Optical to Planar X-ray Mouse Image Mapping in Preclinical Nuclear Medicine Using Conditional Adversarial Networks
Abstract
:1. Introduction
2. Methodology
2.1. Data Collection and Preprocessing
2.2. Modeling and Performance Evaluation
3. Results
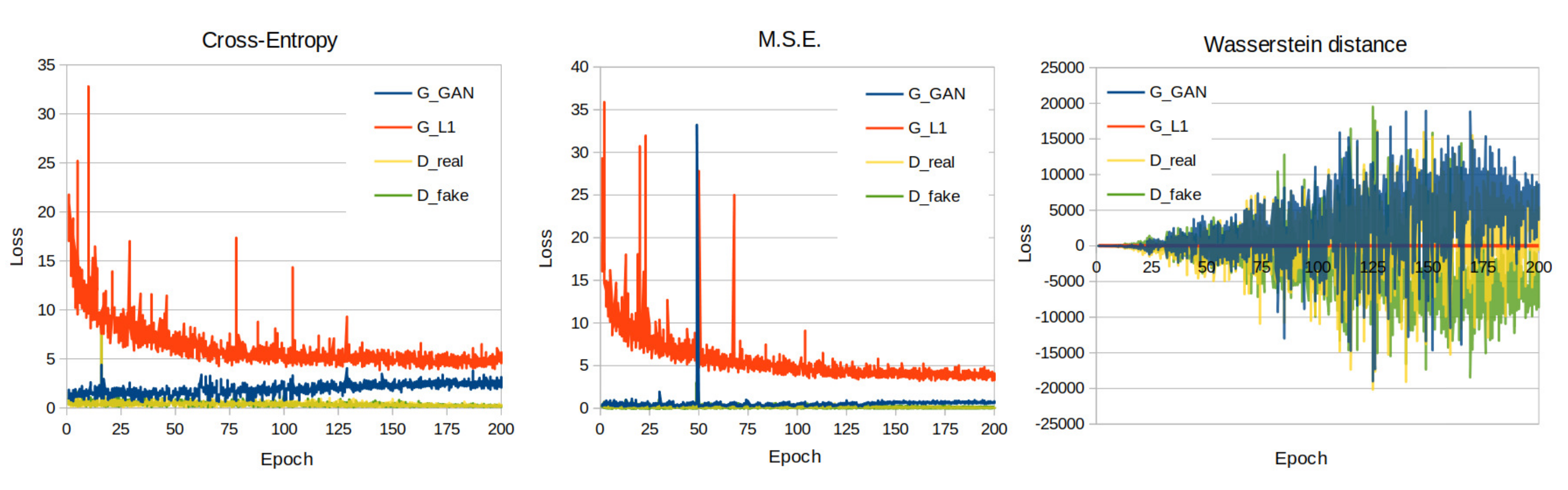
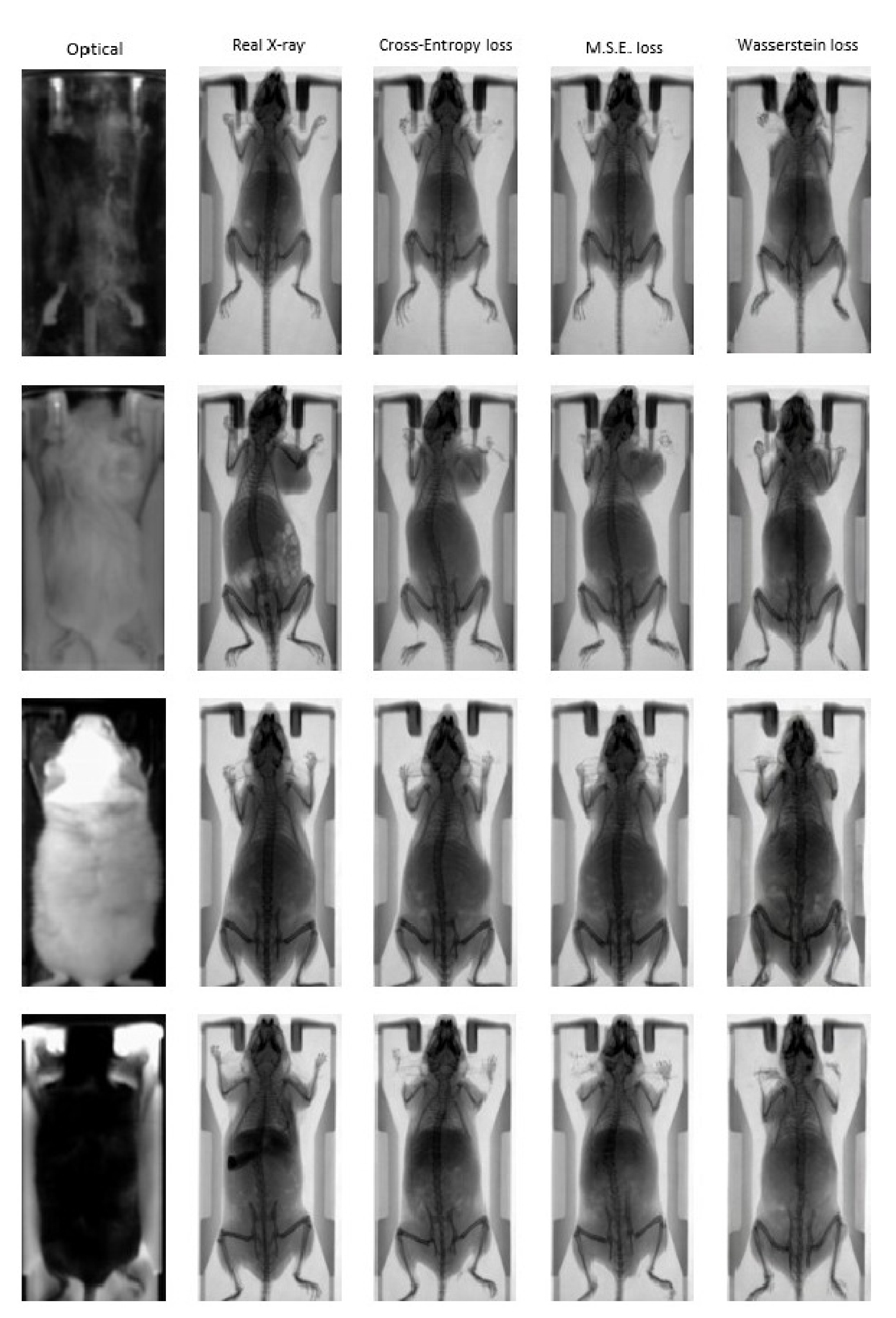
3.1. Quantitative Evaluation
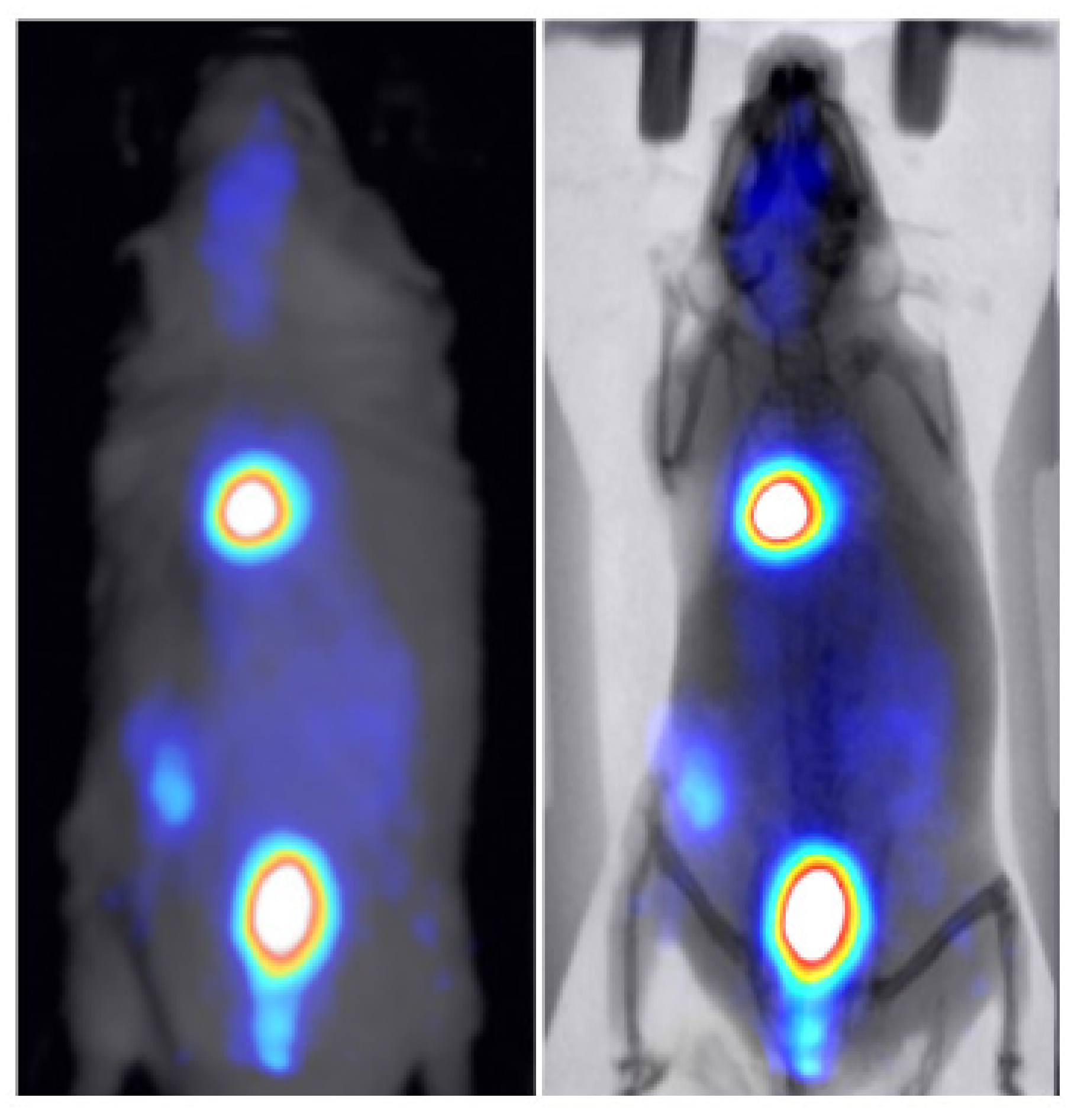
3.2. Animal Mapping during In Vivo Molecular Imaging Experiments
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Willmann, J.; van Bruggen, N.; Dinkelborg, L.; Gambhir, S. Molecular imaging in drug development. Nat. Rev. Drug Discov. 2008, 7, 591–607. [Google Scholar]
- Pysz, M.A.; Gambhir, S.S.; Willmann, J.K. Molecular imaging: Current status and emerging strategies. Clin. Radiol. 2010, 65, 500–516. [Google Scholar]
- Kagadis, G.; Ford, N.; Karbanatidis, D.; Loudos, G. Handbook of Small Animal Imaging; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Paul, S.M.; Mytelka, D.S.; Dunwiddie, C.T.; Persinger, C.C.; Munos, B.H.; Lindborg, S.R. How to improve RD productivity: The pharmaceutical industry’s grand challenge. Nat. Rev. Drug Discov. 2010, 9, 203–214. [Google Scholar]
- US Food and Drug Administration. US Department of Health and Human Services; Estimating the Maximum Safe Starting Dose in Initial Clinical Trials for Therapeutics in Adult Healthy Volunteers; Guidance for Industry: Rockville, MD, USA, 2015.
- Mannheim, J.G.; Kara, F.; Doorduin, J.; Fuchs, K.; Reischl, G.; Liang, S.; Verhoye, M.; Gremse, F.; Mezzanotte, L.; Huisman, M.C. Standardization of Small Animal Imaging-Current Status and Future Prospects. Mol. Imaging Biol. 2018, 20, 716–731. [Google Scholar]
- Stuker, F.; Ripoll, J.; Rudin, M. Fluorescence Molecular Tomography: Principles and Potential for Pharmaceutical Research. Pharmaceutics 2011, 3, 229–274. [Google Scholar]
