A Low-Spin CoII/Nitroxide Complex for Distance Measurements at Q-Band Frequencies
Abstract
:1. Introduction
2. Results and Discussion
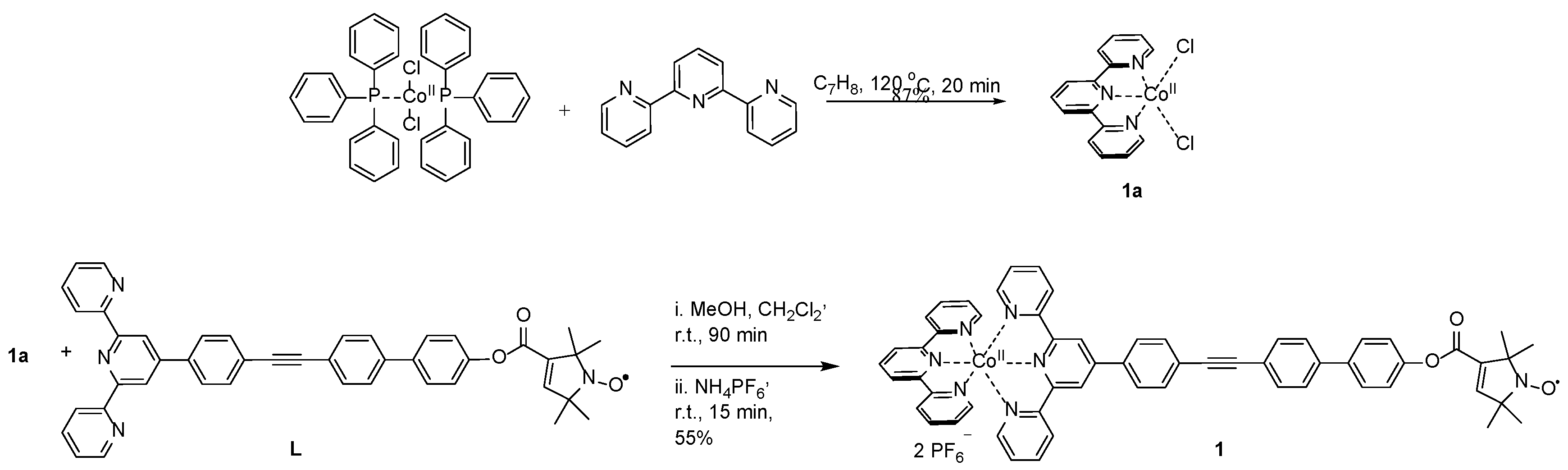
2.1. Synthesis of the Chemical Model
2.2. PDS on 1
3. Conclusions
4. Methods
4.1. General Synthesis Conditions
4.2. Syntheses
4.3. X-ray Crystallography
4.4. EPR Sample Preparation
4.5. EPR Spectroscopy
4.5.1. Echo-Detected EPR Spectrum
4.5.2. Inversion Recovery Data
4.5.3. Phase Memory Time Data
4.5.4. Q-Band DEER and RIDME
4.5.5. Data Analysis
4.5.6. Calculation of Sensitivity
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Milov, A.D.; Ponomarev, A.B.; Tsvetkov, Y.D. Electron Electron Double-Resonance in Electron-Spin Echo—Model Biradical Systems and the Sensitized Photolysis of Decalin. Chem. Phys. Lett. 1984, 110, 67–72. [Google Scholar] [CrossRef]
- Milov, A.D.; Salikhov, K.M.; Shirov, M.D. Application of Eldor in Electron-Spin Echo for Paramagnetic Center Space Distribution in Solids. Fiz. Tverd. Tela 1981, 23, 975–982. [Google Scholar]
- Kulik, L.V.; Dzuba, S.A.; Grigoryev, I.A.; Tsvetkov, Y.D. Electron dipole-dipole interaction in ESEEM of nitroxide biradicals. Chem. Phys. Lett. 2001, 343, 315–324. [Google Scholar] [CrossRef]
- EPR Spectroscopy: Fundamentals and Methods; Goldfarb, D.; Stoll, S. (Eds.) Wiley: Hoboken, NJ, USA, 1988. [Google Scholar]
- Pannier, M.; Veit, S.; Godt, A.; Jeschke, G.; Spiess, H.W. Dead-time free measurement of dipole-dipole interactions between electron spins. J. Magn. Reson. 2000, 142, 331–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, R.G.; Singel, D.J. Double electron-electron resonance spin-echo modulation—Spectroscopic measurement of electron-spin pair separations in orientationally disordered solids. J. Chem. Phys. 1993, 98, 5134–5146. [Google Scholar] [CrossRef] [Green Version]
- Martin, R.E.; Pannier, M.; Diederich, F.; Gramlich, V.; Hubrich, M.; Spiess, H.W. Determination of End-to-End Distances in a Series of TEMPO Diradicals of up to 2.8 nm Length with a New Four-Pulse Double Electron Electron Resonance Experiment. Angew. Chem. Int. Ed. Engl. 1998, 37, 2833–2837. [Google Scholar] [CrossRef]
- Milikisyants, S.; Scarpelli, F.; Finiguerra, M.G.; Ubbink, M.; Huber, M. A pulsed EPR method to determine distances between paramagnetic centers with strong spectral anisotropy and radicals: The dead-time free RIDME sequence. J. Magn. Reson. 2009, 201, 48–56. [Google Scholar] [CrossRef]
- Konov, K.B.; Knyazev, A.A.; Galyametdinov, Y.G.; Isaev, N.P.; Kulik, L.V. Selective Hole-Burning in RIDME Experiment: Dead-Time Free Measurement of Dipolar Modulation. Appl. Magn. Reson. 2013, 44, 949–966. [Google Scholar] [CrossRef]
