Artificial Intelligence (AI) and Nuclear Features from the Fine Needle Aspirated (FNA) Tissue Samples to Recognize Breast Cancer
Abstract
1. Introduction
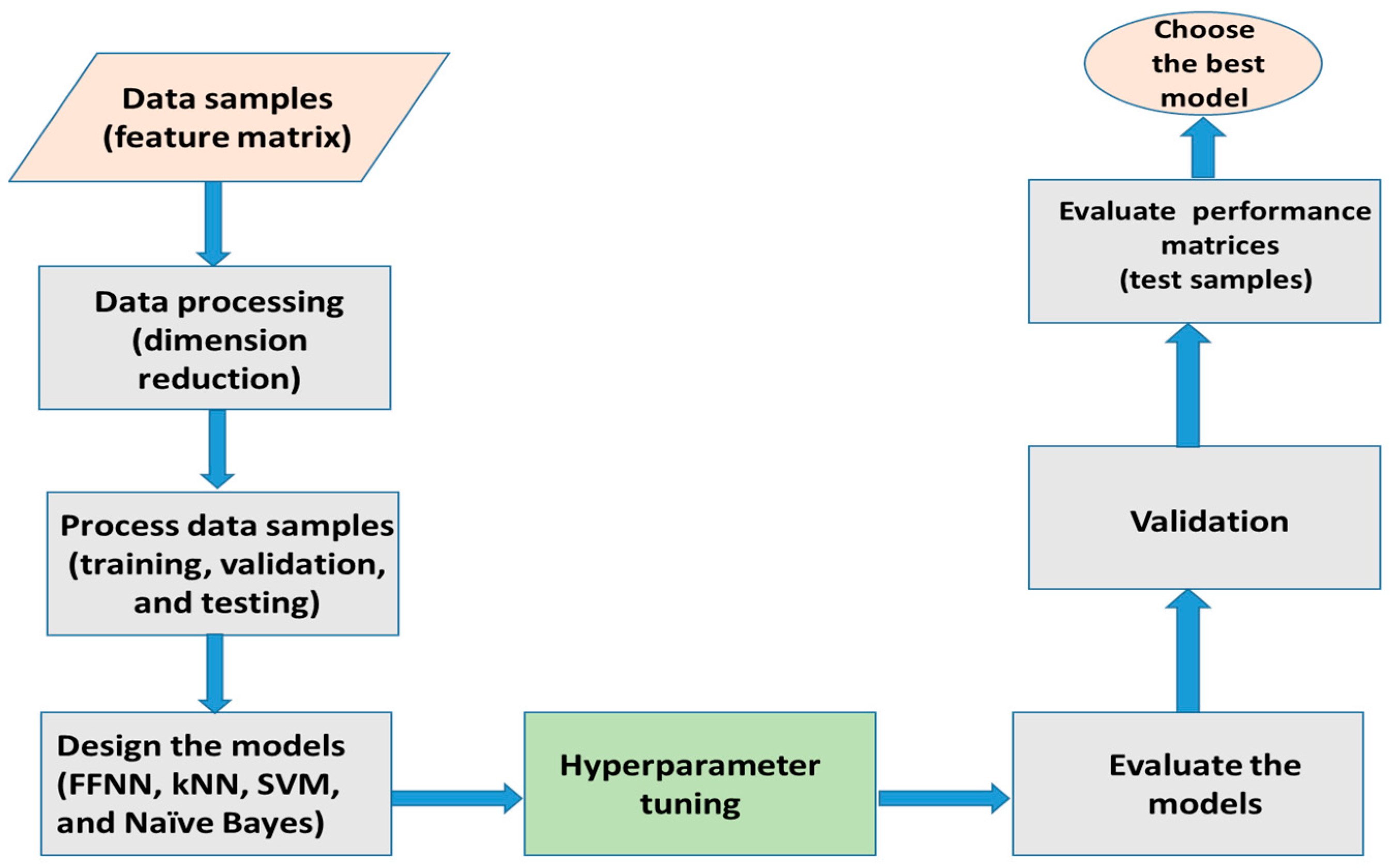
2. Materials and Methods
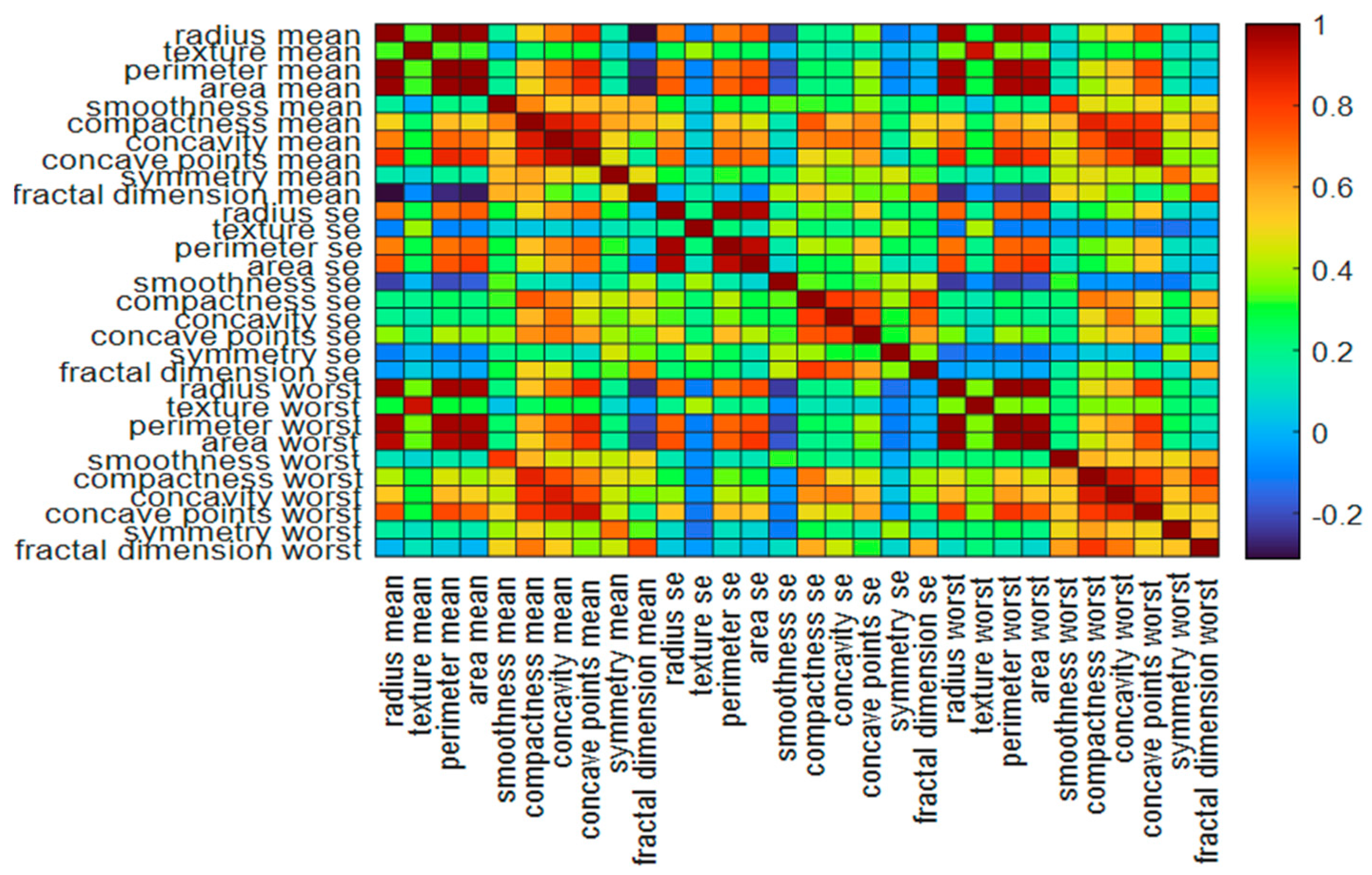
2.1. The Data Samples and Features
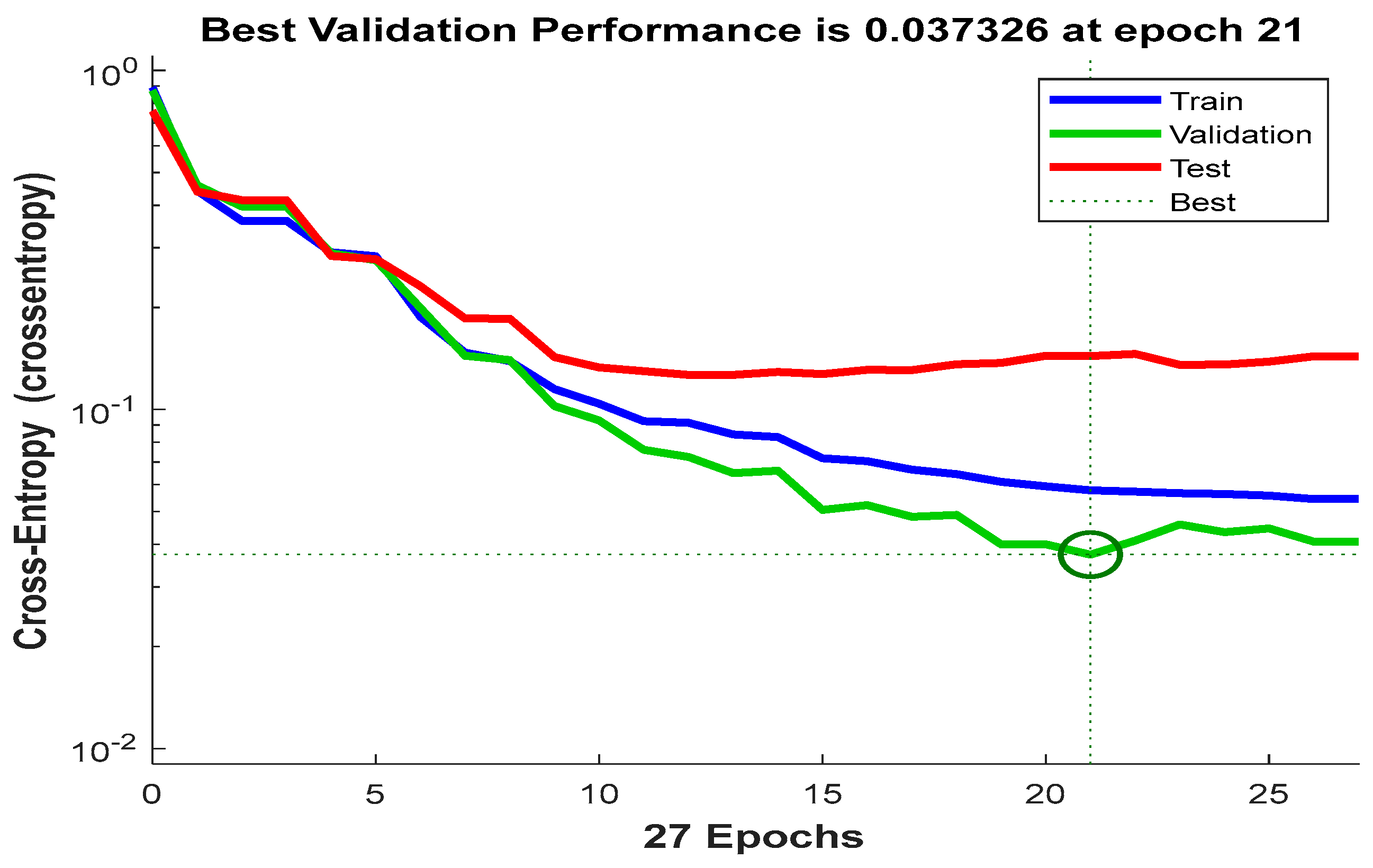
2.2. The Classification Network
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 70, 7–30. [Google Scholar] [CrossRef]
- Nasser, M.; Yusof, U.K. Deep Learning Based Methods for Breast Cancer Diagnosis: A Systematic Review and Future Direction. Diagnostics 2023, 13, 161. [Google Scholar] [CrossRef] [PubMed]
- Khalid, A.; Mehmood, A.; Alabrah, A.; Alkhamees, B.F.; Amin, F.; AlSalman, H.; Choi, G.S. Cancer Detection and Prevention Using Machine Learning. Diagnostics 2023, 13, 3113. [Google Scholar] [CrossRef] [PubMed]
- El Zahra, F.; Hateem, A.; Mohammad, M.; Tarique, M. Fourier transform based early detection of breast cancer by mammogram image processing. J. Biomed. Eng. Med. Imaging 2015, 2, 17–32. [Google Scholar] [CrossRef]
- Jiang, H.; Zhou, Y.; Lin, Y.; Chan, R.C.K.; Liu, J.; Chen, H. Deep Learning for Computational Cytology: A Survey. Med. Image Anal. 2023, 84, 102691. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, P.; Lacambra, M.; Tan, P.H.; Tse, G.M. Fine Needle Aspiration Cytology of the Breast: The Nonmalignant Categories. Pathol. Res. Int. 2011, 2011, 547580. [Google Scholar] [CrossRef] [PubMed]
- Islam, R.; Tarique, M. Chest X-ray Images to Differentiate COVID-19 from Pneumonia with Artificial Intelligence Techniques. Int. J. Biomed. Imaging 2022, 2022, 5318447. [Google Scholar] [CrossRef]