- Debie, P.; Lafont, C.; Defrise, M.; Hansen, I.; van Willigen, D.M.; van Leeuwen, F.; Gijsbers, R.; D’Huyvetter, M.; Devoogdt, N.; Lahoutte, T.; et al. Size and affinity kinetics of nanobodies influence targeting and penetration of solid tumours. J. Control Release 2020, 317, 34–42. [Google Scholar]
- Georgiou, M.; Fysikopoulos, E.; Mikropoulos, K.; Fragogeorgi, E.; Loudos, G. Characterization of g-eye: A low cost benchtop mouse sized gamma camera for dynamic and static imaging studies. Mol. Imaging Biol. 2017, 19, 398. [Google Scholar]
- Zhang, H.; Bao, Q.; Vu, N.; Silverman, R.; Taschereau, R.; Berry-Pusey, B.; Douraghy, A.; Rannou, F.; Stout, D.; Chatziioannou, A. Performance evaluation of PETbox: A low cost bench top preclinical PET scanner. Mol. Imaging Biol. 2011, 13, 949–961. [Google Scholar]
- Rouchota, M.; Georgiou, M.; Fysikopoulos, E.; Fragogeorgi, E.; Mikropoulos, K.; Papadimitroulas, P.; Kagadis, G.; Loudos, G. A prototype PET/SPET/X-rays scanner dedicated for whole body small animal studies. Hell. J. Nucl. Med. 2017, 20, 146–153. [Google Scholar]
- Isola, P.; Zhu, J.-Y.; Zhou, T.; Efros, A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Proceedings, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 8–14 December 2019; Volume 32. [Google Scholar]
- Gong, K.; Yang, J.; Larson, P.E.Z.; Behr, S.C.; Hope, T.A.; Seo, Y.; Li, Q. MR-based Attenuation Correction for Brain PET Using 3D Cycle-Consistent Adversarial Network. IEEE Trans. Radiat. Plasma Med. Sci. 2021, 5, 185–192. [Google Scholar]
- Amyar, A.; Ruan, S.; Vera, P.; Decazes, P.; Modzelewski, R. RADIOGAN: Deep Convolutional Conditional Generative Adversarial Network to Generate PET Images. In Proceedings of the 7th International Conference on Bioinformatics Research and Applications (ICBRA), Berlin, Germany, 13–15 September 2020. [Google Scholar]
- Denck, J.; Guehring, J.; Maier, A.; Rothgang, E. Enhanced Magnetic Resonance Image Synthesis with Contrast-Aware Generative Adversarial Networks. J. Imaging 2021, 7, 133. [Google Scholar]
