Continuous Flow Labeling and In-Line Magnetic Separation of Cells
Abstract
:1. Introduction
2. Materials and Methods
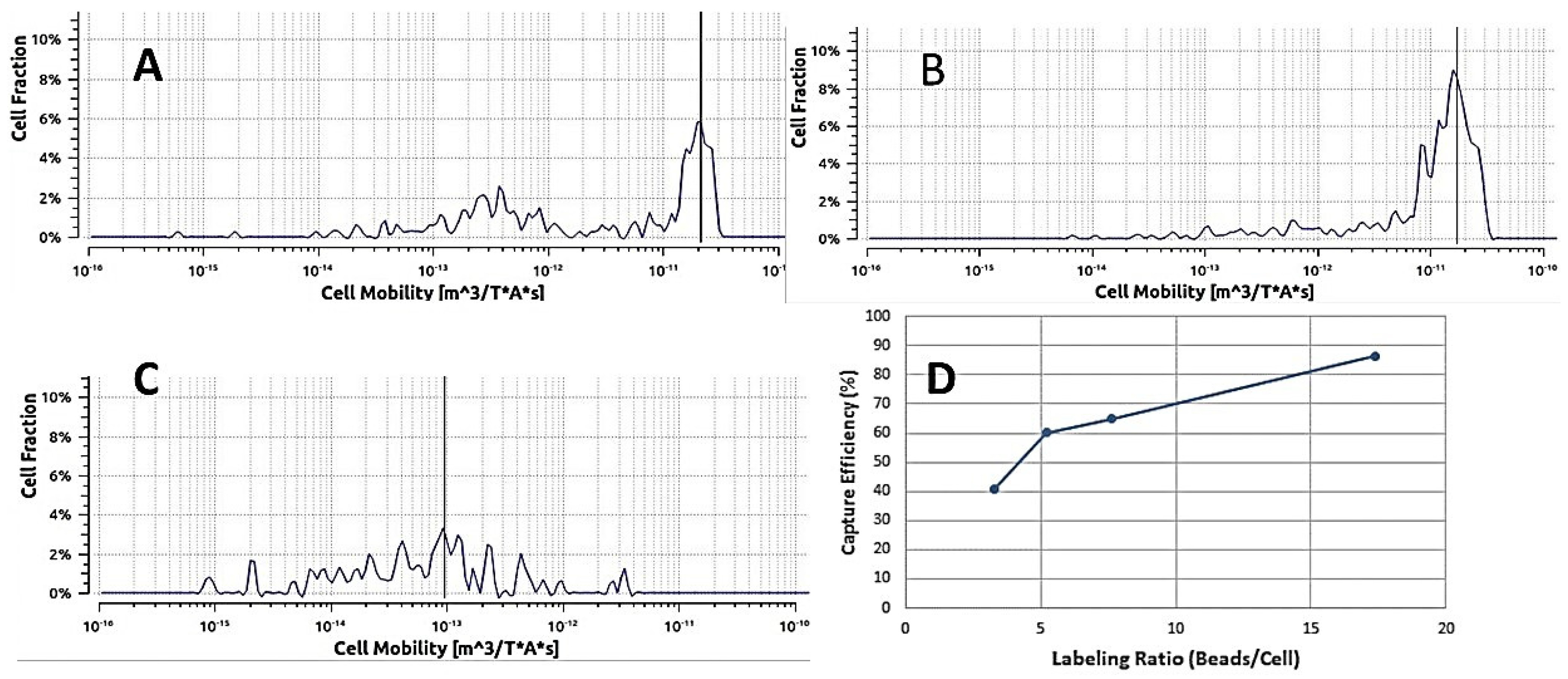
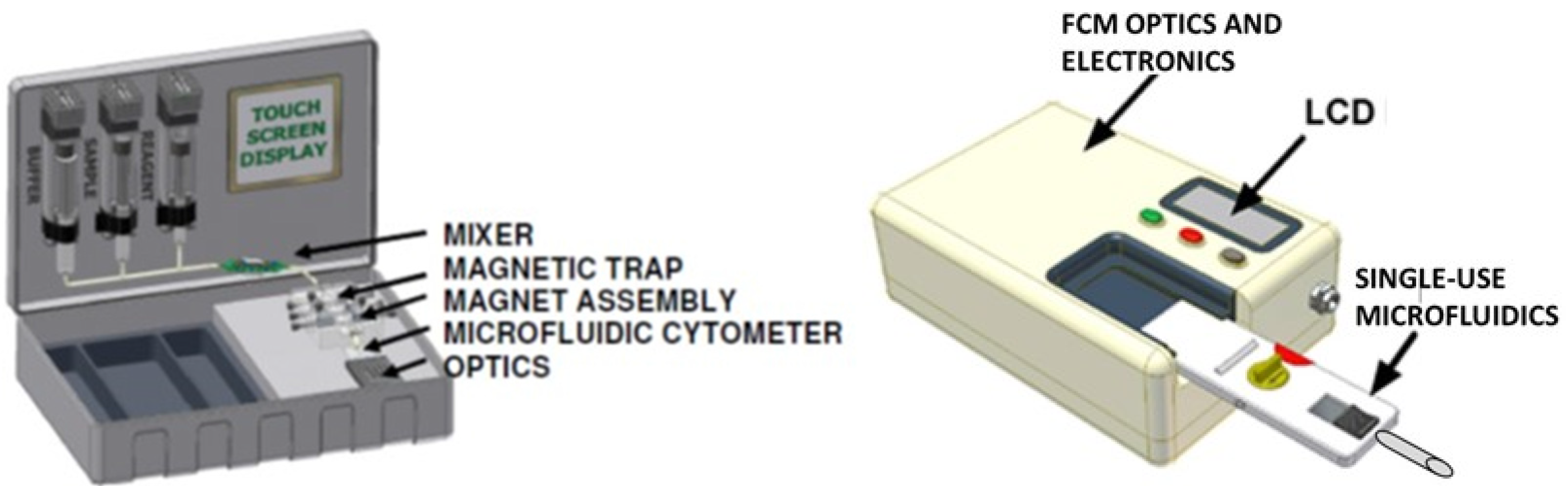
2.1. In-Line Mixing Device
2.2. Cells and Labels
2.3. Magnetic Reagents
2.4. Properties of Components in the Simulated Labeling Mixture
2.5. Magnetophoretic Mobillity Measurement
2.6. Magnetic Trap
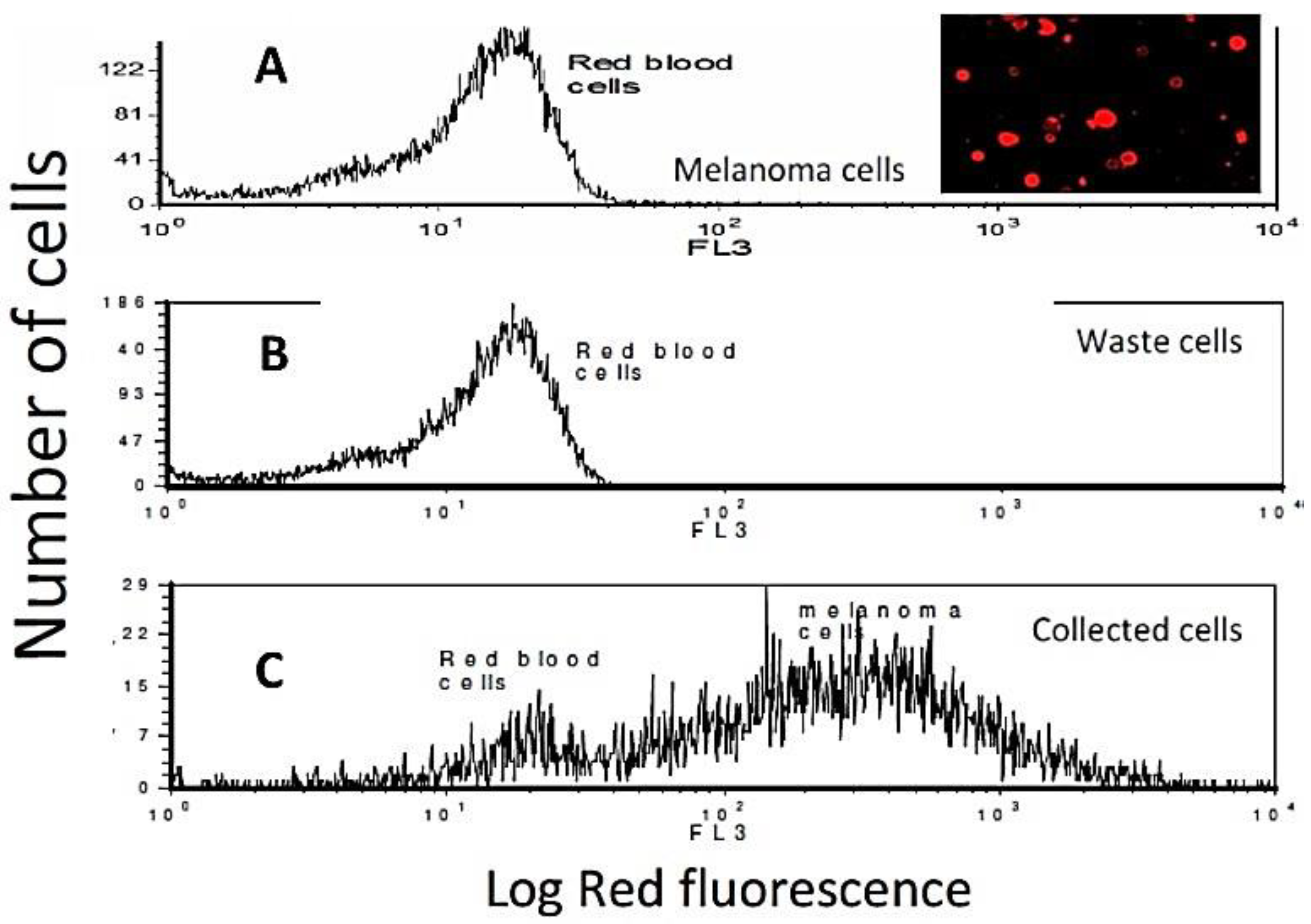
2.7. Fluorescence and Flow Cytometry
3. Theory and Calculations
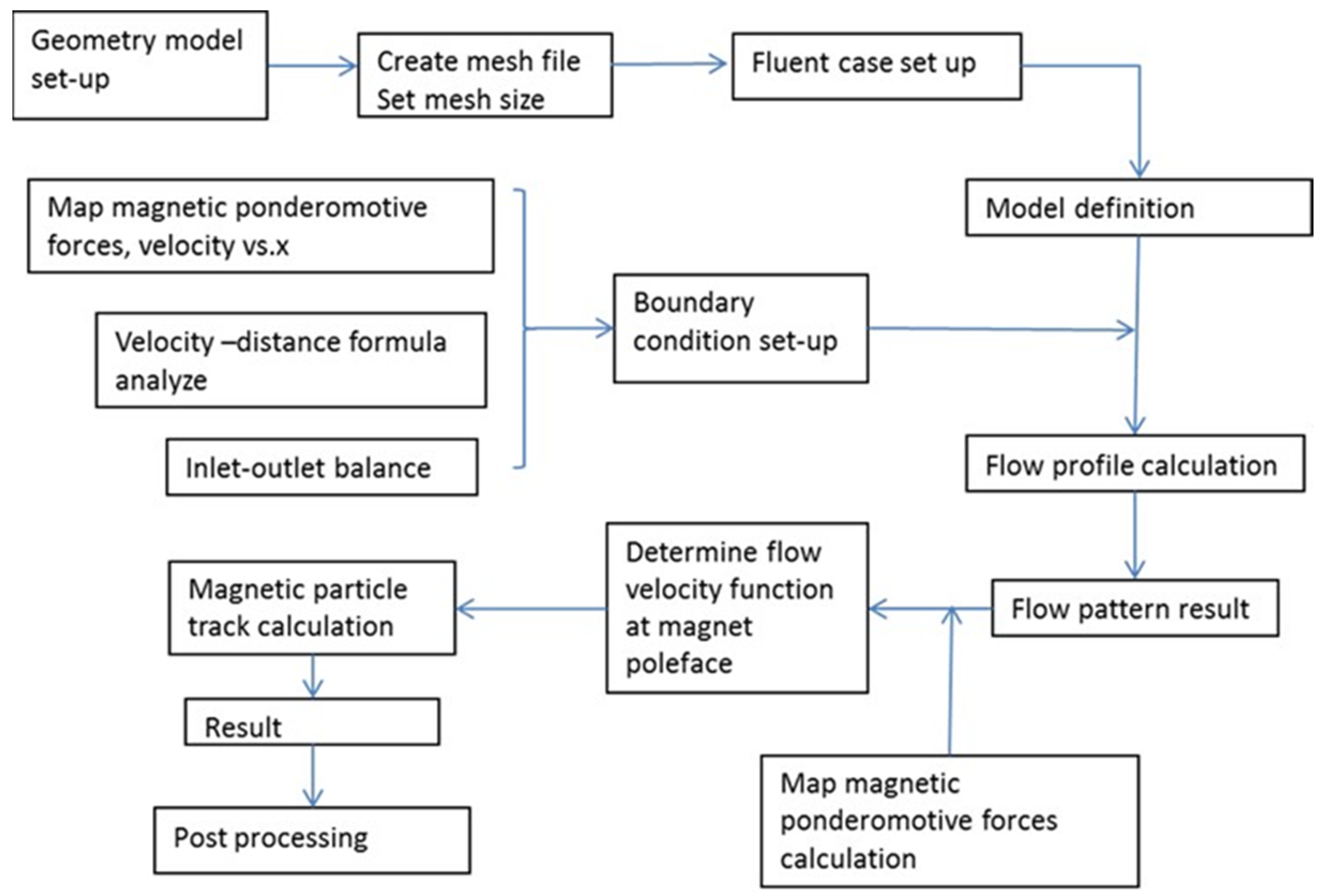
3.1. In-Line Mixer Simulations Using FLUENT®
3.2. Diffusion of Particles and Cells in the Mixer
3.3. The Effect of Sedimentation on Mixing
3.4. Magnetic Trap Simulations
3.5. Calculation of Particle Movement toward Magnets
4. Results and Discussion
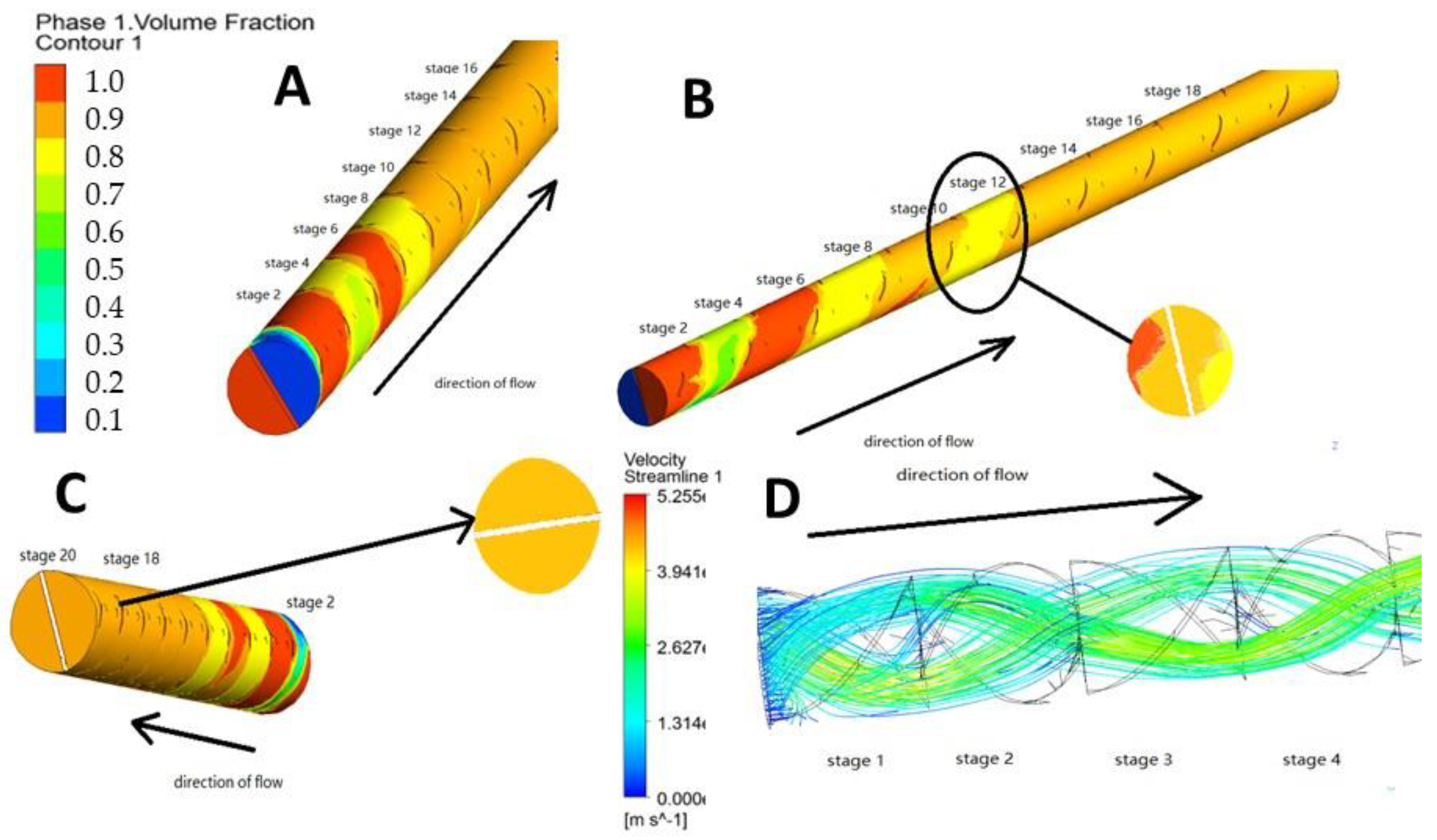
4.1. Results of Static Mixer Simulation
4.1.1. Evaluation of Mixing of Input Fluids