- Islam, R.; Tarique, M. Discriminating COVID-19 from Pneumonia using Machine Learning Algorithms and Chest X-ray Images. In Proceedings of the IEEE International Conference on Industrial Technology (ICIT), Shanghai, China, 22–25 August 2022; pp. 1–6. [Google Scholar] [CrossRef]
- Islam, R.; Abdel-Raheem, E.; Tarique, M. Cochleagram to Recognize Dysphonia: Auditory Perceptual Analysis for Health Informatics. IEEE Access 2024, 12, 59198–59210. [Google Scholar] [CrossRef]
- Islam, R.; Tarique, M. Robust Assessment of Dysarthrophonic Voice with RASTA-PLP Features: A Nonlinear Spectral Measures. In Proceedings of the IEEE International Conference on Mechatronics and Electrical Engineering (MEEE), Abu Dhabi, United Arab Emirates, 10–12 February 2023; pp. 74–78. [Google Scholar] [CrossRef]
- George, Y.M.; Zayed, H.H.; Roushdy, M.I.; Elbagour, B.M. Remote computer-aided breast cancer detection and diagnosis system based on cytological images. IEEE Syst. J. 2014, 8, 949–964. [Google Scholar] [CrossRef]
- Ara, S.; Das, A.; Dey, A. Malignant and Benign Breast Cancer Classification using Machine Learning Algorithms. In Proceedings of the International Conference on Artificial Intelligence (ICAI), Islamabad, Pakistan, 5–7 April 2021; pp. 97–101. [Google Scholar] [CrossRef]
- Khourdifi, Y.; Bahaj, M. Applying best machine learning algorithms for breast cancer prediction and classification. In Proceedings of the International Conference on Electronics, Control, Optimization and Computer Science (ICECOCS), Kenitra, Morocco, 5–7 December 2018; pp. 1–5. [Google Scholar] [CrossRef]