- Ouyang, J.; Chen, K.T.; Gong, E.; Pauly, J.; Zaharchuk, G. Ultra-low-dose PET reconstruction using generative adversarial network with feature matching and task-specific perceptual loss. Med. Phys. 2019, 46, 3555–3564. [Google Scholar]
- Fysikopoulos, E.; Rouchota, M.; Eleftheriadis, V.; Gatsiou, C.-A.; Pilatis, I.; Sarpaki, S.; Loudos, G.; Kostopoulos, S.; Glotsos, D. Photograph to X-ray Image Translation for Anatomical Mouse Mapping in Preclinical Nuclear Molecular Imaging. In Proceedings of the 2021 International Conference on Medical Imaging and Computer-Aided Diagnosis (MICAD 2021), Birmingham, UK, 25–26 March 2021. [Google Scholar]
- Mao, X.; Li, Q.; Xie, H.; Lau, R.Y.K.; Wang, Z.; Smolley, S.P. Least Squares Generative Adversarial Networks. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017. [Google Scholar]
- Arjovsky, M.; Chintala, S.; Bottou, L. Wasserstein GAN. arXiv 2017, arXiv:1701.07875. [Google Scholar]
- Sara, U.; Akter, M.; Uddin, M. Image Quality Assessment through FSIM, SSIM, MSE and PSNR—A Comparative Study. J. Comput. Commun. 2019, 7, 8–18. [Google Scholar]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar]
- Heusel, M.; Ramsauer, H.; Unterthiner, T.; Nessler, B.; Hochreiter, S. GANs Trained by a Two Time-Scale Update Rule Converge to a Local Nash Equilibrium. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Benny, Y.; Galanti, T.; Benaim, S.; Wolf, L. Evaluation Metrics for Conditional Image Generation. Int. J. Comput. Vis. 2021, 129, 1712–1731. [Google Scholar]
- Salimans, T.; Goodfellow, I.; Zaremba, W.; Cheung, V.; Radford, A.; Chen, X. Improved techniques for training gans. In Proceedings of the Advances in Neural Information Processing Systems, Barcelona, Spain, 5–10 December 2016. [Google Scholar]
- Kupyn, O.; Budzan, V.; Mykhailych, M.; Mishkin, D.; Matas, J.; DeblurGAN, J. Blind motion deblurring using conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018. [Google Scholar]