4.1.2. Evaluation of Particle Diffusion and Collision in the Static Mixer
4.1.3. Calculation of the Effects of Sedimentation
4.2. Results of Magnetic Trap Simulations
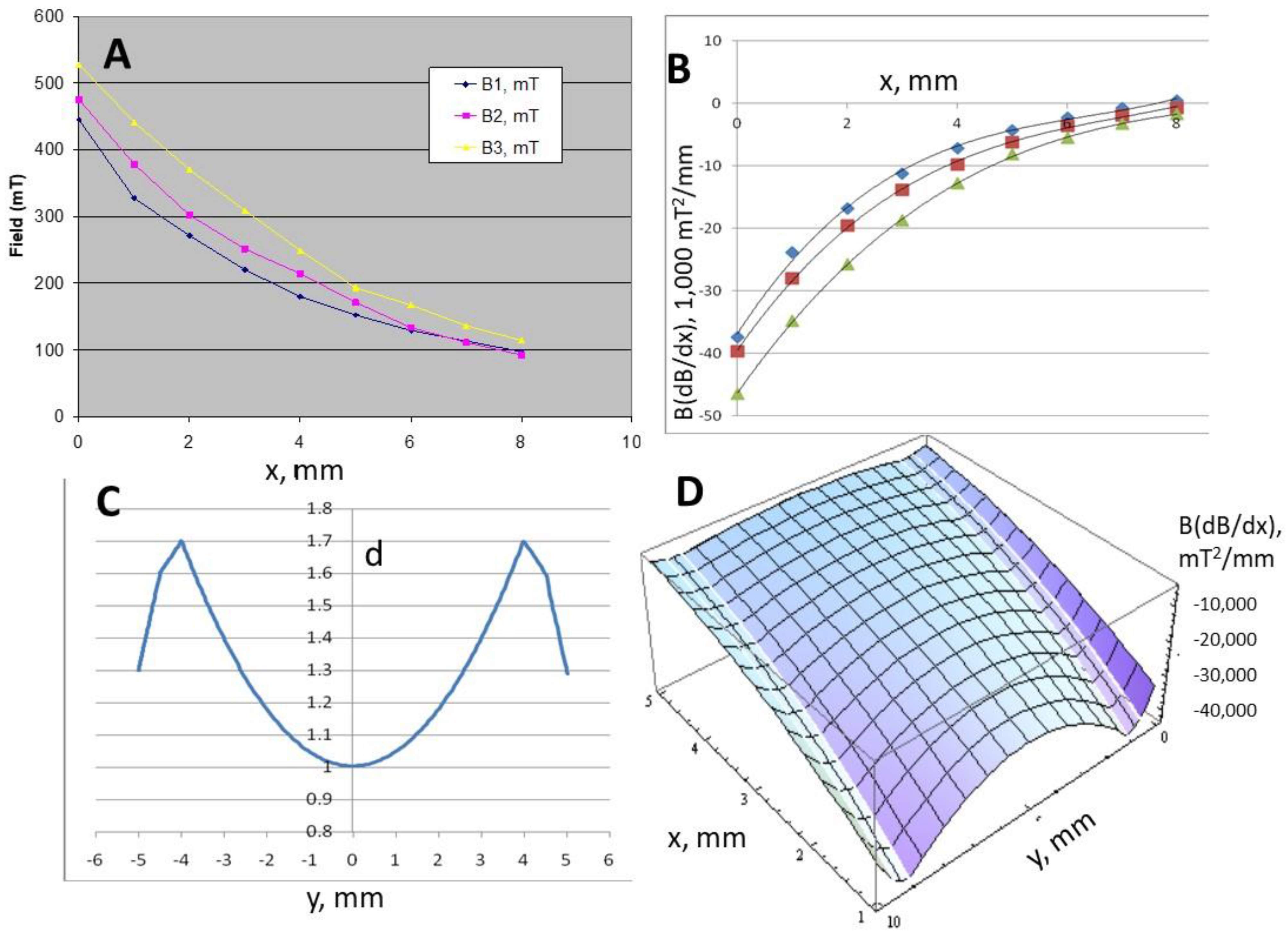
4.2.1. Magnetic Field Analysis
Magnet 2 B = 4.7240x2 − 83.642x + 463.82
Magnet 3 B = 4.5509x2 − 87.957x + 527.23
d = 1.6 + 0.2(y − 0.5) for (0.5 ≤ y ≤ 1)
d = 0.0437y2 − 0.4368y + 2.0952 for (1 ≤ y ≤ 9)
d = 1.7 − 0.2(y − 9) for (9 ≤ y ≤ 9.5)
d = 1.6 − 0.6(y − 9.5) for (9.5 ≤ y ≤ 10)
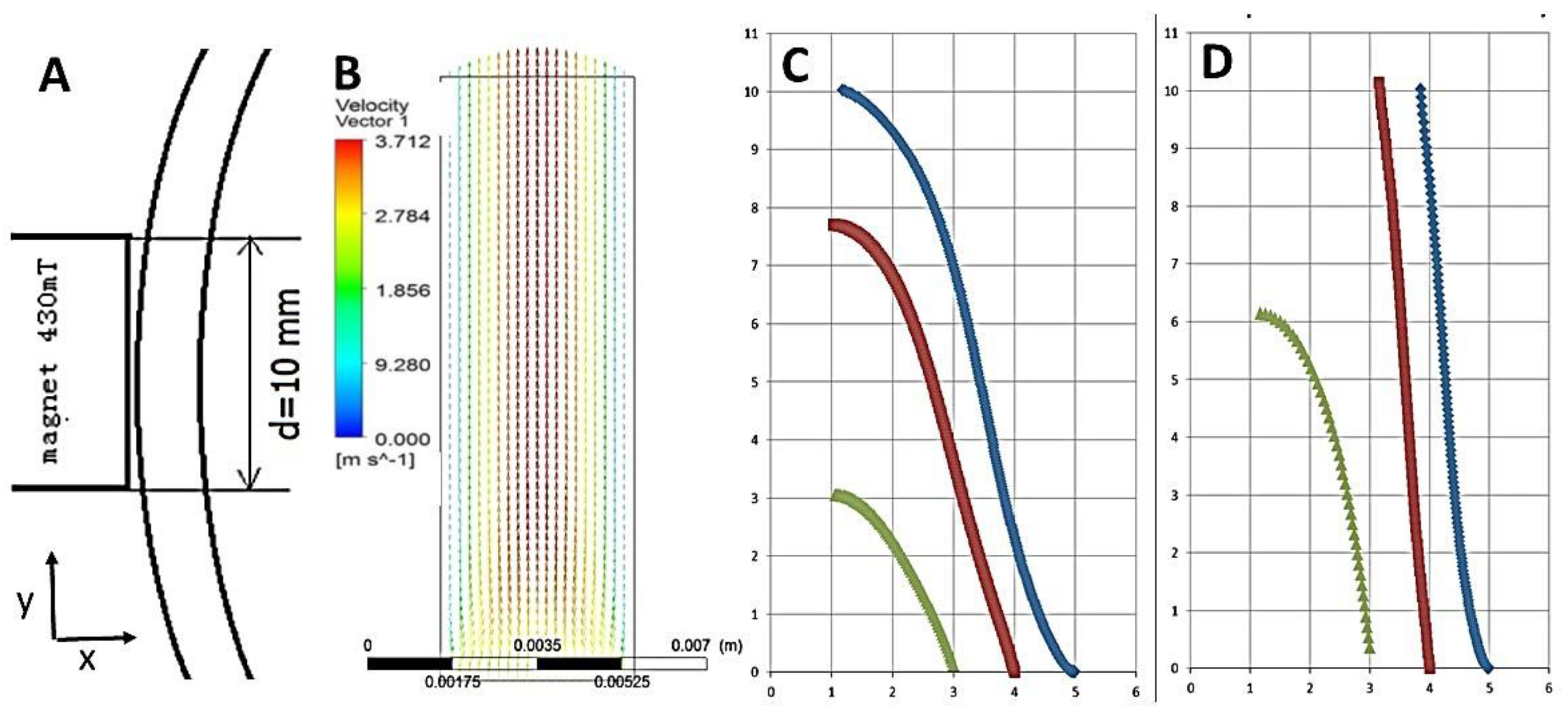
4.2.2. Computation of Magnetic Particle Trajectories
4.3. Experimental Test of In-Line Magnetic Capture
4.3.1. Magnetic Bead Capture Test
4.3.2. Labeled Cell Capture Test
4.3.3. Optimization
4.3.4. Experimental Test of In-Line Labeling Followed by Magnetic Capture
4.3.5. Capture of Target Cells from Whole Blood