- Giannoulis, A.; Motion, C.L.; Oranges, M.; Buhl, M.; Smith, G.M.; Bode, B.E. Orientation selection in high-field RIDME and PELDOR experiments involving low-spin CoII ions. Phys. Chem. Chem. Phys. 2018, 20, 2151–2154. [Google Scholar] [CrossRef] [Green Version]
- Giannoulis, A.; Oranges, M.; Bode, B.E. Monitoring Complex Formation by Relaxation-Induced Pulse Electron Paramagnetic Resonance Distance Measurements. Chemphyschem 2017, 18, 2318–2321. [Google Scholar] [CrossRef] [Green Version]
- Abdullin, D.; Duthie, F.; Meyer, A.; Muller, E.S.; Hagelueken, G.; Schiemann, O. Comparison of PELDOR and RIDME for distance measurements between nitroxides and low-spin Fe(III) ions. J. Phys. Chem. B 2015, 119, 13534–13542. [Google Scholar] [CrossRef] [PubMed]
- Jassoy, J.J.; Berndhauser, A.; Duthie, F.; Kuhn, S.P.; Hagelueken, G.; Schiemann, O. Versatile Trityl Spin Labels for Nanometer Distance Measurements on Biomolecules In Vitro and within Cells. Angew. Chem. Int. Ed. Engl. 2017, 56, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Russell, H.; Stewart, R.; Prior, C.; Oganesyan, V.S.; Gaule, T.G.; Lovett, J.E. DEER and RIDME Measurements of the Nitroxide-Spin Labelled Copper-Bound Amine Oxidase Homodimer from Arthrobacter Globiformis. Appl. Magn. Reson. 2021, 52, 995–1015. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.; Schiemann, O. PELDOR and RIDME Measurements on a High-Spin Manganese(II) Bisnitroxide Model Complex. J. Phys. Chem. A 2016, 120, 3463–3472. [Google Scholar] [CrossRef]
- Ritsch, I.; Hintz, H.; Jeschke, G.; Godt, A.; Yulikov, M. Improving the accuracy of Cu(ΙΙ)-nitroxide RIDME in the presence of orientation correlation in water-soluble Cu(ΙΙ)-nitroxide rulers. Phys. Chem. Chem. Phys. 2019, 21, 9810–9830. [Google Scholar] [CrossRef] [Green Version]
- Abdullin, D.; Brehm, P.; Fleck, N.; Spicher, S.; Grimme, S.; Schiemann, O. Pulsed EPR Dipolar Spectroscopy on Spin Pairs with one Highly Anisotropic Spin Center: The Low-Spin FeIII Case. Chem. Eur. J. 2019, 25, 14388–14398. [Google Scholar] [CrossRef] [Green Version]
- Boulon, M.E.; Fernandez, A.; Pineda, E.M.; Chilton, N.F.; Timco, G.; Fielding, A.J.; Winpenny, R.E.P. Measuring Spin…Spin Interactions between Heterospins in a Hybrid [2] Rotaxane. Angew. Chem. Int. Ed. Engl. 2017, 56, 3876–3879. [Google Scholar] [CrossRef]
- Astashkin, A.V.; Rajapakshe, A.; Cornelison, M.J.; Johnson-Winters, K.; Enemark, J.H. Determination of the Distance between the Mo(V) and Fe(III) Heme Centers of Wild Type Human Sulfite Oxidase by Pulsed EPR Spectroscopy. J. Phys. Chem. B 2012, 116, 1942–1950. [Google Scholar] [CrossRef] [Green Version]
- Azarkh, M.; Bieber, A.; Qi, M.; Fischer, J.W.A.; Yulikov, M.; Godt, A.; Drescher, M. Gd(III)-Gd(III) Relaxation-Induced Dipolar Modulation Enhancement for In-Cell Electron Paramagnetic Resonance Distance Determination. J. Phys. Chem. Lett. 2019, 10, 1477–1481. [Google Scholar] [CrossRef]