- Dhanya, R.; Paul, I.R.; Akula, S.S.; Sivakumar, M.; Nair, J.J. A comparative study for breast cancer prediction using machine learning and feature selection. In Proceedings of the International Conference on Intelligent Computing and Control Systems (ICCCS), Madurai, India, 15–17 May 2019; pp. 1049–1055. [Google Scholar] [CrossRef]
- Islam, M.M.; Iqbal, H.; Haque, M.R.; Hasan, M.K. Prediction of breast cancer using support vector machine and k-nearest neighbors. In Proceedings of the IEEE Region 10 Humanitarian Technology Conference (R10- HTC), Dhaka, Bangladesh, 21–23 December 2017; pp. 226–229. [Google Scholar] [CrossRef]
- Raza, A.; Ullah, N.; Khan, J.A.; Assam, M.; Guzzo, A.; Aljuaid, H. DeepBreastCancerNet: A Novel Deep Learning Model for Breast Cancer Detection Using Ultrasound Images. Appl. Sci. 2023, 13, 2082. [Google Scholar] [CrossRef]
- Reshan, M.S.A.; Amin, S.; Zeb, M.A.; Sulaiman, A.; Alshahrani, H.; Azar, A.T.; Shaikh, A. Enhancing Breast Cancer Detection and Classification Using Advanced Multi-Model Features and Ensemble Machine Learning Techniques. Life 2023, 13, 2093. [Google Scholar] [CrossRef]
- Singh, S.P.; Urooj, S.; Lay-Ekuakille, A. Breast Cancer detection using PCPCET and ADEWNN: A geometric invariant approach to medical X-ray image sensors. IEEE Sens. J. 2016, 16, 4847–4855. [Google Scholar] [CrossRef]
- Guo, R.; Lu, G.; Qin, B.; Fei, B. Ultrasound imaging technologies for breast cancer detection and management: A review. Ultrasound Med. Biol. 2018, 44, 37–70. [Google Scholar] [CrossRef]
- Byra, M.; Jarosik, P.; Szubert, A.; Galperin, M.; Ojeda-Fournier, H.; Olson, L.; O’Boyle, M.; Comstock, C.; Andre, M. Breast mass segmentation in ultrasound with selective kernel U-Net convolutional neural network. Biomed. Signal Process. Control 2020, 61, 102027. [Google Scholar] [CrossRef]
