A Review of Advancements and Challenges in Liver Segmentation
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Public Datasets
3.2. Evaluation Standards
4. Development and Evolution of Liver Segmentation Technology
Early Methods
5. Threshold and Region-Growing Methods
6. Edge- and Shape-Based Methods
7. Deep Learning and Fully Convolutional Networks
8. Advances with U-Net and Variants
9. Integration of Emerging Technologies
10. Key Technological Milestones
11. Discussion
12. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Couinaud, C. Liver anatomy: Portal and suprahepatic or biliary segmentation. Dig. Surg. 1999, 16, 459–467. [Google Scholar] [CrossRef]
- Gonzalez, R.C.; Woods, R.E. Digital Image Process.; Pearson: London, UK, 2018. [Google Scholar]
- Gotra, A.; Sivakumaran, L.; Chartrand, G.; Vu, K.N.; Vandenbroucke-Menu, F.; Kauffmann, C.; Kadoury, S.; Gallix, B.; de Guise, J.A.; Tang, A. Liver segmentation: Indications, techniques and future directions. Insights Imaging 2017, 8, 377–392. [Google Scholar] [CrossRef] [PubMed]
- Bilic, P.; Christ, P.; Li, H.B.; Vorontsov, E.; Ben-Cohen, A.; Kaissis, G.; Szeskin, A.; Jacobs, C.; Mamani, G.E.H.; Chartrand, G.; et al. The Liver Tumor Segmentation Benchmark (LiTS). Med. Image Anal. 2023, 84, 102680. [Google Scholar] [CrossRef] [PubMed]
- Niño, S.B.; Bernardino, J.; Domingues, I. Algorithms for Liver Segmentation in Computed Tomography Scans: A Historical Perspective. Sensors 2024, 24, 1752. [Google Scholar] [CrossRef]
- Alirr, O.I.; Rahni, A.A.A. Survey on Liver Tumour Resection Planning System: Steps, Techniques, and Parameters. J. Digit. Imaging 2020, 33, 304–323. [Google Scholar] [CrossRef]
- Jeong, J.G.; Choi, S.; Kim, Y.J.; Lee, W.S.; Kim, K.G. Deep 3D attention CLSTM U-Net based automated liver segmentation and volumetry for the liver transplantation in abdominal CT volumes. Sci. Rep. 2022, 12, 6370. [Google Scholar] [CrossRef]
- Ansari, M.Y.; Abdalla, A.; Ansari, M.Y.; Ansari, M.I.; Malluhi, B.; Mohanty, S.; Mishra, S.; Singh, S.S.; Abinahed, J.; Al-Ansari, A.; et al. Practical utility of liver segmentation methods in clinical surgeries and interventions. BMC Med. Imaging 2022, 22, 97. [Google Scholar] [CrossRef] [PubMed]
- Bhat, M.; Rabindranath, M.; Chara, B.S.; Simonetto, D.A. Artificial intelligence, machine learning, and deep learning in liver transplantation. J. Hepatol. 2023, 78, 1216–1233. [Google Scholar] [CrossRef]
- Senthilvelan, J.; Jamshidi, N. A pipeline for automated deep learning liver segmentation (PADLLS) from contrast enhanced CT exams. Sci. Rep. 2022, 12, 15794. [Google Scholar] [CrossRef]
- Bilic, P.; Christ, P.; Li, H.B.; Vorontsov, E. LiTS (Liver Tumor Segmentation Challenge). Available online: https://competitions.codalab.org/competitions/17094 (accessed on 10 July 2024).
