Automatic Aortic Valve Cusps Segmentation from CT Images Based on the Cascading Multiple Deep Neural Networks
Abstract
:1. Introduction
- To the best of our knowledge, this is the second study to build a fully automatic method for segmenting aortic valve cusps. Our method constructs a computational flow of cascaded multiple networks for landmark detection and segmentation without transforming the original images. This is expected to improve performance as the input size for the networks is reduced step by step and sub-regions are more accurately located. Each network is based on methods presented in previous studies [18,19], however the method of combining them is reported for the first time.
- We proposed an evaluation method for the impact of segmentation results on measurement values for each measurement item, and evaluated our proposed segmentation method using this evaluation method. Many papers on segmentation methods focus on segmentation accuracy and do not investigate the impact on measurements. To the best of our knowledge, this paper is the first work to investigate the impact of aortic valve cusps segmentation results on various measurements.
2. Materials and Methods
2.1. Data and Annotation
- Volume data in end systolic (ES) and/or end diastolic (ED) phases suitable for acquiring morphological information of the aortic valve were selected for each CT imaging data based on the information available from CT images, which included ECG gating timing, LV volume size, aortic valve motion and blurring. A total of 258 volume data were selected in this step, and the following annotation work was performed only on these volume data.
- The Vitrea workstation (Canon Medical Informatics, inc., Otawara, Tochigi, Japan) was used to semi-automatically generate an initial label of the aortic root by providing seed points within the aortic root region. The initial label was manually modified as needed.
- Each of the eight landmarks were manually annotated one point on CT image.
- The labels for each cusp were separately manually annotated on the internal regions of the aortic root. Each cusp did not have an overlapping area.
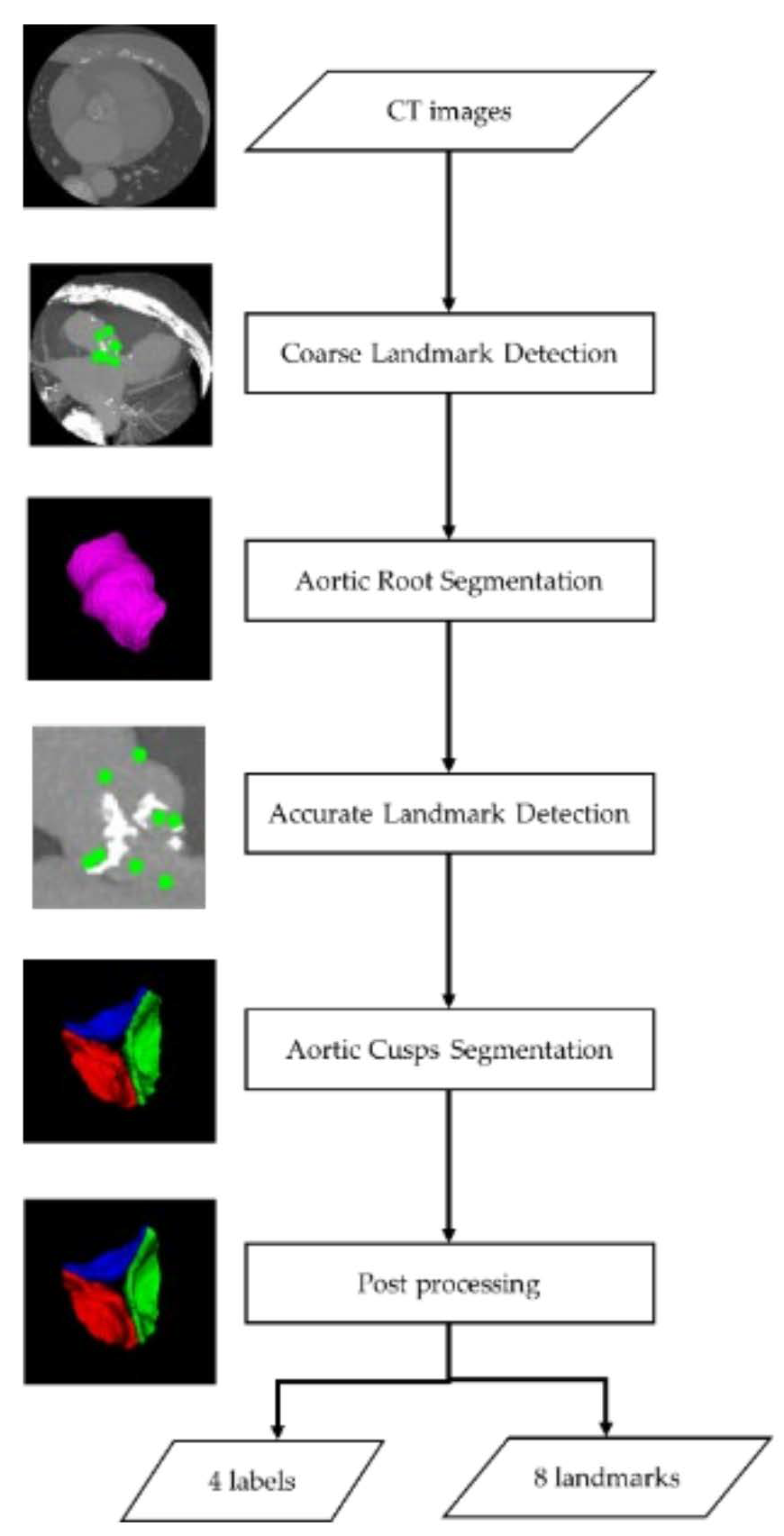
2.2. Aortic Valve Cusps Segmentation Method
2.2.1. Landmark Detection Processing
2.2.2. Segmentation Processing
2.2.3. Post-Processing
2.3. Evaluation
2.3.1. Evaluation Setup
2.3.2. Automation and Processing Time
2.3.3. Landmark Evaluation
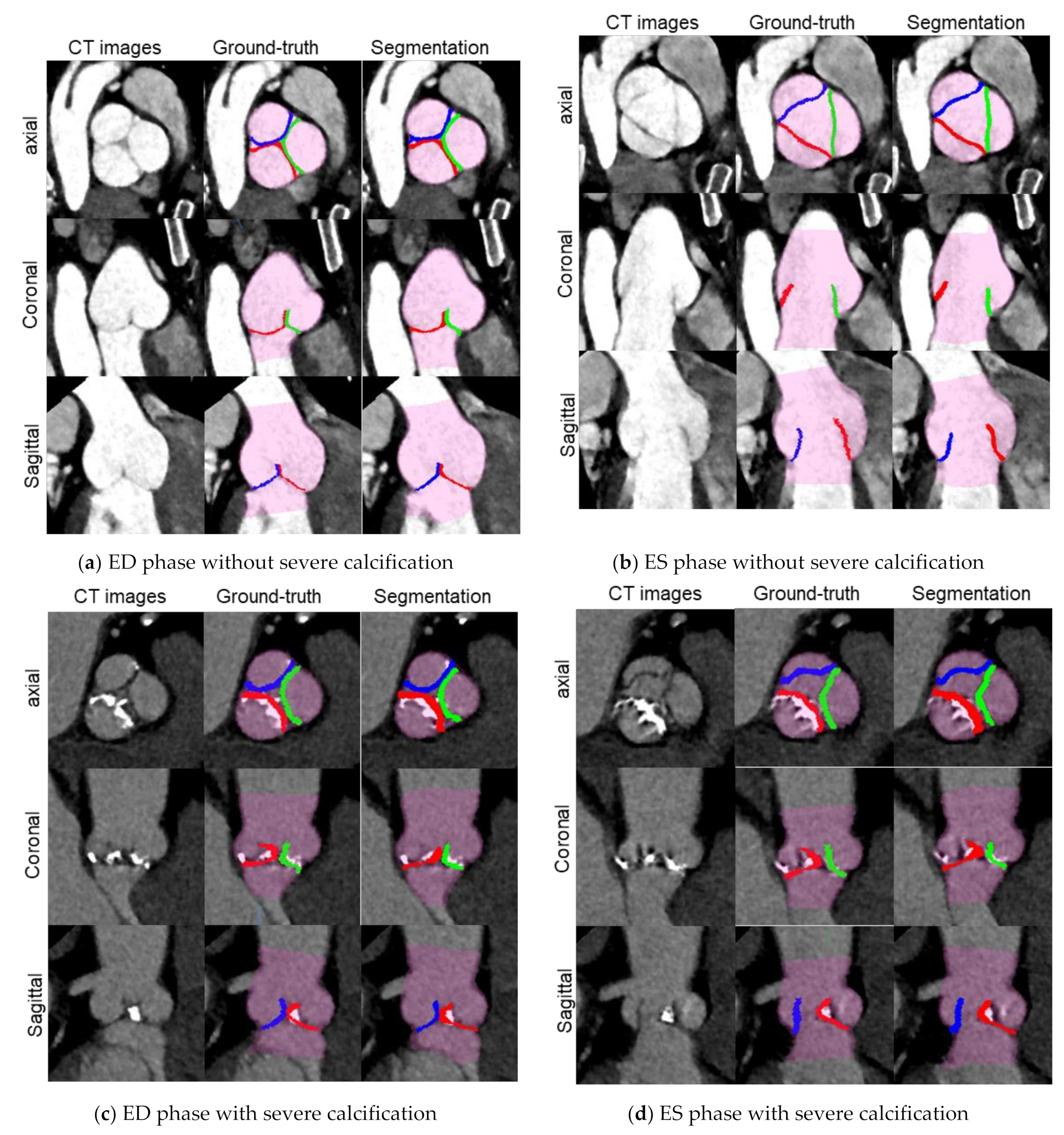
2.3.4. Segmentation Evaluation
- Volumetric overlap: Dice Coefficient (DC) [21] was calculated as volumetric overlap between two sets of voxels G and S, which was defined as follows:
- 2.
- Surface Distance: The Mean symmetric Surface Distance (MSD) is defined as follows:
- 3.