- Kaji, S.; Kida, S. Overview of image-to-image translation by use of deep neural networks: Denoising, super-resolution, modality conversion, and reconstruction in medical imaging. Radiol. Phys. Technol. 2019, 12, 235–248. [Google Scholar]
- Yoo, J.; Eom, H.; Choi, Y. Image-to-image translation using a crossdomain auto-encoder and decoder. Appl. Sci. 2019, 9, 4780. [Google Scholar]
- Cordts, M.; Omran, M.; Ramos, S.; Rehfeld, T.; Enzweiler, M.; Benenson, R.; Franke, U.; Roth, S.; Schiele, B. The cityscapes dataset for semantic urban scene understanding. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26–30 June 2016. [Google Scholar]
- Albahar, B.; Huang, J. Guided Image-to-Image Translation With Bi-Directional Feature Transformation. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Korea, 27–28 October 2019. [Google Scholar]
- Liu, Z.; Luo, P.; Qiu, S.; Wang, X.; Tang, X. DeepFashion: Powering Robust Clothes Recognition and Retrieval with Rich Annotations. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 26–30 June 2016. [Google Scholar]
- Xian, W.; Sangkloy, P.; Agrawal, V.; Raj, A.; Lu, J.; Fang, C.; Yu, F.; Hays, J. TextureGAN: Controlling Deep Image Synthesis with Texture Patches. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–22 June 2018. [Google Scholar]
- Paavilainen, P.; Akram, S.U.; Kannala, J. Bridging the Gap Between Paired and Unpaired Medical Image Translation. In Proceedings of the MICCAI Workshop on Deep Generative Models, Strasbourg, France, 1 October 2021. [Google Scholar]





| Mouse Color | Bed Color | Train | Test | Test/Train (%) |
|---|---|---|---|---|
| White | Black | 260 | 30 | 11.5 |
| White | White | 90 | 10 | 11.1 |
| Black | White | 260 | 30 | 11.5 |
| Black | Black | 90 | 10 | 11.1 |
| Total | 700 | 80 |
| Parameter | Value |
|---|---|
| Learning rate | 0.0002 |
| Beta 1 parameter for the optimizer (adam) | 0.5 |
| Beta 2 parameter for the optimizer (adam) | 0.999 |
| Maximum epochs | 200 |
| Lambda () weight for the L1 loss | 100 |