- Soler, L.; Hostettler, A.; Agnus, V.; Charnoz, A.; Fasquel, J.; Moreau, J.; Osswald, A.; Bouhadjar, M.; Marescaux, J. 3D Image Reconstruction for Comparison of Algorithm Database: A Patient Specific Anatomical and Medical Image Database; Technical Report; IRCAD: Strasbourg, France, 2010. [Google Scholar]
- Wang, J.; Zhang, X.; Guo, L.; Shi, C.; Tamura, S. Multi-scale attention and deep supervision-based 3D UNet for automatic liver segmentation from CT. Math. Biosci. Eng. 2023, 20, 1297–1316. [Google Scholar] [CrossRef]
- Yang, J.; Fu, M.; Hu, Y. Liver vessel segmentation based on inter-scale V-Net. Math. Biosci. Eng. 2021, 18, 4327–4340. [Google Scholar] [CrossRef]
- Chartrand, G.; Cresson, T.; Chav, R.; Gotra, A.; Tang, A.; De Guise, J.A. Liver Segmentation on CT and MR Using Laplacian Mesh Optimization. IEEE Trans. Bio-Med. Eng. 2017, 64, 2110–2121. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Chen, X.; Shi, F.; Zhu, W.; Tian, J.; Xiang, D. Automatic Liver Segmentation Based on Shape Constraints and Deformable Graph Cut in CT Images. IEEE Trans. Image Process. Publ. IEEE Signal Process. Soc. 2015, 24, 5315–5329. [Google Scholar] [CrossRef]
- Zhou, Y.R.; Kong, Q.Q.; Zhu, Y.; Su, Z. MCFA-UNet: Multiscale Cascaded Feature Attention U-Net for Liver Segmentation. IRBM 2023, 44, 100789. [Google Scholar] [CrossRef]
- Quinton, F.; Popoff, R.; Presles, B.; Leclerc, S.; Meriaudeau, F.; Nodari, G.; Lopez, O.; Pellegrinelli, J.; Chevallier, O.; Ginhac, D. A Tumour and Liver Automatic Segmentation (ATLAS) Dataset on Contrast-Enhanced Magnetic Resonance Imaging for Hepatocellular Carcinoma. Data 2023, 8, 79. [Google Scholar] [CrossRef]
- Hossain, M.S.A.; Gul, S.; Chowdhury, M.E.H.; Khan, M.S.; Sumon, M.S.I.; Bhuiyan, E.H.; Khandakar, A.; Hossain, M.; Sadique, A.; Al-Hashimi, I.; et al. Deep Learning Framework for Liver Segmentation from T1-Weighted MRI Images. Sensors 2023, 23, 8890. [Google Scholar] [CrossRef]
- Kavur, A.E.; Gezer, N.S.; Barış, M.; Aslan, S.; Conze, P.H.; Groza, V.; Pham, D.D.; Chatterjee, S.; Ernst, P.; Özkan, S.; et al. CHAOS Challenge—combined (CT-MR) healthy abdominal organ segmentation. Med. Image Anal. 2021, 69, 101950. [Google Scholar] [CrossRef] [PubMed]
- Mark and Mary Stevens Neuroimaging and Informatics Institute, University of Southern California, ATLAS v2.0 Dataset. Used under Creative Commons Attribution 4.0 International License (CC-BY 4.0). Available online: https://opendata.atlas.cern/docs/documentation/overview_data/ (accessed on 10 July 2024).
