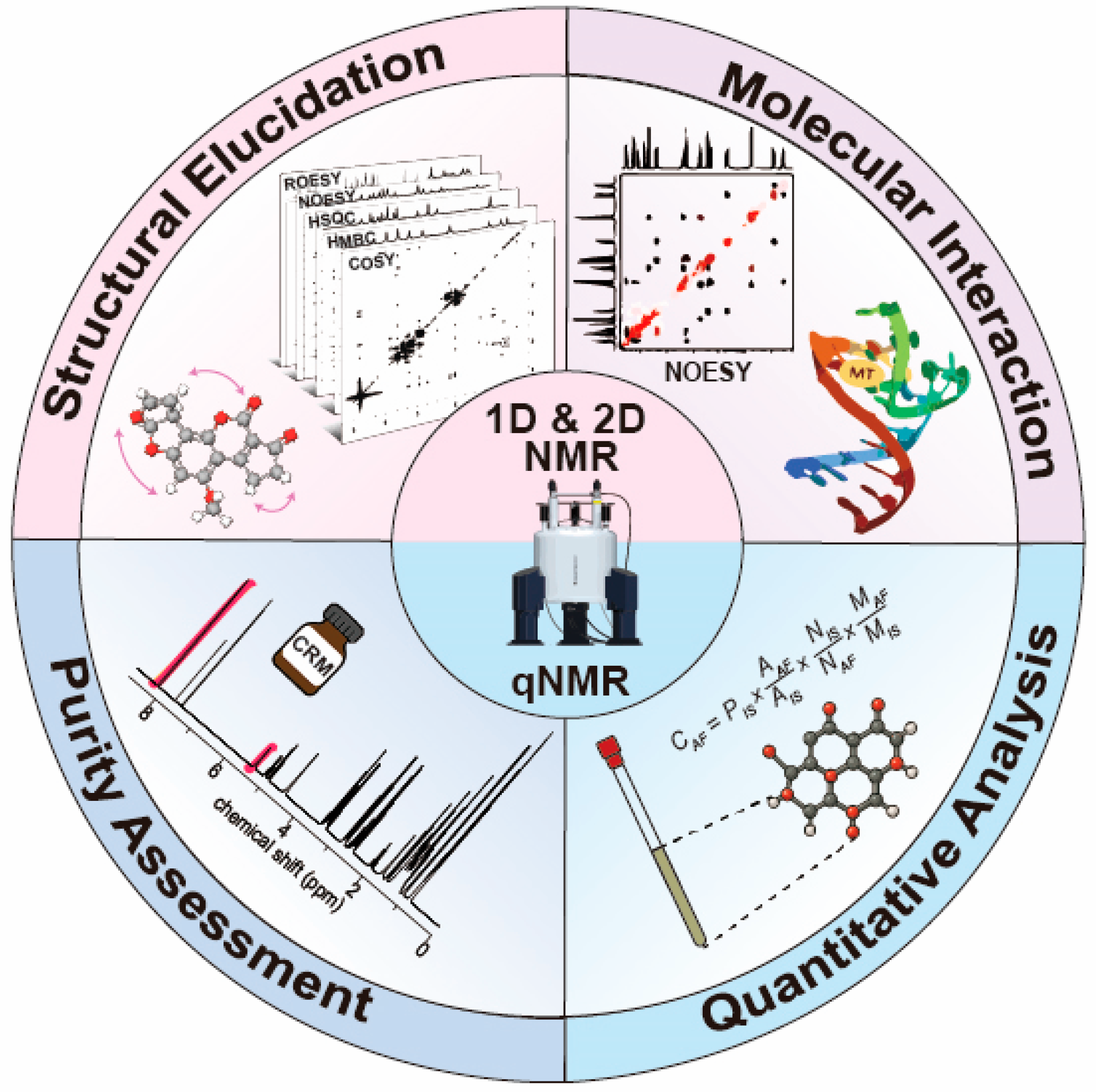
Nuclear Magnetic Resonance-Based Approaches for the Structural and Quantitative Analysis of Mycotoxins
Abstract
1. Introduction
