. Conf. Ser. 2020, 1642, 012018. [Google Scholar] [CrossRef]
- Gon, C.; Souza, J.; Fernandes, H. CNN architecture optimization using bio-inspired algorithms for breast cancer detection in infrared images. Comput. Biol. Med. 2022, 142, 105205. [Google Scholar] [CrossRef]
- Mouelhi, A.; Rmili, H.; Ali, J.; Sayadi, M.; Doghri, R.; Mrad, K. Fast unsupervised nuclear segmentation and classification scheme for automatic allred cancer scoring in immunohistochemical breast tissue images. Comput. Methods Programs Biomed. 2018, 165, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Ammar, M.B.; Ayachi, F.L.; Ksantini, R.; Mahjoubi, H. Data warehouse for machine learning: Application to breast cancer diagnosis. Procedia Comput. Sci. 2022, 196, 692–698. [Google Scholar] [CrossRef]
- Dumont, L.; Levacher, N.; Schapman, D.; Rives-Feraille, A.; Moutard, L.; Delessard, M.; Saulnier, J.; Rondanino, C.; Rives, N. IHC_Tool: An open-source Fiji procedure for quantitative evaluation of cross sections of testicular explants. Reprod. Biol. 2021, 21, 100507. [Google Scholar] [CrossRef]
- Yoshizawa, K.; Ando, H.; Kimura, Y.; Kawashiri, S.; Moroi, A.; Ueki, K. Automatic Machine-Learning Classification of the Mode of Invasion of Oral Squamous Cell Carcinoma Using Digital Microscopic Images: A Retrospective Study. Review 2020, preprint. [Google Scholar] [CrossRef]
- Evangeline, I.K.; Precious, J.G.; Pazhanivel, N.; Kirubha, S.A. Automatic Detection and Counting of Lymphocytes from Immunohistochemistry Cancer Images Using Deep Learning. J. Med. Biol. Eng. 2020, 40, 735–747. [Google Scholar] [CrossRef]
- Mokni, R.; Gargouri, N.; Damak, A.; Sellami, D.; Feki, W.; Mnif, Z. An automatic Computer-Aided Diagnosis system based on the Multimodal fusion of Breast Cancer (MF-CAD. Biomed. Signal Process. Control. 2021, 69, 102914. [Google Scholar] [CrossRef]
- Shu, J.; Liu, J.; Zhang, Y.; Fu, H.; Ilyas, M.; Faraci, G.; Della Mea, V.; Liu, B.; Qiu, G. Marker controlled superpixel nuclei segmentation and automatic counting on immunohistochemistry staining images. Bioinformatics 2020, 36, 3225–3233. [Google Scholar] [CrossRef]
- Tan, W.; Nerurkar, S.; Cai, H.; Ng, H.; Wu, D.; Wee, Y.; Lim, J.; Yeong, J.; Lim, T. Overview of multiplex immunohistochemistry/immunofluorescence techniques in the era of cancer immunotherapy. Cancer Commun. 2020, 40, 135–153. [Google Scholar] [CrossRef] [PubMed]
- Fassler, D.; Abousamra, S.; Gupta, R.; Chen, C.; Zhao, M.; Paredes, D.; Batool, S.; Knudsen, B.; Escobar-Hoyos, L.; Shroyer, K.; et al. Deep learning-based image analysis methods for brightfield-acquired multiplex immunohistochemistry images. Diagn. Pathol. 2020, 15, 100. [Google Scholar] [CrossRef] [PubMed]
- Elazab, N.; Soliman, H.; El-Sappagh, S.; Islam, S.; Elmogy, M. Objective Diagnosis for Histopathological Images Based on Machine Learning Techniques: Classical Approaches and New Trends. Mathematics 2020, 8, 1863. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, J.; Che, S.; Huang, J.; Han, X.; Yuan, Y. Diagnose Like A Pathologist: Weakly-Supervised Pathologist-Tree Network for Slide-Level Immunohistochemical Scoring. Proc. Conf. AAAI Artif. Intell. 2021, 35, 47–54. [Google Scholar] [CrossRef]
- Morrison, L.; Lefever, M.; Behman, L.; Leibold, T.; Roberts, E.; Horchner, U.; Bauer, D. Brightfield multiplex immunohistochemistry with multispectral imaging. Lab. Investig. 2020, 100, 1124–1136. [Google Scholar] [CrossRef] [PubMed]