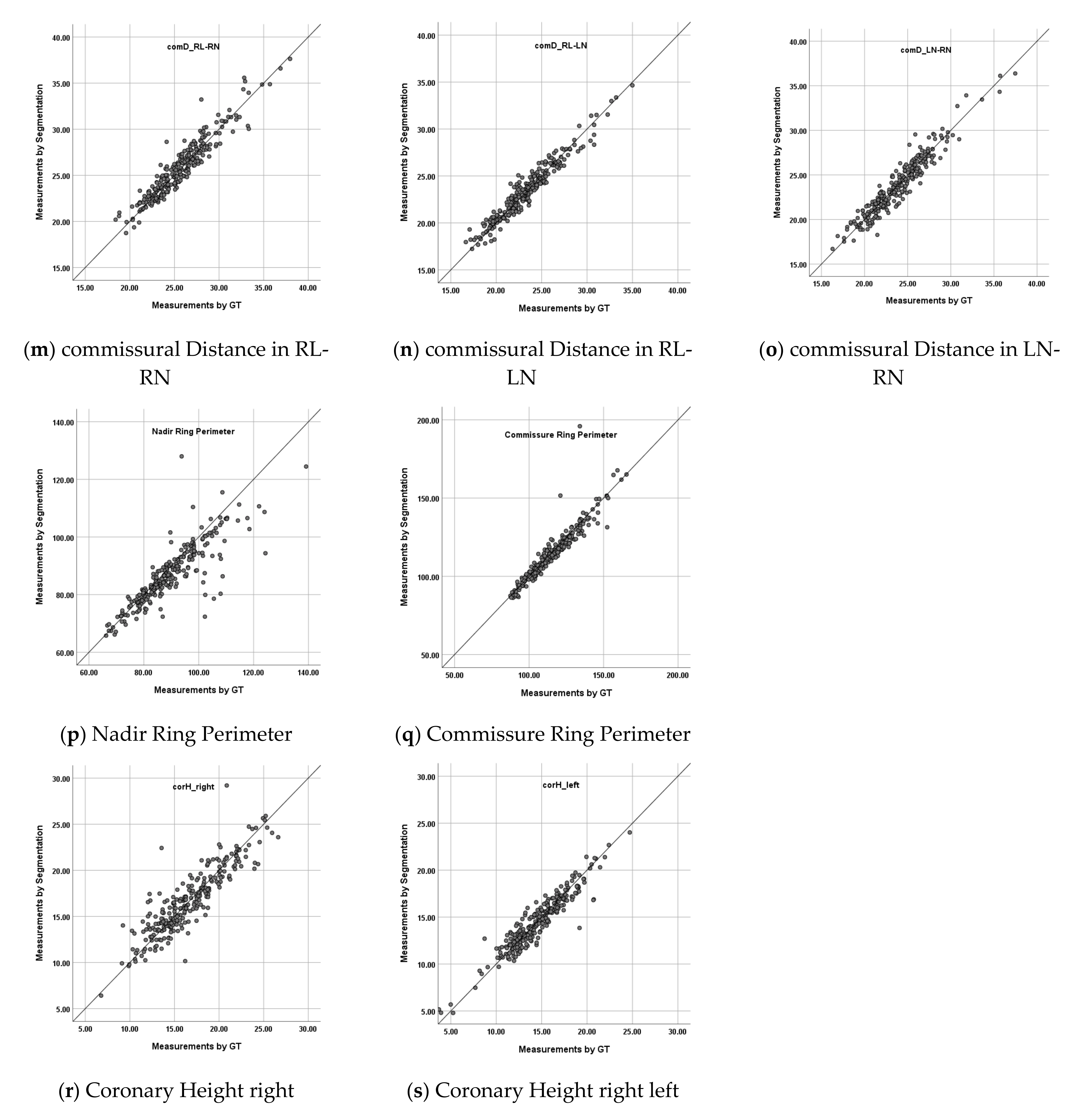
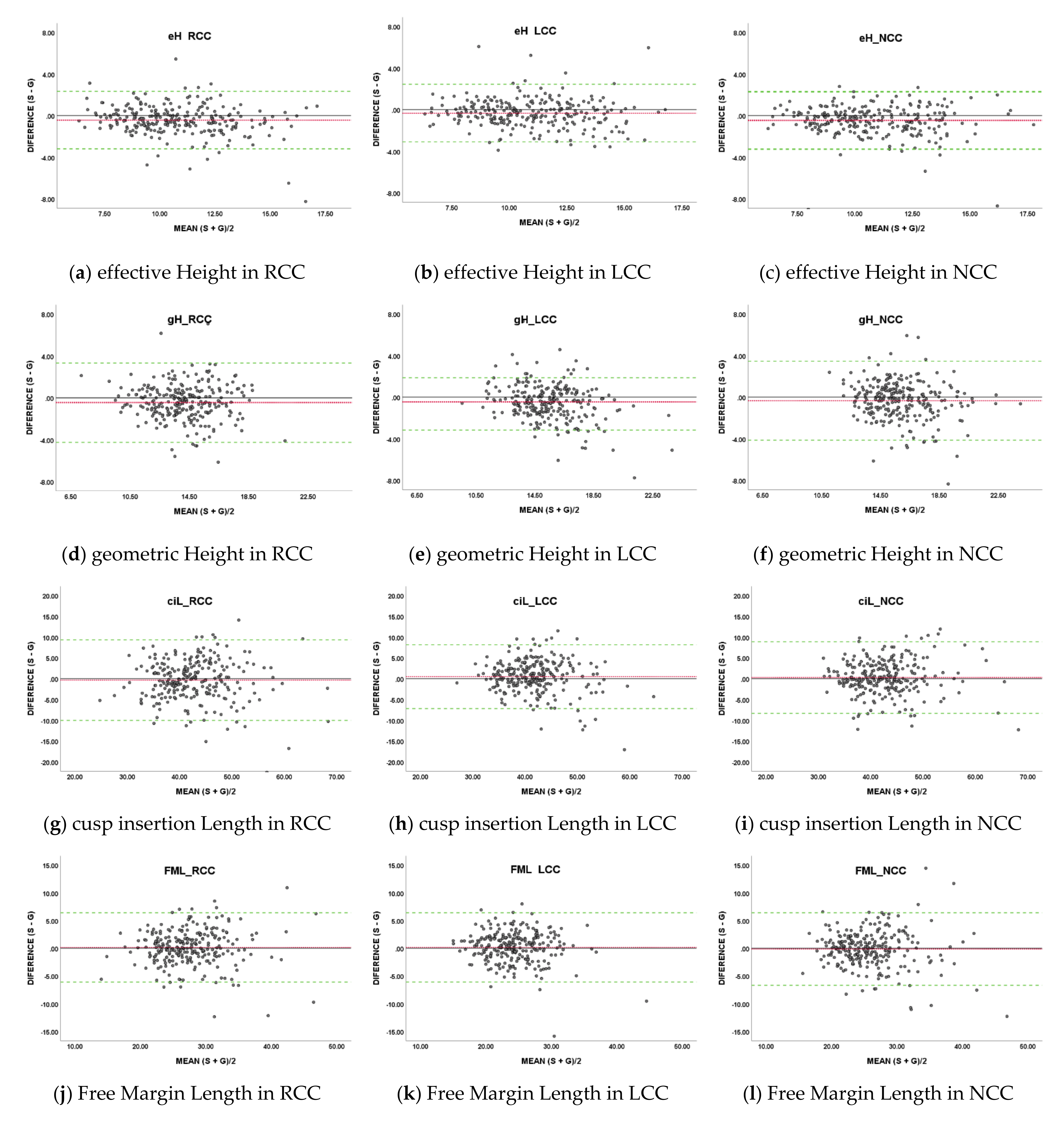
- Impact on measurements: For the eight measurement items, the differences between the values measured based on G and measured based on S were calculated using the same measurement algorithm, as the impacts for each measurement item (Figure 4). Each measurement item was defined in this paper as follows:
- effective Height (eH): Straight line distance from Arantius body to Nadir plane;
- geometric Height (gH): Shortest curved line distance along the surface of the cusp, from Arantius body to Hinge point;
- cusp insertion Length (ciL): Curved line distance between commissure points on the contour of the cusp in contact with the Sinus of Valsalva;
- Free Margin Length (FML): Curved line distance between commissure points on the contour of the cusp not in contact with the Sinus of Valsalva;
- commissural Distance (comD): Straight line distance between commissure points;
- Nadir Ring Perimeter (NRP): Perimeter of the contour of the Aortic root region on the Nadir plane;
- Commissure Ring Perimeter (CRP): Perimeter of the contour of the Aortic root region on the Commissure plane;
- coronary Height (corH): Straight line distance from coronary ostium point to Nadir plane.