- Breitgoff, F.D.; Keller, K.; Qi, M.; Klose, D.; Yulikov, M.; Godt, A.; Jeschke, G. UWB DEER and RIDME distance measurements in Cu(II)-Cu(II) spin pairs. J. Magn. Reson. 2019, 308, 106560. [Google Scholar] [CrossRef]
- Ackermann, K.; Wort, J.L.; Bode, B.E. Nanomolar Pulse Dipolar EPR Spectroscopy in Proteins: CuII-CuII and Nitroxide-Nitroxide Cases. J. Phys. Chem. B. 2021, 125, 5358–5364. [Google Scholar] [CrossRef] [PubMed]
- Razzaghi, S.; Qi, M.; Nalepa, A.I.; Godt, A.; Jeschke, G.; Savitsky, A.; Yulikov, M. RIDME Spectroscopy with Gd(III) Centers. J. Phys. Chem. Lett. 2014, 5, 3970–3975. [Google Scholar] [CrossRef] [PubMed]
- Akhmetzyanov, D.; Ching, H.Y.; Denysenkov, V.; Demay-Drouhard, P.; Bertrand, H.C.; Tabares, L.C.; Policar, C.; Prisner, T.F.; Un, S. RIDME spectroscopy on high-spin Mn2+ centers. Phys. Chem. Chem. Phys. 2016, 18, 30857–30866. [Google Scholar] [CrossRef] [Green Version]
- Collauto, A.; Frydman, V.; Lee, M.D.; Abdelkader, E.H.; Feintuch, A.; Swarbrick, J.D.; Graham, B.; Otting, G.; Goldfarb, D. RIDME distance measurements using Gd(III) tags with a narrow central transition. Phys. Chem. Chem. Phys. 2016, 18, 19037–19049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller, K.; Mertens, V.; Qi, M.; Nalepa, A.I.; Godt, A.; Savitsky, A.; Jeschke, G.; Yulikov, M. Computing distance distributions from dipolar evolution data with overtones: RIDME spectroscopy with Gd(III)-based spin labels. Phys. Chem. Chem. Phys. 2017, 19, 17856–17876. [Google Scholar] [CrossRef] [PubMed]
- Salvadori, E.; Fusco, E.; Chiesa, M. Long-Range Spatial Distribution of Single Aluminum Sites in Zeolites. J. Phys. Chem. Lett. 2022, 13, 1283–1289. [Google Scholar] [CrossRef]
- Kuzhelev, A.A.; Krumkacheva, O.A.; Shevelev, G.Y.; Yulikov, M.; Fedin, M.V.; Bagryanskaya, E.G. Room-temperature distance measurements using RIDME and the orthogonal spin labels trityl/nitroxide. Phys. Chem. Chem. Phys. 2018, 20, 10224–10230. [Google Scholar] [CrossRef]
- Savitsky, A.; Dubinskii, A.A.; Zimmermann, H.; Lubitz, W.; Mobius, K. High-Field Dipolar Electron Paramagnetic Resonance (EPR) Spectroscopy of Nitroxide Biradicals for Determining Three-Dimensional Structures of Biomacromolecules in Disordered Solids. J. Phys. Chem. B 2011, 115, 11950–11963. [Google Scholar] [CrossRef]
- Abdullin, D.; Suchatzki, M.; Schiemann, O. Six-Pulse RIDME Sequence to Avoid Background Artifacts. Appl. Magn. Reson. 2021, 1–16. [Google Scholar] [CrossRef]
- Astashkin, A.V. Mapping the Structure of Metalloproteins with RIDME. Methods Enzymol. 2015, 563, 251–284. [Google Scholar]
- Keller, K.; Qi, M.; Gmeiner, C.; Ritsch, I.; Godt, A.; Jeschke, G.; Savitsky, A.; Yulikov, M. Intermolecular background decay in RIDME experiments. Phys. Chem. Chem. Phys. 2019, 21, 8228–8245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldfarb, D. Metal-Based Spin Labeling for Distance Determination. Struct. Inf. Spin Labels Intrinsic Paramagn. Cent. Biosci. 2012, 152, 163–204. [Google Scholar]