- Priego-Torres, B.; Lobato-Delgado, B.; Atienza-Cuevas, L. Deep learning-based instance segmentation for the precise automated quantification of digital breast cancer immunohistochemistry images. Expert Syst. Appl. 2022, 193, 116471. [Google Scholar] [CrossRef]
- Berezsky, O.; Pitsun, O.; Derysh, B.; Pazdriy, I.; Melnyk, G.; Batko, Y. Automatic Segmentation of Immunohistochemical Images Based on U-net Architecture. In Proceedings of the 2021 IEEE 16th International Conference on Computer Sciences and Information Technologies (CSIT), IEEE, Lviv, Ukraine, 22–25 September 2021; pp. 29–32. [Google Scholar] [CrossRef]
- Li, D.; Chu, X.; Cui, Y.; Zhao, J.; Zhang, K.; Yang, X. Improved U-Net based on contour prediction for efficient segmentation of rectal cancer. Comput. Methods Programs Biomed. 2021, 213, 106493. [Google Scholar] [CrossRef] [PubMed]
- Berezsky, O.; Pitsun, O.; Derish, B.; Berezska, K.; Melnyk, G.; Batko, Y. Adaptive Immunohistochemical Image Pre-processing Method. In Proceedings of the 2020 10th International Conference on Advanced Computer Information Technologies (ACIT), IEEE, Deggendorf, Germany, 16–18 September 2020; pp. 820–823. [Google Scholar] [CrossRef]
- Peleshko, D.; Rak, T.; Izonin, I. Image Superresolution via Divergence Matrix and Automatic Detection of Crossover. Int. J. Intell. Syst. Appl. 2016, 8, 1–8. [Google Scholar] [CrossRef]
- Berezsky, O.; Pitsun, O.; Dubchak, L.; Berezka, K.; Dolynyuk, T.; Derish, B. Cytological Images Clustering of Breast Pathologies. In Proceedings of the 2020 IEEE 15th International Conference on Computer Sciences and Information Technologies (CSIT), IEEE, Zbarazh, Ukraine, 23–26 September; pp. 62–65. [CrossRef]
- Berezsky, O.; Dubchak, L.; Batryn, N.; Datsko, T.; Berezska, K.; Pitsun, O.; Batko, Y. Fuzzy System For Breast Disease Diagnosing Based On Image Analysis. Inform. Data-Driven Med. 2019, 2488, 69–83. [Google Scholar]
- Hu, Z.; Mashtalir, S.V.; Tyshchenko, O.K.; Stolbovyi, M.I. Video Shots‘ Matching via Various Length of Multidimensional Time Sequences. Int. J. Intell. Syst. Appl. 2017, 9, 10–16. [Google Scholar] [CrossRef]
- Berezsky, O.; Verbovyy, S.; Datsko, T. The intelligent system for diagnosing breast cancers based on image analysis. In Proceedings of the 2015 Information Technologies in Innovation Business Conference (ITIB), IEEE, Kharkiv, Ukraine, 7–9 October 2015; pp. 27–30. [Google Scholar] [CrossRef]
- Hore, A.; Ziou, D. Image Quality Metrics: PSNR vs. SSIM. In Proceedings of the 2010 20th International Conference on Pattern Recognition, IEEE, Istanbul, Turkey, 23–26 August 2010; pp. 2366–2369. [Google Scholar] [CrossRef]
- Vasuki, P.; Kanimozhi, J.; Devi, M. A survey on image preprocessing techniques for diverse fields of medical imagery. In Proceedings of the 2017 IEEE International Conference on Electrical, Instrumentation and Communication Engineering (ICEICE), IEEE, Karur, India, 27–28 April 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Berezsky, O.; Pitsun, O.; Datsko, T.; Derish, B.; Melnyk, G. Breast cancer immunohistological imaging database. Comput. Syst. Inf. Technol. 2022, 1, 75–82. [Google Scholar] [CrossRef]
- Hu, Z.; Bodyanskiy, Y.V.; Kulishova, N.Y.; Tyshchenko, O.K. A Multidimensional Extended Neo-Fuzzy Neuron for Facial Expression Recognition. Int. J. Intell. Syst. Appl. 2017, 9, 29–36. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).