- Ali Emre Kavur, M.; Alper, S.; Oğuz, D.; Mustafa, B.; Sinem Gezer, N. (CHAOS—Combined (CT-MR) Healthy Abdominal Organ Segmentation Challenge Data (Version v1.03) [Data set]. Zenodo 2019. [Google Scholar] [CrossRef]
- Rafiei, S.; Karimi, N.; Mirmahboub, B.; Najarian, K.; Felfeliyan, B.; Samavi, S.; Reza Soroushmehr, S.M. Liver Segmentation in Abdominal CT Images Using Probabilistic Atlas and Adaptive 3D Region Growing. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 6310–6313. [Google Scholar] [CrossRef]
- Özcan, F.; Uçan, O.N.; Karaçam, S.; Tunçman, D. Fully Automatic Liver and Tumor Segmentation from CT Image Using an AIM-Unet. Bioengineering 2023, 10, 215. [Google Scholar] [CrossRef]
- Shelhamer, E.; Long, J.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 640–651. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Proceedings of the 18th International Conference, Munich, Germany, 5–9 October 2015; Navab, N., Hornegger, J., Wells, W., Frangi, A., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2015; Volume 9351, p. 9351. [Google Scholar] [CrossRef]
- Wang, K.; Mamidipalli, A.; Retson, T.; Bahrami, N.; Hasenstab, K.; Blansit, K.; Bass, E.; Delgado, T.; Cunha, G.; Middleton, M.S.; et al. Automated CT and MRI Liver Segmentation and Biometry Using a Generalized Convolutional Neural Network. Radiology Artif. Intell. 2019, 1, 180022. [Google Scholar] [CrossRef] [PubMed]
- Ahn, Y.; Yoon, J.S.; Lee, S.S.; Suk, H.I.; Son, J.H.; Sung, Y.S.; Lee, Y.; Kang, B.K.; Kim, H.S. Deep Learning Algorithm for Automated Segmentation and Volume Measurement of the Liver and Spleen Using Portal Venous Phase Computed Tomography Images. Korean J. Radiol. 2020, 21, 987–997. [Google Scholar] [CrossRef]
- Ahn, S.H.; Yeo, A.U.; Kim, K.H.; Kim, C.; Goh, Y.; Cho, S.; Lee, S.B.; Lim, Y.K.; Kim, H.; Shin, D.; et al. Comparative clinical evaluation of atlas and deep-learning-based auto-segmentation of organ structures in liver cancer. Radiat. Oncol. 2019, 14, 213. [Google Scholar] [CrossRef]
- Ayalew, Y.A.; Fante, K.A.; Mohammed, M.A. Modified U-Net for liver cancer segmentation from computed tomography images with a new class balancing method. BMC Biomed. Eng. 2021, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Liu, Z.; Van Der, M.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef]
- Li, X.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.W.; Heng, P.A. H-DenseUNet: Hybrid Densely Connected UNet for Liver and Tumor Segmentation From CT Volumes. IEEE Trans. Med. Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef]
- Luan, S.; Xue, X.; Ding, Y.; Wei, W.; Zhu, B. Adaptive Attention Convolutional Neural Network for Liver Tumor Segmentation. Front. Oncol. 2021, 11, 680807. [Google Scholar] [CrossRef] [PubMed]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Gibson, E.; Giganti, F.; Hu, Y.; Bonmati, E.; Bandula, S.; Gurusamy, K.; Davidson, B.; Pereira, S.P.; Clarkson, M.J.; Barratt, D.C. Automatic Multi-Organ Segmentation on Abdominal CT with Dense V-Networks. IEEE Trans. Med. Imaging 2018, 37, 1822–1834. [Google Scholar] [CrossRef] [PubMed]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016, Proceedings of the 9th International Conference, Athens, Greece, 17–21 October 2016; Ourselin, S., Joskowicz, L., Sabuncu, M., Unal, G., Wells, W., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2016; Volume 9901, p. 9901. [Google Scholar] [CrossRef]