- Ji, M.; Ruthstein, S.; Saxena, S. Paramagnetic metal ions in pulsed ESR distance distribution measurements. Acc. Chem. Res. 2014, 47, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Schiemann, O.; Prisner, T.F. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Q. Rev. Biophys. 2007, 40, 1–53. [Google Scholar] [CrossRef] [PubMed]
- Abdullin, D.; Schiemann, O. Pulsed Dipolar EPR Spectroscopy and Metal Ions: Methodology and Biological Applications. Chempluschem 2020, 85, 353–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giannoulis, A.; Feintuch, A.; Unger, T.; Amir, S.; Goldfarb, D. Monitoring the Conformation of the Sba1/Hsp90 Complex in the Presence of Nucleotides with Mn(II)-Based Double Electron-Electron Resonance. J. Phys. Chem. Lett. 2021, 12, 12235–12241. [Google Scholar] [CrossRef]
- Keller, K.; Doll, A.; Qi, M.A.; Godt, A.; Jeschke, G.; Yulikov, M. Averaging of nuclear modulation artefacts in RIDME experiments. J. Magn. Reson. 2016, 272, 108–113. [Google Scholar] [CrossRef] [Green Version]
- Meyer, A.; Abdullin, D.; Schnakenburg, G.; Schiemann, O. Single and double nitroxide labeled bis(terpyridine)-copper(II): Influence of orientation selectivity and multispin effects on PELDOR and RIDME. Phys. Chem. Chem. Phys. 2016, 18, 9262–9271. [Google Scholar] [CrossRef]
- Meyer, A.; Jassoy, J.J.; Spicher, S.; Berndhauser, A.; Schiemann, O. Performance of PELDOR, RIDME, SIFTER, and DQC in measuring distances in trityl based bi- and triradicals: Exchange coupling, pseudosecular coupling and multi-spin effects. Phys. Chem. Chem. Phys. 2018, 20, 13858–13869. [Google Scholar] [CrossRef]
- Dal Farra, M.G.; Richert, S.; Martin, C.; Larminie, C.; Gobbo, M.; Bergantino, E.; Timmel, C.R.; Bowen, A.M.; Di Valentin, M. Light-Induced Pulsed EPR Dipolar Spectroscopy on a Paradigmatic Hemeprotein. Chemphyschem 2019, 20, 931–935. [Google Scholar] [CrossRef] [Green Version]
- Doll, A.; Qi, M.; Godt, A.; Jeschke, G. CIDME: Short distances measured with long chirp pulses. J. Magn. Reson. 2016, 273, 73–82. [Google Scholar] [CrossRef] [Green Version]
- Wort, J.L.; Arya, S.; Ackermann, K.; Stewart, A.J.; Bode, B.E. Pulse Dipolar EPR Reveals Double-Histidine Motif Cu(II)-NTA Spin-Labeling Robustness against Competitor Ions. J. Phys. Chem. Lett. 2021, 12, 2815–2819. [Google Scholar] [CrossRef] [PubMed]