2.3.5. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A


References
- Nkomo, V.T.; Gardin, J.M.; Skelton, T.N.; Gottdiener, J.S.; Scott, C.; Enriquez-Sarano, M. Burden of valvular heart diseases: A population-based study. Lancet 2006, 368, 1005–1011. [Google Scholar] [CrossRef]
- Siddiqui, R.F.; Abraham, J.R.; Butany, J. Bioprosthetic heart valves: Modes of fail-ure. Histopathology 2009, 55, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Vahanian, A.; Beyersdorf, F.; Praz, F.; Milojevic, M.; Baldus, S.; Bauersachs, J. 2021 ESC/EACTS Guidelines for the management of valvular heart disease: Developed by the Task Force for the management of valvular heart disease of the European Society of Cardiology (ESC) and the European Association for Cardio-Thoracic Surgery (EACTS). Eur. Heart J. 2021, 60, 727–800. [Google Scholar]
- Tian, Z.; Liu, L.; Zhang, Z.; Fei, B. Superpixel-Based Segmentation for 3D Prostate MR Images. IEEE Trans. Med. Imaging 2015, 35, 791–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen DC, T.; Benameur, S.; Mignotte, M.; Lavoie, F. Superpixel and multi-atlas based fusion entropic model for the segmentation of X-ray images. Med. Image Anal. 2018, 48, 58–74. [Google Scholar] [CrossRef]
- Huang, Y.-L.; Chen, D.-R. Watershed segmentation for breast tumor in 2-D sonography. Ultrasound Med. Biol. 2004, 30, 625–632. [Google Scholar] [CrossRef]
- Masoumi, H.; Behrad, A.; Pourmina, M.A.; Roosta, A. Automatic liver segmentation in MRI images using an iterative watershed algorithm and artificial neural network. Biomed. Signal Process. Control. 2012, 7, 429–437. [Google Scholar] [CrossRef]
- Ciecholewski, M.; Spodnik, J.H. Semi-Automatic Corpus Callosum Segmentation and 3D Visualization Using Active Contour Methods. Symmetry 2018, 10, 589. [Google Scholar] [CrossRef] [Green Version]
- Nguyen DC, T.; Benameur, S.; Mignotte, M.; Lavoie, F. Automated vessel segmentation using infinite perimeter active contour model with hybrid region infor-mation with application to retinal images. IEEE Trans. Med. Imaging 2015, 34, 1797–1807. [Google Scholar]
- Zhu, W.; Huang, Y.; Zeng, L.; Chen, X.; Liu, Y.; Qian, Z.; Du, N.; Fan, W.; Xie, X. AnatomyNet: Deep learning for fast and fully automated whole-volume segmentation of head and neck anatomy. Med. Phys. 2018, 46, 576–589. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-like Pure Transformer for Medical Image Segmentation. Arxiv 2021, Arxiv preprint. arXiv:2105.05537. [Google Scholar]
- Zheng, Y.; John, M.; Liao, R.; Boese, J.; Kirschstein, U.; Georgescu, B.; Zhou, S.K.; Kempfert, J.; Walther, T.; Brockmann, G.; et al. Automatic Aorta Segmentation and Valve Landmark Detection in C-Arm CT: Application to Aortic Valve Implantation; Springer: Berlin/Heidelberg, Germany, 2010; pp. 476–483. [Google Scholar] [CrossRef]
- Elattar, M.A.; Wiegerinck, E.M.; Planken, R.N.; Van Bavel, E.; Van Assen, H.C.; Baan, J.; Marquering, H. Automatic segmentation of the aortic root in CT angiography of candidate patients for transcatheter aortic valve implantation. Med. Biol. Eng. Comput. 2014, 52, 611–618. [Google Scholar] [CrossRef]
- Ravichandran, S.R.; Nataraj, B.; Huang, S.; Qin, Z.; Lu, Z.; Katsuki, A. 3D Inception U-Net for Aorta Segmentation using Computed Tomography Cardiac An-giography. In Proceedings of the 2019 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), Chicago, IL, USA, 19–22 May 2019. [Google Scholar]
- Pouch, A.M.; Wang, H.; Takabe, M.; Jackson, B.M.; Sehgal, C.M.; Gorman, J.H. Automated segmentation and geometrical modeling of the tricuspid aortic valve in 3D echocardio-graphic images. In Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Fan, B.; Tomii, N.; Tsukihara, H.; Maeda, E.; Yamauchi, H.; Nawata, K.; Ono, M. Attention-guided decoder in dilated residual network for accurate aortic valve segmentation in 3D CT scans. In Machine Learning and Medical Engineering for Cardiovascular Health and Intravascular Imaging and Computer Assisted Stenting; Springer: Cham, Switzerland, 2019; pp. 121–129. [Google Scholar]
- Pak, D.H.; Caballero, A.; Sun, W.; Duncan, J.S. Efficient Aortic Valve Multilabel Segmentation Using a Spatial Transformer Network; IEEE: Piscataway, NJ, USA, 2020; pp. 1738–1742. [Google Scholar] [CrossRef]
- Payer, C.; Štern, D.; Bischof, H.; Urschler, M. Integrating spatial configuration into heatmap regression based CNNs for landmark localization. Med. Image Anal. 2019, 54, 207–219. [Google Scholar] [CrossRef]
- Isensee, F.; Jäger, P.F.; Kohl, S.A.; Petersen, J.; Maier-Hein, K.H. Automated design of deep learning methods for biomedical image segmentation. arXiv 2019, arXiv:1904.08128. [Google Scholar]
- Aktouf, Z.; Bertrand, G.; Perroton, L. A three-dimensional holes closing algorithm. Pattern Recognit. Lett. 2002, 23, 523–531. [Google Scholar] [CrossRef]
- Dice, L.R. Measures of the Amount of Ecologic Association Between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Sapien, E. SAPIEN Transcatheter Heart Valve with the Edwards Commander Delivery System. 2018. Available online: https://www.accessdata.fda.gov/cdrh_docs/pdf14/P140031S085D.pdf (accessed on 20 November 2021).
- Medtronic. CoreValveTM EvolutTM R Transcatheter Aortic Valve Delivery Catheter System Loading System. 2019. Available online: https://www.accessdata.fda.gov/cdrh_docs/pdf13/P130021S058D.pdf (accessed on 20 November 2021).