- Dou, Q.; Chen, H.; Jin, Y.; Yu, L.; Qin, J.; Heng, P.A. 3D Deeply Supervised Network for Automatic Liver Segmentation from CT Volumes. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016, Proceedings of the 9th International Conference, Athens, Greece, 17–21 October 2016; Ourselin, S., Joskowicz, L., Sabuncu, M., Unal, G., Wells, W., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2016; Volume 9901, p. 9901. [Google Scholar] [CrossRef]
- Alom, M.Z.; Yakopcic, C.; Hasan, M.; Taha, T.M.; Asari, V.K. Recurrent residual U-Net for medical image segmentation. J. Med. Imaging 2019, 6, 014006. [Google Scholar] [CrossRef]
- Chen, X.; Wang, X.; Zhang, K.; Fung, K.M.; Thai, T.C.; Moore, K.; Mannel, R.S.; Liu, H.; Zheng, B.; Qiu, Y. Recent advances and clinical applications of deep learning in medical image analysis. Med. Image Anal. 2022, 79, 102444. [Google Scholar] [CrossRef]
- Siam, A.; Alsaify, A.R.; Mohammad, B.; Biswas, M.R.; Ali, H.; Shah, Z. Multimodal deep learning for liver cancer applications: A scoping review. Front. Artif. Intell. 2023, 6, 1247195. [Google Scholar] [CrossRef] [PubMed]
- Jafargholi Rangraz, E.; Coudyzer, W.; Maleux, G.; Baete, K.; Deroose, C.M.; Nuyts, J. Multi-modal image analysis for semi-automatic segmentation of the total liver and liver arterial perfusion territories for radioembolization. EJNMMI Res. 2019, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Roth, H.R.; Oda, H.; Zhou, X.; Shimizu, N.; Yang, Y.; Hayashi, Y.; Oda, M.; Fujiwara, M.; Misawa, K.; Mori, K. An application of cascaded 3D fully convolutional networks for medical image segmentation. Comput. Med. Imaging Graph. Off. J. Comput. Med. Imaging Soc. 2018, 66, 90–99. [Google Scholar] [CrossRef] [PubMed]
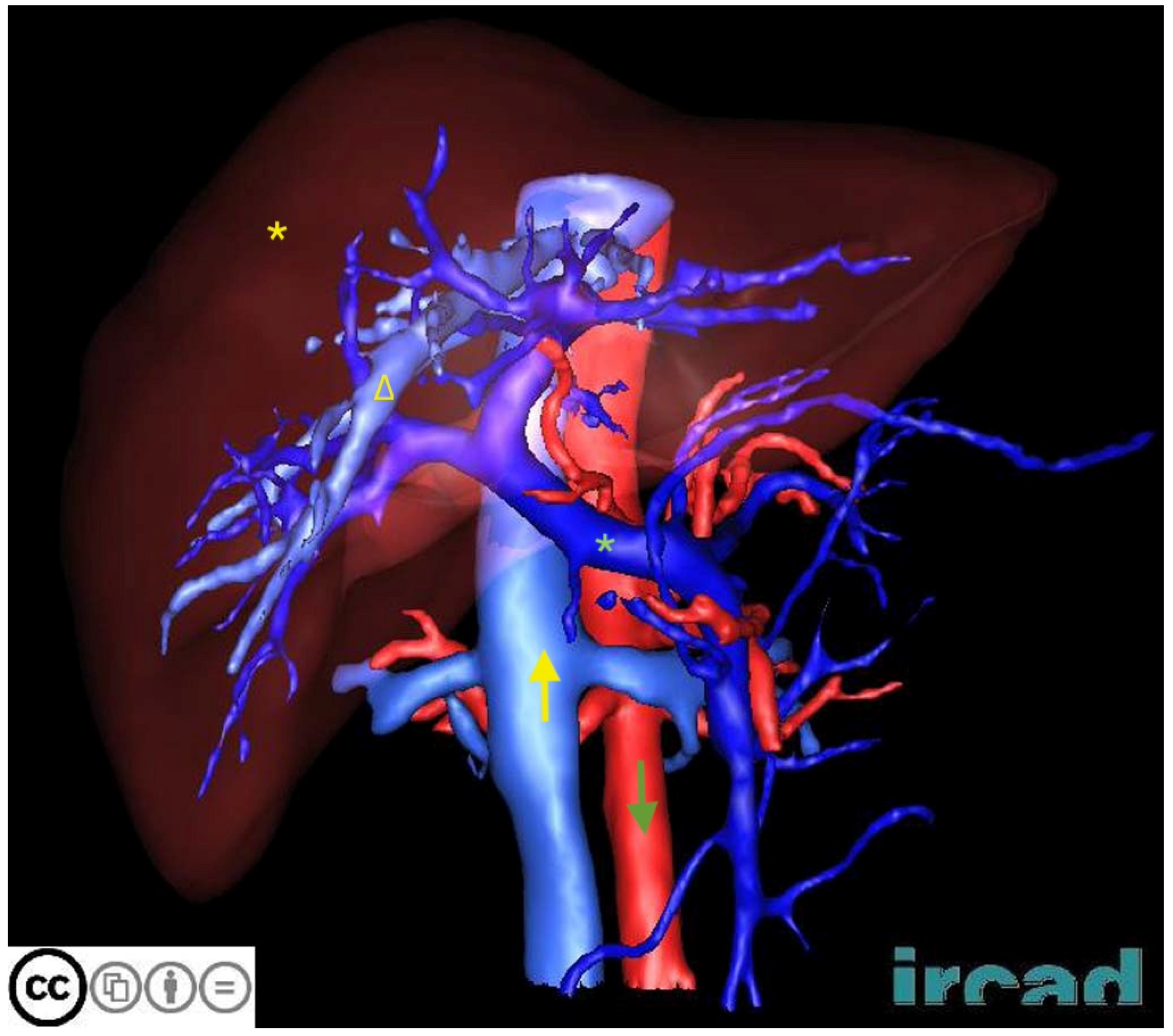
| Dataset | Content | Main Advantages | Main Disadvantages |
|---|---|---|---|
| LiTS | 201 abdominal CT scans with annotations for liver and liver tumor segmentation | Rich data, especially suitable for complex cases | Diversity of liver lesions represented may complicate algorithm development |
| 3DIRCADb | CT scans from 20 patients with annotations for liver and liver tumor segmentation | Detailed 3D reconstruction data aid the development of segmentation algorithms for complex liver structures | Small sample with limited case types |
| SLIVER07 | CT images of diseased livers | Useful for algorithm evaluation and comparison | Older datasets that may lack recently recognized lesion types and technologically up-to-date images |
| ATLAS | Annotated CE-MRI data, particularly for inoperable HCC | First dataset of its kind, suitable for the optimization of contouring in liver cancer treatment planning | Newer datasets requiring validation of compatibility for widespread use |
| CHAOS | Abdominal (kidney, liver, and spleen) CT and MRI scans from 80 patients in DICOM format with ground-truth masks annotated by certified radiologists | Promotes multi-modality imaging research and provides data on healthy organs that are useful for benchmarking | Small sample and lack of pathological information, may be insufficient for model training for pathology detection |
| Metric | Description | Usage | Limitations |
|---|---|---|---|