- Wort, J.L.; Ackermann, K.; Giannoulis, A.; Stewart, A.J.; Norman, D.G.; Bode, B.E. Sub-Micromolar Pulse Dipolar EPR Spectroscopy Reveals Increasing CuII-labelling of Double-Histidine Motifs with Lower Temperature. Angew. Chem. Int. Ed. Engl. 2019, 58, 11681–11685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Astashkin, A.V.; Elmore, B.O.; Fan, W.H.; Guillemette, J.G.; Feng, C.J. Pulsed EPR Determination of the Distance between Heme Iron and FMN Centers in a Human Inducible Nitric Oxide Synthase. J. Am. Chem. Soc. 2010, 132, 12059–12067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giannoulis, A.; Ackermann, K.; Spindler, P.E.; Higgins, C.; Cordes, D.B.; Slawin, A.M.Z.; Prisner, T.F.; Bode, B.E. Nitroxide-nitroxide and nitroxide-metal distance measurements in transition metal complexes with two or three paramagnetic centres give access to thermodynamic and kinetic stabilities. Phys. Chem. Chem. Phys. 2018, 20, 11196–11205. [Google Scholar] [CrossRef] [Green Version]
- Wort, J.L.; Ackermann, K.; Norman, D.G.; Bode, B.E. A general model to optimise Cu(II) labelling efficiency of double-histidine motifs for pulse dipolar EPR applications. Phys. Chem. Chem. Phys. 2021, 23, 3810–3819. [Google Scholar] [CrossRef]
- Cunningham, T.F.; Putterman, M.R.; Desai, A.; Horne, W.S.; Saxena, S. The double-histidine Cu2+-binding motif: A highly rigid, site-specific spin probe for electron spin resonance distance measurements. Angew. Chem. Int. Ed. Engl. 2015, 54, 6330–6334. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.; Lawless, M.J.; Rule, G.S.; Saxena, S. The Cu2+-nitrilotriacetic acid complex improves loading of alpha-helical double histidine site for precise distance measurements by pulsed ESR. J. Magn. Reson. 2018, 286, 163–171. [Google Scholar] [CrossRef]
- Yang, Y.; Gong, Y.J.; Litvinov, A.; Liu, H.K.; Yang, F.; Su, X.C.; Goldfarb, D. Generic tags for Mn(II) and Gd(III) spin labels for distance measurements in proteins. Phys. Chem. Chem. Phys. 2017, 19, 26944–26956. [Google Scholar] [CrossRef]
- Martorana, A.; Yang, Y.; Zhao, Y.; Li, Q.F.; Su, X.C.; Goldfarb, D. Mn(II) tags for DEER distance measurements in proteins via C-S attachment. Dalton Trans. 2015, 44, 20812–20816. [Google Scholar] [CrossRef]
- Pfrunder, M.C.; Whittaker, J.J.; Parsons, S.; Moubaraki, B.; Murray, K.S.; Moggach, S.A.; Sharma, N.; Micallef, A.S.; Clegg, J.; McMurtrie, J.C. Controlling Spin Switching with Anionic Supramolecular Frameworks. Chem. Mater. 2020, 32, 3229–3234. [Google Scholar] [CrossRef]
- Pavlov, A.A.; Denisov, G.L.; Kiskin, M.A.; Nelyubina, Y.V.; Novikov, V.V. Probing Spin Crossover in a Solution by Paramagnetic NMR Spectroscopy. Inorg. Chem. 2017, 56, 14759–14762. [Google Scholar] [CrossRef] [PubMed]
- Spindler, P.E.; Glaser, S.J.; Skinner, T.E.; Prisner, T.F. Broadband inversion PELDOR spectroscopy with partially adiabatic shaped pulses. Angew. Chem. Int. Ed. Engl. 2013, 52, 3425–3429. [Google Scholar] [CrossRef] [PubMed]
- Gaponenko, V.; Altieri, A.S.; Li, J.; Byrd, R.A. Breaking symmetry in the structure determination of (large) symmetric protein dimers. J. Biomol. NMR 2002, 24, 143–148. [Google Scholar] [CrossRef]