| Generator layers | 8 |
| Discriminator layers | 3 |
| Load size | 512 |
| Mini batch size | 1 |
| cGAN Loss Function | PSNR ↑ | SSIM ↑ | FID ↓ |
|---|---|---|---|
| Cross entropy | 21.923 | 0.771 | 85.428 |
| MSE | 21.954 | 0.770 | 90.824 |
| Wasserstein distance | 17.952 | 0.682 | 162.015 |
| Mouse Color | Bed Color | PSNR ↑ | SSIM ↑ | FID ↓ |
|---|---|---|---|---|
| black | white | 22.808 | 0.791 | 112.948 |
| black | black | 22.894 | 0.794 | 151.006 |
| white | black | 21.196 | 0.750 | 109.116 |
| white | white | 20.270 | 0.743 | 163.056 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fysikopoulos, E.; Rouchota, M.; Eleftheriadis, V.; Gatsiou, C.-A.; Pilatis, I.; Sarpaki, S.; Loudos, G.; Kostopoulos, S.; Glotsos, D. Optical to Planar X-ray Mouse Image Mapping in Preclinical Nuclear Medicine Using Conditional Adversarial Networks. J. Imaging 2021, 7, 262. https://doi.org/10.3390/jimaging7120262
Fysikopoulos E, Rouchota M, Eleftheriadis V, Gatsiou C-A, Pilatis I, Sarpaki S, Loudos G, Kostopoulos S, Glotsos D. Optical to Planar X-ray Mouse Image Mapping in Preclinical Nuclear Medicine Using Conditional Adversarial Networks. Journal of Imaging. 2021; 7(12):262. https://doi.org/10.3390/jimaging7120262
Chicago/Turabian StyleFysikopoulos, Eleftherios, Maritina Rouchota, Vasilis Eleftheriadis, Christina-Anna Gatsiou, Irinaios Pilatis, Sophia Sarpaki, George Loudos, Spiros Kostopoulos, and Dimitrios Glotsos. 2021. "Optical to Planar X-ray Mouse Image Mapping in Preclinical Nuclear Medicine Using Conditional Adversarial Networks" Journal of Imaging 7, no. 12: 262. https://doi.org/10.3390/jimaging7120262
APA StyleFysikopoulos, E., Rouchota, M., Eleftheriadis, V., Gatsiou, C.-A., Pilatis, I., Sarpaki, S., Loudos, G., Kostopoulos, S., & Glotsos, D. (2021). Optical to Planar X-ray Mouse Image Mapping in Preclinical Nuclear Medicine Using Conditional Adversarial Networks. Journal of Imaging, 7(12), 262. https://doi.org/10.3390/jimaging7120262