| Dice similarity coefficient | Measure of similarity between predicted and ground-truth segmentations [0–1 (perfect similarity)] | Ideal for tracking model performance improvements | May be misleading due to imbalanced classes, heavier penalization of errors in smaller regions |
| Jaccard index | Ratio of intersection to union between predicted and actual segmentations | Commonly used to assess overlap and similarity | Sensitive to noise and minor boundary deviations, resulting in fluctuations |
| Accuracy | Proportion of correctly classified voxels out of the total number of voxels | Measures overall classification correctness | Insufficient alone for full evaluation of algorithm performance |
| Sensitivity | Ability to identify true-positive samples | Used to evaluate detection capability in relevant areas | Insufficient alone for full evaluation of algorithm performance |
| Specificity | Ability to correctly identify true-negative samples | Used to evaluate exclusion capability in irrelevant areas | Insufficient alone for full evaluation of algorithm performance |
| Volume overlap error | Quantification of error between predicted and ground-truth segmentations | Suitable for the evaluation of large-volume structures | May be inaccurate for small volumes and sensitive to incidental errors |
| Relative volume difference | Measure of the relative difference between predicted and actual volumes | Focuses on overall volume accuracy | Less effective for complex shapes and sensitive to noise |
| Average symmetric surface distance | Average distance between the boundaries of predicted and actual segmentations | Used to assess boundary accuracy and detail quality | May overemphasize minor boundary errors, neglecting overall segmentation accuracy |
| Maximum symmetric surface distance | Maximum distance between the boundaries of predicted and actual segmentations | Used to assess maximum boundary deviation | May overemphasize minor boundary errors, neglecting overall segmentation accuracy |
| Method | Technique | Main Features | Advantages | Disadvantages |
|---|---|---|---|---|
| Manual segmentation | Liver contours delineated manually by radiologists or technicians | High accuracy, clinically accepted | Time consuming, operator dependent | |
| Semi-automatic segmentation | Algorithm-based segmentation with manual input | Reduces manual workload, adapts to complex structures | Requires manual intervention for complex or atypical anatomy | |
| Threshold segmentation | Liver and non-liver tissues distinguished based on intensity threshold | Simple and fast, easy to implement | Sensitive to threshold setting, image quality, and noise | |
| Region-growing algorithm | Expansion from a seed point to include similar neighboring pixels | Adapts to local image variations, improves segmentation accuracy | Sensitive to initial point selection, limited for complex structures | |
| Edge-based segmentation | Edge detection algorithms used to identify liver boundaries | Effective for images with clear boundaries | Sensitive to noise and blurry edges, struggles with complex shapes | |
| Fully automatic segmentation | Algorithm-driven segmentation with no manual intervention | Significantly reduces manual work, improves efficiency | Dependent on image quality and algorithm performance | |