- Maragiannis, D.; Jackson, M.S.; Igo, S.R.; Schutt, R.C.; Connell, P.; Grande-Allen, J. Replicating patient-specific severe aortic valve stenosis with functional 3D model-ing. Circ. Cardiovasc. Imaging 2015, 8, e003626. [Google Scholar] [CrossRef] [Green Version]
- Hosny, A.; Dilley, J.D.; Kelil, T.; Mathur, M.; Dean, M.N.; Weaver, J.C.; Ripley, B. Pre-procedural fit-testing of TAVR valves using parametric modeling and 3D printing. J. Cardiovasc. Comput. Tomogr. 2018, 13, 21–30. [Google Scholar] [CrossRef]
- Chen, Y.; Luo, H. A computational study of the three-dimensional fluid–structure interaction of aortic valve. J. Fluids Struct. 2018, 80, 332–349. [Google Scholar] [CrossRef]
- Sillesen, A.-S.; Vøgg, O.; Pihl, C.; Raja, A.A.; Sundberg, K.; Vedel, C.; Zingenberg, H.; Jørgensen, F.S.; Vejlstrup, N.; Iversen, K.; et al. Prevalence of Bicuspid Aortic Valve and Associated Aortopathy in Newborns in Copenhagen, Denmark. JAMA 2021, 325, 561–567. [Google Scholar] [CrossRef]
- Zheng, Y.; Barbu, A.; Georgescu, B.; Scheuering, M.; Comaniciu, D. Four-Chamber Heart Modeling and Automatic Segmentation for 3-D Cardiac CT Volumes Using Marginal Space Learning and Steerable Features. IEEE Trans. Med. Imaging 2008, 27, 1668–1681. [Google Scholar] [CrossRef]
- Dormer, J.D.; Fei, B.; Halicek, M.; Ma, L.; Reilly, C.M.; Schreibmann, E. Heart chamber segmentation from CT using convolutional neural networks. Biomed. Appl. Mol. Struct. Funct. Imaging 2018, 10578, 105782S. [Google Scholar] [CrossRef]
- Le Couteulx, S.; Caudron, J.; Dubourg, B.; Cauchois, G.; Dupré, M.; Michelin, P. Multidetector computed tomography sizing of aortic annulus prior to transcatheter aortic valve re-placement (TAVR): Variability and impact of observer experience. Diagn. Interv. Imaging 2018, 99, 279–289. [Google Scholar] [CrossRef]
- Schmidkonz, C.; Marwan, M.; Klinghammer, L.; Mitschke, M.; Schuhbaeck, A.; Arnold, M.; Lell, M.; Achenbach, S.; Pflederer, T. Interobserver variability of CT angiography for evaluation of aortic annulus dimensions prior to transcatheter aortic valve implantation (TAVI). Eur. J. Radiol. 2014, 83, 1672–1678. [Google Scholar] [CrossRef]




| Coarse Landmark Detection | Nadir RCC [mm] | Nadir LCC [mm] | Nadir NCC [mm] | Commissure RL [mm] | Commissure RN [mm] | Commissure LN [mm] | Coronary Ostium R [mm] | Coronary Ostium L [mm] |
|---|---|---|---|---|---|---|---|---|
| ES w/o sCa | 2.08 ± 1.12 | 1.62 ± 0.93 | 1.68 ± 1.08 | 2.08 ± 1.14 | 2.15 ± 1.47 | 1.98 ± 1.23 | 1.61 ± 1.51 | 1.54 ± 1.32 |
| ED w/o sCa | 1.72 ± 0.91 | 1.68 ± 0.77 | 1.72 ± 1.11 | 1.84 ± 1.24 | 2.17 ± 1.27 | 1.74 ± 1.21 | 1.46 ± 1.41 | 1.37 ± 0.98 |
| ES w/sCa | 1.57 ± 0.87 | 1.46 ± 0.83 | 2.18 ± 1.43 | 1.34 ± 0.62 | 1.76 ± 1.11 | 1.73 ± 1.53 | 1.53 ± 1.16 | 1.31 ± 0.81 |
| ED w/sCa | 1.37 ± 0.74 | 2.13 ± 3.29 | 1.54 ± 0.99 | 1.34 ± 0.90 | 1.63 ± 1.04 | 1.41 ± 1.08 | 1.29 ± 0.84 | 1.33 ± 0.73 |