- Togaçar, M.; Özkurt, K.B.; Ergen, B.; Cömert, Z. BreastNet: A novel convolutional neural network model through histopathological images for the diagnosis of breast cancer. Phys. A Stat. Mech. Its Appl. 2020, 545, 123592. [Google Scholar] [CrossRef]
- Khan, S.; Islam, N.; Jan, Z.; Din, I.U.; Rodrigues, J.J.C. A novel deep learning based framework for the detection and classification of breast cancer using transfer learning. Pattern Recognit. Lett. 2019, 125, 1–6. [Google Scholar] [CrossRef]
- Nahid, A.A.; Mehrabi, M.A.; Kong, Y. Histopathological BC image classification by deep neural network techniques guided by local clustering. BioMed Res. Int. 2018, 2018, 2362108. [Google Scholar] [CrossRef]
- Mansouri, S.; Alhadidi, T.; Azouz, M.B. Breast Cancer Detection Using Low-Frequency Bioimpedance Device. Breast Cancer (Dove Med. Press) 2020, 12, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.; Pant, S.; Srivatzen, S.; Asokan, S. A Non-Invasive Breast Cancer Detection System Using FBG Thermal Sensor Array: A Feasibility Study. IEEE Sens. J. 2021, 21, 24106–24113. [Google Scholar] [CrossRef]
- Ertosun, M.G.; Rubin, D.L. Probabilistic visual search for masses within mammography images using deep learning. In Proceedings of the IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Washington, DC, USA, 9–12 November 2015; pp. 1310–1315. [Google Scholar] [CrossRef]
- Kumar, P.; Srivastava, S.; Mishra, R.K.; Sai, Y.P. End-to-end improved convolutional neural network model for breast cancer detection using mammographic data. J. Def. Model. Simul. 2022, 19, 375–384. [Google Scholar] [CrossRef]