5. Summary and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Labib, M.; Kelley, S.O. Circulating tumor cell profiling for precision oncology. Mol. Oncol. 2021, 15, 1622–1646. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; Guan, G.; Bhagat, A.A. ClearCell® FX, a label-free microfluidics technology for enrichment of viable circulating tumor cells. Cytom. A 2018, 93, 1251–1254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grafton, M.M.G.; Maleki, T.; Zordan, M.D.; Reece, L.M.; Byrnes, R.; Jones, A.; Todd, P.; Leary, J.F. Microfluidic MEMS Hand-Held Flow Cytometer. Proc. SPIE 2011, 7929, 79290C. [Google Scholar]
- Maleki, T.; Fricke, T.; Quesenberry, J.T.; Todd, P.W.; Leary, J.F. Point-of-care, portable microfluidic blood analyzer system. Proc. SPIE 2012, 8251, 82510C. [Google Scholar] [CrossRef]
- Plouffe, B.D.; Lewis, L.H.; Murthy, S.K. Computational design optimization for microfluidic magnetophoresis. Biomicrofluidics 2011, 5, 013413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohanty, S.; Baier, T.; Schönfeld, F. Three-dimensional CFD modelling of a continuous immunomagnetophoretic cell capture in BioMEMs. Biochem. Eng. J. 2010, 51, 110–116. [Google Scholar] [CrossRef]
- Habibi, M.R.; Ghasemi, M. Numerical study of magnetic nanoparticles concentration in biofluid (blood) under influence of high gradient magnetic field. J. Magn. Magn. Mater. 2011, 323, 32–38. [Google Scholar] [CrossRef]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.J.; Uhr, J.W.; Terstappen, L.W.M.M. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, E.L. Handbook of Industrial Mixing-Science and Practice; John Wiley & Sons: Hoboken, NJ, USA, 2004; p. 399. [Google Scholar]
- Todd, P.W.; Fricke, T.; Moyers, S. Cell Processing Cartridge for Miniature Cytometer. U.S. Patent 9,753,026 B1, 5 September 2017. [Google Scholar]