| Total | 1.76 ± 0.99 | 1.70 ± 1.54 | 1.75 ± 1.15 | 1.76 ± 1.11 | 2.01 ± 1.30 | 1.77 ± 1.26 | 1.49 ± 1.33 | 1.41 ± 1.05 |
| Accurate Landmark Detection | Nadir RCC [mm] | Nadir LCC [mm] | Nadir NCC [mm] | Commissure RL [mm] | Commissure RN [mm] | Commissure LN [mm] | Coronary Ostium R [mm] | Coronary Ostium L [mm] |
| ES w/o sCa | 1.62 ± 1.05 | 1.46 ± 0.72 | 1.50 ± 0.96 | 1.96 ± 1.26 | 1.92 ± 1.47 | 1.69 ± 1.22 | 1.72 ± 3.42 | 1.35 ± 0.91 |
| ED w/o sCa | 1.18 ± 0.60 | 1.38 ± 0.74 | 1.51 ± 1.00 | 1.73 ± 1.25 | 2.16 ± 2.86 | 1.51 ± 1.09 | 1.36 ± 2.37 | 0.99 ± 0.56 |
| ES w/sCa | 1.42 ± 0.86 | 1.36 ± 0.84 | 1.92 ± 1.42 | 1.09 ± 0.62 | 1.71 ± 1.04 | 1.47 ± 1.35 | 1.47 ± 1.60 | 1.06 ± 0.69 |
| ED w/sCa | 1.07 ± 0.60 | 1.91 ± 3.34 | 1.42 ± 0.91 | 1.16 ± 0.79 | 1.49 ± 1.13 | 1.29 ± 1.12 | 1.12 ± 1.04 | 0.99 ± 0.57 |
| Total | 1.35 ± 0.84 | 1.49 ± 1.51 | 1.56 ± 1.06 | 1.61 ± 1.16 | 1.90 ± 1.97 | 1.53 ± 1.19 | 1.46 ± 2.54 | 1.12 ± 0.74 |
| Aortic Root | RCC | LCC | NCC | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DC | MSD [mm] | HD [mm] | DC | MSD [mm] | HD [mm] | DC | MSD [mm] | HD [mm] | DC | MSD [mm] | HD [mm] | |
| ES w/o sCa | 0.96 ± 0.05 | 0.43 ± 0.93 | 1.46 ± 3.81 | 0.70 ± 0.07 | 0.36 ± 0.13 | 1.43 ± 1.09 | 0.67 ± 0.10 | 0.43 ± 0.17 | 1.46 ± 1.13 | 0.69 ± 0.07 | 0.37 ± 0.12 | 1.38 ± 1.19 |
| ED w/o sCa | 0.95 ± 0.11 | 0.39 ± 0.49 | 1.88 ± 4.76 | 0.71 ± 0.05 | 0.34 ± 0.12 | 0.93 ± 0.27 | 0.70 ± 0.04 | 0.34 ± 0.07 | 0.96 ± 0.33 | 0.69 ± 0.05 | 0.34 ± 0.08 | 0.99 ± 0.34 |
| ES w/sCa | 0.94 ± 0.09 | 0.63 ± 1.70 | 1.80 ± 5.10 | 0.69 ± 0.06 | 0.40 ± 0.11 | 1.00 ± 0.25 | 0.68 ± 0.08 | 0.41 ± 0.10 | 1.01 ± 0.29 | 0.62 ± 0.08 | 0.45 ± 0.13 | 1.25 ± 0.52 |
| ED w/sCa | 0.96 ± 0.03 | 0.52 ± 1.40 | 1.40 ± 3.88 | 0.71 ± 0.05 | 0.36 ± 0.08 | 0.87 ± 0.16 | 0.69 ± 0.05 | 0.37 ± 0.08 | 0.92 ± 0.19 | 0.63 ± 0.08 | 0.43 ± 0.11 | 1.10 ± 0.27 |
| Total | 0.95 ± 0.08 | 0.46 ± 1.02 | 1.64 ± 4.36 | 0.70 ± 0.06 | 0.36 ± 0.12 | 1.10 ± 0.70 | 0.69 ± 0.07 | 0.38 ± 0.13 | 1.13 ± 0.85 | 0.67 ± 0.07 | 0.39 ± 0.11 | 1.18 ± 0.77 |
| Measurement Items | Ground Truth Label Based [mm] | Segmentation Result Based [mm] | Difference [mm] | Absolute Error [mm] (SE) | Error Rate [%] (SE) | CC b/w G and S r | Fixed Errorp-Value | Proportional Error rp | |
|---|---|---|---|---|---|---|---|---|---|
| effective Height | RCC | 10.97 ± 2.43 | 10.54 ± 2.14 | −0.43 ± 1.41 | 1.04 (0.07) | 9.47 (0.55) | 0.82 * | <0.001 * | −0.22 * |
| LCC | 11.12 ± 2.44 | 10.78 ± 2.25 | −0.34 ± 1.42 | 1.05 (0.06) | 9.53 (0.63) | 0.82 * | <0.001 * | −0.14 | |
| NCC | 11.06 ± 2.39 | 10.56 ± 2.27 | −0.50 ± 1.41 | 1.04 (0.07) | 9.17 (0.50) | 0.82 * | <0.001 * | −0.09 | |
| geometric Height | RCC | 14.42 ± 2.40 | 13.95 ± 2.21 | −0.47 ± 1.93 | 1.40 (0.09) | 9.58 (0.55) | 0.65 * | <0.001 * | −0.11 |
| LCC | 15.73 ± 2.56 | 15.24 ± 2.05 | −0.49 ± 1.87 | 1.38 (0.08) | 8.99 (0.67) | 0.69 * | <0.001 * | −0.30 * | |
| NCC | 16.05 ± 2.47 | 15.71 ± 2.16 | −0.34 ± 1.94 | 1.39 (0.09) | 8.57 (0.48) | 0.65 * | 0.006 | −0.20 * | |