- Gupta, K.G.; Sharma, D.K.; Ahmed, S.; Gupta, H.; Gupta, D.; Hsu, C.H. A Novel Lightweight Deep Learning-Based Histopathological Image Classification Model for IoMT. Neural Process. Lett. 2023, 55, 205–228. [Google Scholar] [CrossRef]
- Wang, Z.; Li, M.; Wang, H.; Jiang, H.; Yao, Y.; Zhang, H.; Xin, J. Breast Cancer Detection Using Extreme Learning Machine Based on Feature Fusion with CNN Deep Features. IEEE Access 2019, 7, 105146–105158. [Google Scholar] [CrossRef]
- Saidin, N.; Ngah, U.K.; Sakim, H.A.M.; Siong, D.N.; Hoe, M.K. Density based breast segmentation for mammograms using graph cut techniques. In Proceedings of the IEEE TENCON Region 10 Conference, Singapore, 23–26 January 2009; pp. 1–5. [Google Scholar] [CrossRef]
- Islam, R.; Tarique, M.; Abdel-Raheem, E. A Survey on Signal Processing Based Pathological Voice Detection Techniques. IEEE Access 2020, 8, 66749–66776. [Google Scholar] [CrossRef]
- Suvradeep, M.; Pranab, D. Fine Needle aspiration and core biopsy in the diagnosis of breast lesions: A comparison and review of the literature. Cytojournal 2016, 13, 18. [Google Scholar] [CrossRef]
- Zakhour, H.; Wells, C.; Perry, N.M. Diagnostic Cytopathology of the Breast, 1st ed.; Churchill Livingstone: London, UK, 1999. [Google Scholar]
- Zarbo, R.J.; Howanitz, P.J.; Bachner, P. Interinstitutional comparison of performance in breast fine-needle aspiration cytology. A Q-probe quality indicator study. Arch. Pathol. Lab. Med. 1991, 115, 743–750. [Google Scholar] [PubMed]
- Wolberg, W.; Mangasarian, O.; Street, N.; Street, W. Breast Cancer Wisconsin (Diagnostic); UCI Machine Learning Repository. 1995. Available online: https://archive.ics.uci.edu/dataset/17/breast+cancer+wisconsin+diagnostic (accessed on 11 August 2024).