| Edge-based segmentation | Edge detection algorithms used to identify liver boundaries | Effective for images with clear boundaries | Sensitive to noise and blurry edges, struggles with complex shapes | |
| Shape model segmentation | Statistical-shape or active-contour models used to guide segmentation | Enhances the recognition of complex structures, robust | High computational complexity, requires significant resources | |
| Fully convolutional network | Accepts arbitrarily sized images as input, makes pixel-level predictions via learned features | Handles complex image features | Less effective for small objects and detailed images | |
| U-Net | Symmetrical contracting and expanding paths used to capture context and fine details | Performs well on small datasets, captures context and details | Less effective for large images and complex background differentiation | |
| ResNet | Deep network models trained using residual connections | Improves model performance, supports deeper networks | Complex network structure, major hardware requirements, prone to overfitting on small datasets | |
| SegNet | Encoder–decoder structure, retains spatial information | Fewer parameters, suitable for fine segmentation tasks | Less accurate than U-Net and DenseNet for intricate medical images | |
| DenseNet | Efficient parameters, feature extraction improved with dense connections | Suitable for large 3D datasets | High computational cost, memory consumption | |
| V-Net | Specifically designed for 3D image data, segmentation accuracy enhanced through multiscale feature extraction | Adapts to 3D medical imaging data, captures spatial features | High computational resource requirement, long training times | |
| 3D U-Net | Handles 3D image data, 3D contextual information utilized | Improves 3D image segmentation accuracy | High computational resource requirement, prone to overfitting on small datasets | |
| R2U-Net | U-Net, residual connections, and recurrent neural networks combined, enhancing feature extraction capabilities | Improves fine segmentation of complex organs, adapts to various liver sizes and shapes | Complex network structure, high computational resource demand | |
| Multimodal deep learning | Data from different imaging modalities integrated, enhancing diagnostic information | Improves diagnostic accuracy and reliability | Complex data alignment and processing, high computational resource requirement |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, D.; Jiang, Y.; Zhou, X.; Wu, D.; Feng, X. A Review of Advancements and Challenges in Liver Segmentation. J. Imaging 2024, 10, 202. https://doi.org/10.3390/jimaging10080202
Wei D, Jiang Y, Zhou X, Wu D, Feng X. A Review of Advancements and Challenges in Liver Segmentation. Journal of Imaging. 2024; 10(8):202. https://doi.org/10.3390/jimaging10080202
Chicago/Turabian StyleWei, Di, Yundan Jiang, Xuhui Zhou, Di Wu, and Xiaorong Feng. 2024. "A Review of Advancements and Challenges in Liver Segmentation" Journal of Imaging 10, no. 8: 202. https://doi.org/10.3390/jimaging10080202
APA StyleWei, D., Jiang, Y., Zhou, X., Wu, D., & Feng, X. (2024). A Review of Advancements and Challenges in Liver Segmentation. Journal of Imaging, 10(8), 202. https://doi.org/10.3390/jimaging10080202