- Zhou, C.; Boland, E.D.; Todd, P.W.; Hanley, T.R. Magnetic particle characterization—Magnetophoretic mobility and particle size. Cytom. Part A 2016, 89, 585–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, C.; Qian, Z.; Choi, Y.S.; David, A.E.; Todd, P.; Hanley, T.R. Application of magnetic carriers to two examples of quantitative cell analysis. J. Magn. Magn. Mater. 2017, 427, 25–28. [Google Scholar] [CrossRef] [Green Version]
- Zborowski, M.; Chalmers, J.J. (Eds.) Magnetic Cell Separation; Elsevier: Amsterdam, The Netherlands, 2008. [Google Scholar]
- Richardson, J.F.; Zaki, W.N. Sedimentation and fluidisation: Part I. Trans. Inst. Chem. Eng. 1954, 38, 33. [Google Scholar] [CrossRef]
- Leary, J.F. Design of portable microfluidic cytometry devices for rapid medical diagnostics in the field. Imaging Manip. Anal. Biomol. Cells Tissues XVII 2019, 10881, 108810B. [Google Scholar] [CrossRef]
- Liberti, P.A.; Rao, C.; Terstappen, L.W.M.M. Optimisation of ferrofluids and protocols for the enrichment of breast tumor cells in blood. J. Magn. Magn. Mater. 2001, 225, 301–307. [Google Scholar] [CrossRef]



| Component | Diameter, µm | Density, g/cm3 | Concentration, mL−1 | Adheres to |
|---|---|---|---|---|
| Red blood cell | 8.0 | 1.09 | 2 × 109 | Nothing |
| White blood cell | 10.0 | 1.07 | 2 × 106 | Nothing |
| Target cell | 10.0 | 1.07 | 100 | MP and QD |
| Magnetic Particle (MP) | 2.8 | 1.6 | 106 | Target cell |
| Quantum Dot (QD) | 0.02 | 2.0 | 107 | Target cell |
| n (Stage) | W (mm) | t (qdot) | t (Mag Particle) |
|---|---|---|---|
| 1.00 | 2.5 | 4.98 × 105 | 6.98 × 107 |
| 2.00 | 1.25 | 1.24 × 104 | 1.74 × 107 |
| 3.00 | 0.625 | 3.11 × 104 | 4.36 × 106 |
| 4.00 | 0.312 | 7.78 × 103 | 1.09 × 106 |
| 5.00 | 0.156 | 1.94 × 103 | 2.72 × 105 |
| 6.00 | 0.0781 | 486 | 6.81 × 104 |
| 7.00 | 0.039 | 121 | 1.70 × 104 |
| 8.00 | 0.0195 | 30.4 | 4.26 × 103 |