- Street, W.N.; Wolberg, W.H.; Mangasarian, O.L. Nuclear feature extraction for breast tumor diagnosis. In Proceedings of the SPIE, San Jose, CA, USA, 11–16 July 1993; pp. 861–870. [Google Scholar] [CrossRef]
- Mandelbrot, B.B. Fractal Geometry of Nature; W. H. Freeman and Company: San Francisco, CA, USA, 1977; pp. 247–256. [Google Scholar]
- Islam, R.; Tarique, M. Blind Source Separation of Fetal ECG Using Fast Independent Component Analysis and Principle Component Analysis. Int. J. Sci. Technol. Res. 2020, 9, 80–95. [Google Scholar]
- Islam, R.; Abdel-Raheem, E.; Tarique, M. A study of using cough sounds and deep neural networks for the early detection of COVID-19. Biomed. Eng. Adv. 2022, 3, 100025. [Google Scholar] [CrossRef]
- NCCN Guidelines. Available online: https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1419 (accessed on 11 August 2024).






| Features | Definition | Malignant Mean (Mean, se, and Worst) | Benign Mean (Mean, se, and Worst) |
|---|---|---|---|
| Radius | Radius is measured by averaging the length of the radial line segments defined by the centroid of the snake and the individual snake points. | (17.46, 0.6, 21.13) | (13.61, 0.37, 15.48) |
| Texture | The texture of the cell nucleus is measured by finding the standard deviation or variance of the gray scale intensities in the component pixels of each image. | (21.60, 1.21, 29.31) | (19.14, 1.24, 25.37) |
| Perimeter | The total distance between the snake points constitutes the nuclear perimeter. | (115.36, 4.32, 141.37) | (88.35, 2.64, 101.72) |
| Nuclear area | Nuclear area is measured by counting the number of pixels on the interior of the snake and adding one-half of the pixels in the perimeter. | (978.37, 72.67, 1422.29) | (606.38, 35.3, 795.16) |
| Smoothness | Smoothness of a nuclear contour is quantified by measuring the difference between the length of a radial line and the mean length of the lines surrounding it. | (0.10, 0.0068, 0.145) | (0.095, 0.007, 0.123) |
| Compactness | Compactness is defined by combining the perimeter and area to give a measure of the compactness of the cell nuclei using the formula: perimeter2/area. | (0.015, 0.032, 0.375) | (0.097, 0.024, 0.23) |
| Concavity | Concavity is defined by drawing chords between non-adjacent snake points and measuring the extent to which the actual boundary of the nucleus lies on the inside of each chord. | (0.16, 0.042,0.45) | (0.078, 0.031, 0.024) |
| Concave points | Concave points are like the concavity but measure only the number, rather than the magnitude of contour concavities. | (0.088, 0.015, 0.182) | (0.042, 0.011, 0.012) |
| Symmetry | The symmetry is computed by measuring the major axis through the center and then measuring the length difference between lines perpendicular to the major axis to the cell boundary in both directions. | (0.190, 02, 0.323) | (0.18, 0.02, 0.284) |
| Fractal Dimension | The fractal dimension of a cell is approximated using the “coastline approximation-1” described by Mandelbrot [37]. | (0.062, 0.004, 0.09) | (0.062, 0.0038, 0.082) |
| Performance Measures | (%) |
|---|---|
| accuracy | 98.10 ± 1.01 |
| precision | 98.60 ± 1.01 |
| recall/sensitivity | 96.20 ± 1.02 |
| F1 Score | 97.40 ± 1.03 |
| npv | 97.80 ± 1.02 |
| specificity | 99.20 ± 1.02 |
| fnr | 1.45 ± 0.02 |
| fdr | 2.21 ± 0.01 |