- Dvoretsky, A.; Gaponenko, V.; Rosevear, P.R. Derivation of structural restraints using a thiol-reactive chelator. FEBS Lett. 2002, 528, 189–192. [Google Scholar] [CrossRef] [Green Version]
- Bahramzadeh, A.; Huber, T.; Otting, G. Three-Dimensional Protein Structure Determination Using Pseudocontact Shifts of Backbone Amide Protons Generated by Double-Histidine Co2+-Binding Motifs at Multiple Sites. Biochemistry 2019, 58, 3243–3250. [Google Scholar] [CrossRef] [PubMed]
- Seebald, L.M.; DeMott, C.M.; Ranganathan, S.; Asare-Okai, P.N.; Glazunova, A.; Chen, A.; Shekhtman, A.; Royzen, M. Cobalt-based paramagnetic probe to study RNA-protein interactions by NMR. J. Inorg. Biochem. 2017, 170, 202–208. [Google Scholar] [CrossRef] [Green Version]
- Zehnder, J.; Cadalbert, R.; Terradot, L.; Ernst, M.; Bockmann, A.; Guntert, P.; Meier, B.H.; Wiegand, T. Paramagnetic Solid-State NMR to Localize the Metal-Ion Cofactor in an Oligomeric DnaB Helicase. Chem. Eur. J. 2021, 27, 7745–7755. [Google Scholar] [CrossRef]
- Balayssac, S.; Bertini, I.; Bhaumik, A.; Lelli, M.; Luchinat, C. Paramagnetic shifts in solid-state NMR of proteins to elicit structural information. Proc. Natl. Acad. Sci. USA 2008, 105, 17284–17289. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Nakayama, Y.; Yasuda, H. Controlled vinyl-addition-type polymerization of norbornene initiated by several cobalt complexes having substituted terpyridine ligands. J. Organomet. Chem. 2004, 689, 744–750. [Google Scholar] [CrossRef]
- Ackermann, K.; Giannoulis, A.; Cordes, D.B.; Slawin, A.M.Z.; Bode, B.E. Assessing dimerisation degree and cooperativity in a biomimetic small-molecule model by pulsed EPR. Chem. Commun. 2015, 51, 5257–5260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bode, B.E.; Plackmeyer, J.; Prisner, T.F.; Schiemann, O. PELDOR measurements on a nitroxide-labeled Cu(II) porphyrin: Orientation selection, spin-density distribution, and conformational flexibility. J. Phys. Chem. A 2008, 112, 5064–5073. [Google Scholar] [CrossRef] [PubMed]
- Eaton, G.R.; Eaton, S.S.; Barr, D.P.; Weber, R.T. Quantitative EPR. Quant. Epr. 2010, 1–185. [Google Scholar] [CrossRef]
- Diaz-Torres, R.; Alvarez, S. Coordinating ability of anions and solvents towards transition metals and lanthanides. Dalton Trans. 2011, 40, 10742–10750. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, G.; Chechik, V.; Ionita, P.; Godt, A.; Zimmermann, H.; Banham, J.; Timmel, C.R.; Hilger, D.; Jung, H. DeerAnalysis2006—A comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 2006, 30, 473–498. [Google Scholar] [CrossRef]
- Ackermann, K.; Chapman, A.; Bode, B.E. A Comparison of Cysteine-Conjugated Nitroxide Spin Labels for Pulse Dipolar EPR Spectroscopy. Molecules 2021, 26, 7534. [Google Scholar] [CrossRef]