| cusp insertion Length | RCC | 42.82 ± 7.13 | 42.49 ± 6.87 | −0.34 ± 4.94 | 3.80 (0.20) | 8.86 (0.42) | 0.75 * | 0.276 | −0.06 |
| LCC | 41.55 ± 6.14 | 42.05 ± 5.61 | 0.49 ± 3.92 | 2.94 (0.16) | 7.03 (0.37) | 0.78 * | 0.044 | −0.14 | |
| NCC | 42.59 ± 6.53 | 42.88 ± 6.45 | 0.29 ± 4.39 | 3.19 (0.19) | 7.41 (0.41) | 0.77 * | 0.290 | −0.02 | |
| Free Margin Length | RCC | 27.47 ± 5.09 | 27.62 ± 5.18 | 0.16 ± 3.19 | 2.37 (0.13) | 8.60 (0.45) | 0.81 * | 0.431 | 0.03 |
| LCC | 24.30 ± 4.68 | 24.46 ± 4.16 | 0.17 ± 3.19 | 2.21 (0.14) | 9.06 (0.48) | 0.75 * | 0.394 | −0.17 | |
| NCC | 25.96 ± 5.20 | 25.83 ± 4.95 | −0.13 ± 3.34 | 2.42 (0.14) | 9.18 (0.50) | 0.79 * | 0.522 | −0.08 | |
| commissural Distance | RL-RN | 25.62 ± 3.20 | 25.78 ± 3.23 | 0.15 ± 1.07 | 0.84 (0.04) | 3.30 (0.16) | 0.94 * | 0.023 | 0.02 |
| RL-LN | 23.32 ± 3.12 | 23.42 ± 3.00 | 0.10 ± 0.80 | 0.63 (0.03) | 2.76 (0.14) | 0.97 * | 0.043 | −0.16 | |
| LN-RN | 23.86 ± 3.26 | 23.89 ± 3.20 | 0.04 ± 1.00 | 0.78 (0.04) | 3.33 (0.16) | 0.95 * | 0.562 | −0.05 | |
| Nadir Ring Perimeter | 89.44 ± 11.59 | 87.16 ± 11.61 | −2.28 ± 8.78 | 4.39 (0.49) | 4.73 (0.59) | 0.71 * | <0.001 * | 0.002 | |
| Commissure Ring Perimeter | 114.84 ± 15.56 | 114.54 ± 17.83 | −0.30 ± 8.44 | 3.31 (0.48) | 2.77 (0.37) | 0.88 * | 0.569 | 0.28 * | |
| Coronary Height | right | 16.62 ± 3.69 | 16.73 ± 3.58 | 0.11 ± 1.71 | 1.22 (0.07) | 7.80 (0.53) | 0.89 * | 0.306 | −0.07 |
| left | 14.55 ± 3.05 | 14.51 ± 2.88 | −0.04 ± 1.03 | 0.77 (0.04) | 5.69 (0.35) | 0.94 * | 0.512 | −0.17 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aoyama, G.; Zhao, L.; Zhao, S.; Xue, X.; Zhong, Y.; Yamauchi, H.; Tsukihara, H.; Maeda, E.; Ino, K.; Tomii, N.; et al. Automatic Aortic Valve Cusps Segmentation from CT Images Based on the Cascading Multiple Deep Neural Networks. J. Imaging 2022, 8, 11. https://doi.org/10.3390/jimaging8010011
Aoyama G, Zhao L, Zhao S, Xue X, Zhong Y, Yamauchi H, Tsukihara H, Maeda E, Ino K, Tomii N, et al. Automatic Aortic Valve Cusps Segmentation from CT Images Based on the Cascading Multiple Deep Neural Networks. Journal of Imaging. 2022; 8(1):11. https://doi.org/10.3390/jimaging8010011
Chicago/Turabian StyleAoyama, Gakuto, Longfei Zhao, Shun Zhao, Xiao Xue, Yunxin Zhong, Haruo Yamauchi, Hiroyuki Tsukihara, Eriko Maeda, Kenji Ino, Naoki Tomii, and et al. 2022. "Automatic Aortic Valve Cusps Segmentation from CT Images Based on the Cascading Multiple Deep Neural Networks" Journal of Imaging 8, no. 1: 11. https://doi.org/10.3390/jimaging8010011
APA StyleAoyama, G., Zhao, L., Zhao, S., Xue, X., Zhong, Y., Yamauchi, H., Tsukihara, H., Maeda, E., Ino, K., Tomii, N., Takagi, S., Sakuma, I., Ono, M., & Sakaguchi, T. (2022). Automatic Aortic Valve Cusps Segmentation from CT Images Based on the Cascading Multiple Deep Neural Networks. Journal of Imaging, 8(1), 11. https://doi.org/10.3390/jimaging8010011