| 9.00 | 9.77 × 10−3 | 7.59 | 1.06 × 103 |
| 10.00 | 4.88 × 10−3 | 1.90 | 266 |
| 11.00 | 2.44 × 10−3 | 0.475 | 66.5 |
| 12.00 | 1.22 × 10−3 | 0.119 | 16.6 |
| 13.00 | 6.10 × 10−4 | 0.0297 | 4.16 |
| 14.00 | 3.05 × 10−4 | 7.41 × 10−3 | 1.04 |
| 15.00 | 1.53 × 10−4 | 1.85 × 10−3 | 0.260 |
| 16.00 | 7.63 × 10−5 | 4.63 × 10−4 | 0.0650 |
| 17.00 | 3.81 × 10−5 | 1.16 × 10−4 | 0.0162 |
| 18.00 | 1.91 × 10−5 | 2.90 × 10−5 | 4.06 × 10−3 |
| 19.00 | 9.54 × 10−6 | 7.24 × 10−6 | 1.02 × 10−3 |
| 20.00 | 4.77 × 10−6 | 1.81 × 10−6 | 2.54 × 10−4 |
| Condition | Spiking Ratio | % Labeled, Captured | % to Waste |
|---|---|---|---|
| Pure, 4.5 μm beads | 1:0 | 60 | 40 |
| Pure, 1.5 μm beads | 1:0 | 90 ± 7 | 10 ± 3 |
| In blood, 1.5 μm beads | 1:10,000 | 35 (?) | 0–1 |
| In blood, 1.5 μm beads | 1:1,000,000 | 25–100 (range) | 0–10 (range) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qian, Z.; Hanley, T.R.; Reece, L.M.; Leary, J.F.; Boland, E.D.; Todd, P. Continuous Flow Labeling and In-Line Magnetic Separation of Cells. Magnetochemistry 2022, 8, 5. https://doi.org/10.3390/magnetochemistry8010005
Qian Z, Hanley TR, Reece LM, Leary JF, Boland ED, Todd P. Continuous Flow Labeling and In-Line Magnetic Separation of Cells. Magnetochemistry. 2022; 8(1):5. https://doi.org/10.3390/magnetochemistry8010005
Chicago/Turabian StyleQian, Zhixi, Thomas R. Hanley, Lisa M. Reece, James F. Leary, Eugene D. Boland, and Paul Todd. 2022. "Continuous Flow Labeling and In-Line Magnetic Separation of Cells" Magnetochemistry 8, no. 1: 5. https://doi.org/10.3390/magnetochemistry8010005
APA StyleQian, Z., Hanley, T. R., Reece, L. M., Leary, J. F., Boland, E. D., & Todd, P. (2022). Continuous Flow Labeling and In-Line Magnetic Separation of Cells. Magnetochemistry, 8(1), 5. https://doi.org/10.3390/magnetochemistry8010005