| G-mean | 97.70 ± 1.01 |
| MCC | 95.90 ± 1.02 |
| DSc | 97.40 ± 1.03 |
| Performance Measures | Cubic SVM (%) | Weighted kNN (%) | Gaussian Naive Bayes (%) |
|---|---|---|---|
| accuracy | 97.72 ± 1.03 | 97.01 ± 1.13 | 94.02 ± 1.01 |
| precision | 98.54 ± 1.01 | 98.99 ± 1.14 | 92.38 ± 1.02 |
| recall/sensitivity | 95.28 ± 1.11 | 92.92 ± 1.12 | 91.51 ± 1.03 |
| F1 Score | 96.88 ± 1.01 | 95.86 ± 1.11 | 91.94 ± 1.03 |
| npv | 97.25 ± 1.01 | 95.95 ± 1.11 | 94.99 ± 1.11 |
| specificity | 99.16 ± 1.02 | 99.44 ± 1.12 | 95.52 ± 1.02 |
| fnr | 1.46 ± 0.11 | 1.01 ± 0.03 | 7.62 ± 0.01 |
| fdr | 2.75 ± 0.02 | 4.05 ± 0.02 | 5.01 ± 0.02 |
| G-mean | 97.20 ± 1.12 | 96.13 ± 1.13 | 93.49 ± 1.11 |
| MCC | 95.11 ± 1.11 | 93.64 ± 1.11 | 87.20 ± 1.12 |
| Dice Score | 96.88 ± 1.13 | 95.86 ± 1.15 | 91.94 ± 1.13 |
| Research Works | Samples | Features | Tools | Best Accuracy (%) |
|---|---|---|---|---|
| George Y. M. [11] | FNAC | Cell nuclei | Multilayer perceptron, PNN, learning vector quantization (LVQ), and SVM | 95.56 |
| Ara, S. [12] | FNAC | Cell nuclei | Random Forest, Logistic Regression, Decision Tree, Naive Bayes, SVM, and kNN | 96.50 |
| Khourdifi, Y [13] | FNAC | Cell nuclei | Random Forest, Naïve Bayes, SVM, and kNN | 97.90 |
| Islam M. [15] | FNAC | Morphological Features | SVM and kNN | 98.57 |
| Raza, A. [16] | Ultrasound | Breast lesions | DeepBraestCancerNet | 99.35 |
| Reshan, MSA [17] | FNAC | Morphological features | Ensemble machine learning | 99.89 |
| Singh, S. P [18] | Mammographic images | PCET moments | Adaptive Differential Evolution Wavelet Neural Network (ADEWNN) | 97.96 |
| Byra, M. [20] | Ultrasound | Segmentation | Selective kernel (SK) U-Net CNN | 97.90 |
| Togacar, M. [21] | Histopathological images | Original image | BreastNet | 98.80 |
| Nahid, A. [23] | Histopathological images | k-means and Mean-Shift clustering algorithm | CNN-LSTM | 91 |
| Ertosun, M. G [26] | Mammogram | In built feature extractor | CNN | 85% |
| Gupta, K. G [28] | Histopathological images | Image Enhancement | ReducedFireNet | 96.88 |
| Wang, Z. [29] | Mammogram | Deep features, morphological features, texture features, density features | CNN and unsupervised Extreme learning machine (ELM) | 86.50 |
| Proposed work | FNAC | Nuclear features | FFNN, SVM, kNN, and Naïve Bayes | 98.10 (FFNN) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, R.; Tarique, M. Artificial Intelligence (AI) and Nuclear Features from the Fine Needle Aspirated (FNA) Tissue Samples to Recognize Breast Cancer. J. Imaging 2024, 10, 201. https://doi.org/10.3390/jimaging10080201
Islam R, Tarique M. Artificial Intelligence (AI) and Nuclear Features from the Fine Needle Aspirated (FNA) Tissue Samples to Recognize Breast Cancer. Journal of Imaging. 2024; 10(8):201. https://doi.org/10.3390/jimaging10080201
Chicago/Turabian StyleIslam, Rumana, and Mohammed Tarique. 2024. "Artificial Intelligence (AI) and Nuclear Features from the Fine Needle Aspirated (FNA) Tissue Samples to Recognize Breast Cancer" Journal of Imaging 10, no. 8: 201. https://doi.org/10.3390/jimaging10080201
APA StyleIslam, R., & Tarique, M. (2024). Artificial Intelligence (AI) and Nuclear Features from the Fine Needle Aspirated (FNA) Tissue Samples to Recognize Breast Cancer. Journal of Imaging, 10(8), 201. https://doi.org/10.3390/jimaging10080201