- Jeschke, G. DEER distance measurements on proteins. Annu. Rev. Phys. Chem. 2012, 63, 419–446. [Google Scholar] [CrossRef] [Green Version]
- Jeschke, G.; Polyhach, Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys. Chem. Chem. Phys. 2007, 9, 1895–1910. [Google Scholar] [CrossRef]
- Siemeling, U.; Vor der Bruggen, J.; Vorfeld, U.; Stammler, A.; Stammler, H.G. Large scale synthesis of 4′-(4-bromophenyl)-2,2′: 6′,2″-terpyridine and nature of the mysterious green by-product. Z. Naturforsch. B 2003, 58, 443–446. [Google Scholar] [CrossRef]
- Mutai, T.; Arita, S.; Araki, K. Phenyl-substituted 2,2′: 6′,2″-terpyridine as a new series of fluorescent compounds—Their photophysical properties and fluorescence tuning. J. Chem. Soc. Perkin Trans. 2 2001, 7, 1045–1050. [Google Scholar] [CrossRef]
- CrystalClear-SM Expert v2.1; Rigaku Americas: The Woodlands, TX, USA; Rigaku Corporation: Tokyo, Japan, 2015.
- Palatinus, L.; Chapuis, G. SUPERFLIP—A computer program for the solution of crystal structures by charge flipping in arbitrary dimensions. J. Appl. Crystallogr. 2007, 40, 786–790. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- CrystalStructure v4.3.0; Rigaku Americas: The Woodlands, TX, USA; Rigaku Corporation: Tokyo, Japan, 2018.
- Chiang, Y.W.; Borbat, P.P.; Freed, J.H. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J. Magn. Reson. 2005, 172, 279–295. [Google Scholar] [CrossRef] [PubMed]
- Worswick, S.G.; Spencer, J.A.; Jeschke, G.; Kuprov, I. Deep neural network processing of DEER data. Sci. Adv. 2018, 4, eaat5218. [Google Scholar] [CrossRef] [Green Version]
- Hogben, H.J.; Krzystyniak, M.; Charnock, G.T.; Hore, P.J.; Kuprov, I. Spinach-a software library for simulation of spin dynamics in large spin systems. J. Magn. Reson. 2011, 208, 179–194. [Google Scholar] [CrossRef]
- Giannoulis, A.; Cordes, D.B.; Slawin, A.M.Z.; Bode, B.E. A Low-Spin Co(II)/Nitroxide Complex for Distance Measurements at Q-Band Frequencies (Dataset); University of St Andrews Research Portal: St Andrews, UK, 2022. [Google Scholar] [CrossRef]
- Scarpelli, F.; Bartucci, R.; Sportelli, L.; Guzzi, R. Solvent effect on librational dynamics of spin-labelled haemoglobin by ED- and CW-EPR. Eur. Biophys. J. 2011, 40, 273–279. [Google Scholar] [CrossRef]
- Kuzhelev, A.A.; Krumkacheva, O.A.; Ivanov, M.Y.; Prikhod’ko, S.A.; Adonin, N.Y.; Tormyshev, V.M.; Bowman, M.K.; Fedin, M.V.; Bagryanskaya, E.G. Pulse EPR of Triarylmethyl Probes: A New Approach for the Investigation of Molecular Motions in Soft Matter. J. Phys. Chem. B 2018, 122, 8624–8630. [Google Scholar] [CrossRef]
- Bahrenberg, T.; Rosenski, Y.; Carmieli, R.; Zibzener, K.; Qi, M.; Frydman, V.; Godt, A.; Goldfarb, D.; Feintuch, A. Improved sensitivity for W-band Gd(III)-Gd(III) and nitroxide-nitroxide DEER measurements with shaped pulses. J. Magn. Reson. 2017, 283, 1–13. [Google Scholar] [CrossRef]
- Bretschneider, M.; Spindler, P.E.; Rogozhnikova, O.Y.; Trukhin, D.V.; Endeward, B.; Kuzhelev, A.A.; Bagryanskaya, E.; Tormyshev, V.M.; Prisner, T.F. Multiquantum Counting of Trityl Radicals. J. Phys. Chem. Lett. 2020, 11, 6286–6290. [Google Scholar] [CrossRef]
- Reginsson, G.W.; Kunjir, N.C.; Sigurdsson, S.T.; Schiemann, O. Trityl Radicals: Spin Labels for Nanometer-Distance Measurements. Chem. Eur. J. 2012, 18, 13580–13584. [Google Scholar] [CrossRef]
- Akhmetzyanov, D.; Schops, P.; Marko, A.; Kunjir, N.C.; Sigurdsson, S.T.; Prisner, T.F. Pulsed EPR dipolar spectroscopy at Q- and G-band on a trityl biradical. Phys. Chem. Chem. Phys. 2015, 17, 24446–24451. [Google Scholar] [CrossRef] [PubMed]
- Giannoulis, A.; Yang, Y.; Gong, Y.J.; Tan, X.; Feintuch, A.; Carmieli, R.; Bahrenberg, T.; Liu, Y.; Su, X.C.; Goldfarb, D. DEER distance measurements on trityl/trityl and Gd(III)/trityl labelled proteins. Phys. Chem. Chem. Phys. 2019, 21, 10217–10227. [Google Scholar] [CrossRef] [PubMed]
- van Amsterdam, I.M.; Ubbink, M.; Canters, G.W.; Huber, M. Measurement of a Cu-Cu distance of 26 A by a pulsed EPR method. Angew. Chem. Int. Ed. Engl. 2003, 42, 62–64. [Google Scholar] [CrossRef] [PubMed]
- Sameach, H.; Ghosh, S.; Gevorkyan-Airapetov, L.; Saxena, S.; Ruthstein, S. EPR Spectroscopy Detects Various Active State Conformations of the Transcriptional Regulator CueR. Angew. Chem. Int. Ed. Engl. 2019, 58, 3053–3056. [Google Scholar] [CrossRef] [Green Version]
- Henderson, I.M.; Hayward, R.C. Kinetic stabilities of bis-terpyridine complexes with iron(II) and cobalt(II) in organic solvent environments. J. Mater. Chem. 2012, 22, 21366–21369. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giannoulis, A.; Cordes, D.B.; Slawin, A.M.Z.; Bode, B.E. A Low-Spin CoII/Nitroxide Complex for Distance Measurements at Q-Band Frequencies. Magnetochemistry 2022, 8, 43. https://doi.org/10.3390/magnetochemistry8040043
Giannoulis A, Cordes DB, Slawin AMZ, Bode BE. A Low-Spin CoII/Nitroxide Complex for Distance Measurements at Q-Band Frequencies. Magnetochemistry. 2022; 8(4):43. https://doi.org/10.3390/magnetochemistry8040043
Chicago/Turabian StyleGiannoulis, Angeliki, David B. Cordes, Alexandra M. Z. Slawin, and Bela E. Bode. 2022. "A Low-Spin CoII/Nitroxide Complex for Distance Measurements at Q-Band Frequencies" Magnetochemistry 8, no. 4: 43. https://doi.org/10.3390/magnetochemistry8040043
APA StyleGiannoulis, A., Cordes, D. B., Slawin, A. M. Z., & Bode, B. E. (2022). A Low-Spin CoII/Nitroxide Complex for Distance Measurements at Q-Band Frequencies. Magnetochemistry, 8(4), 43. https://doi.org/10.3390/magnetochemistry8040043