2. NMR Techniques for Mycotoxin and Modified Mycotoxin Analysis
2.1. One-Dimensional (1D) NMR Techniques
2.2. Two-Dimensional (2D) NMR Techniques
2.3. Quantitative NMR (qNMR)
2.4. Advantages of NMR in Mycotoxin Analysis
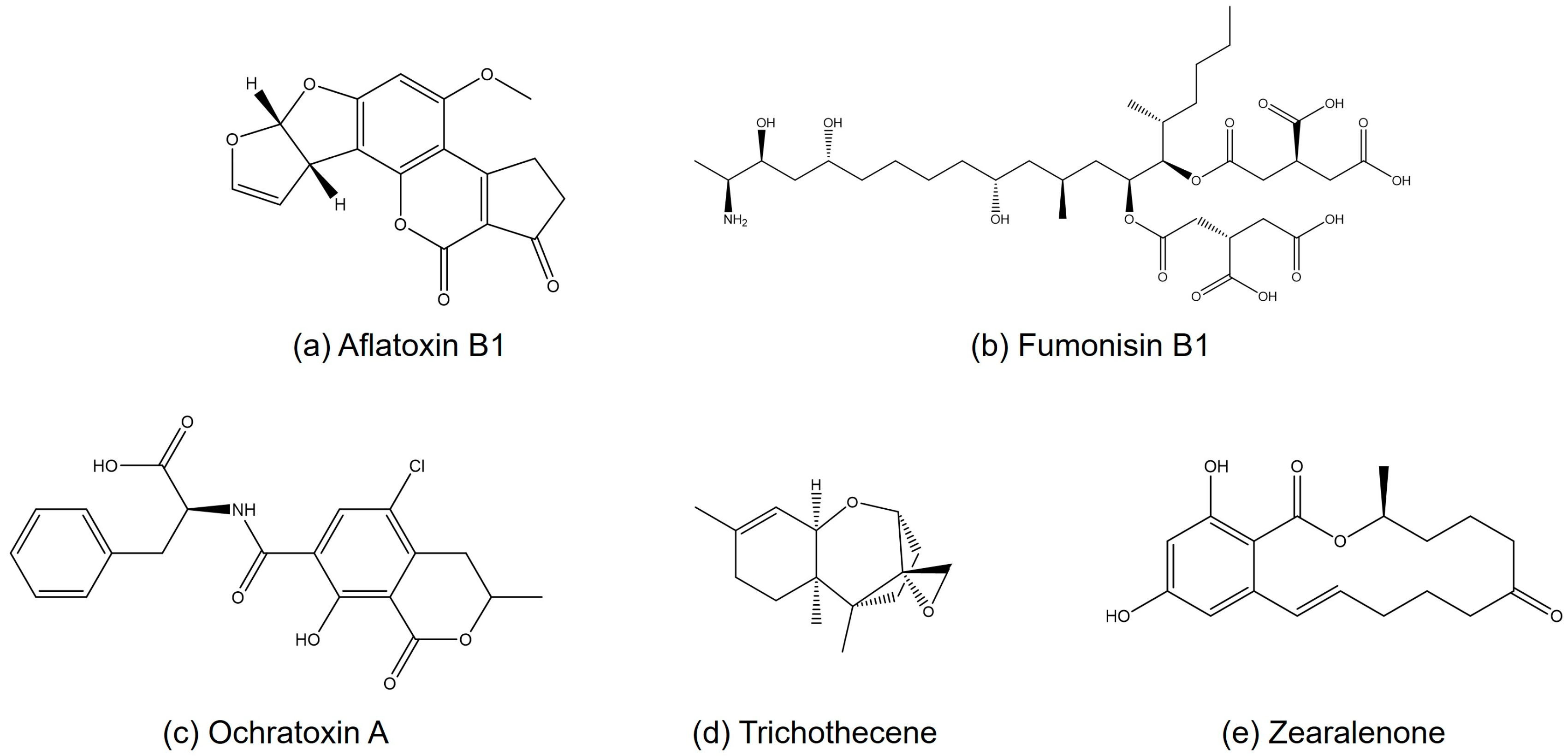
3. Applications of NMR in Structural Analysis of Mycotoxins
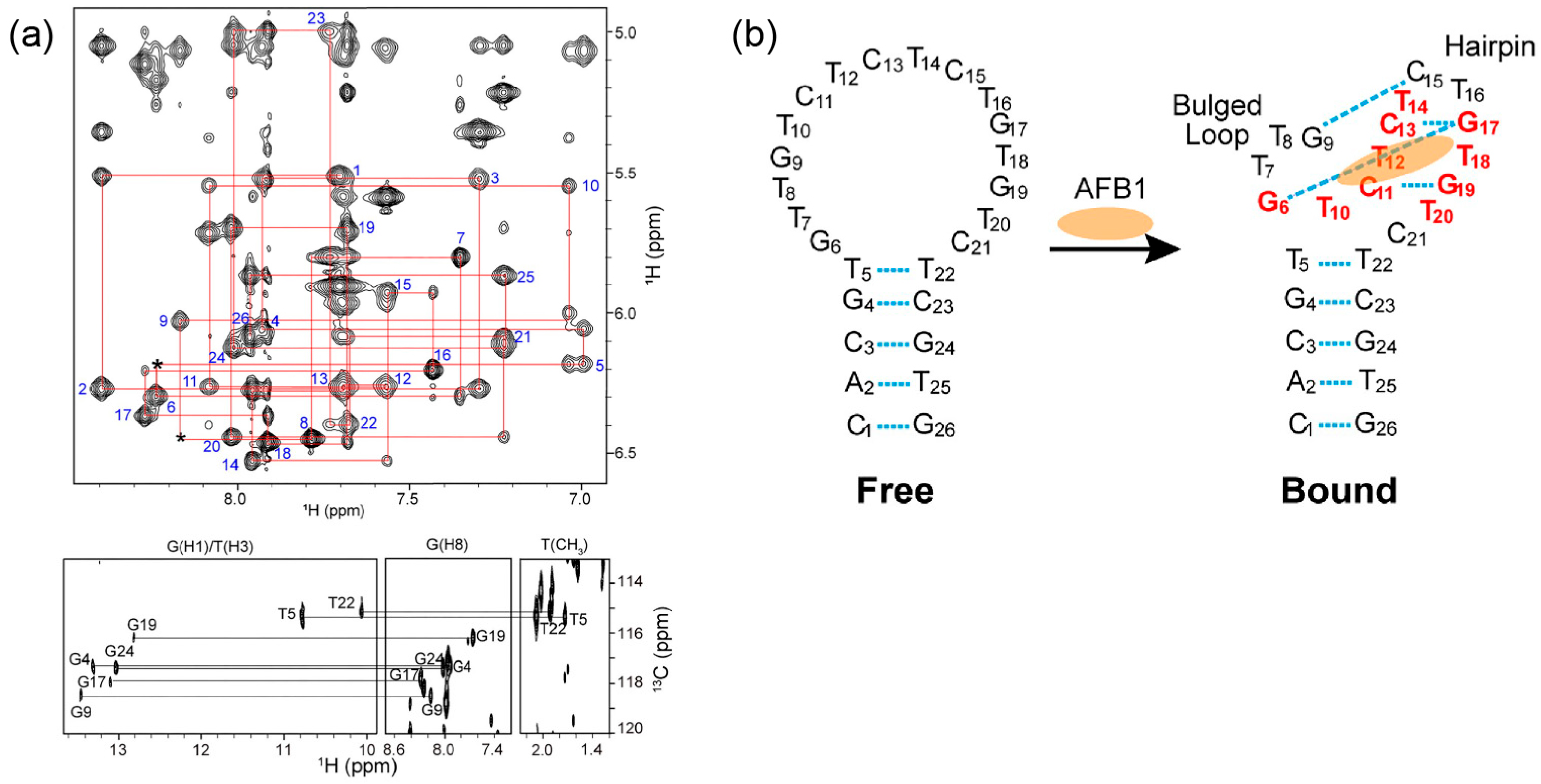
3.1. Aflatoxins
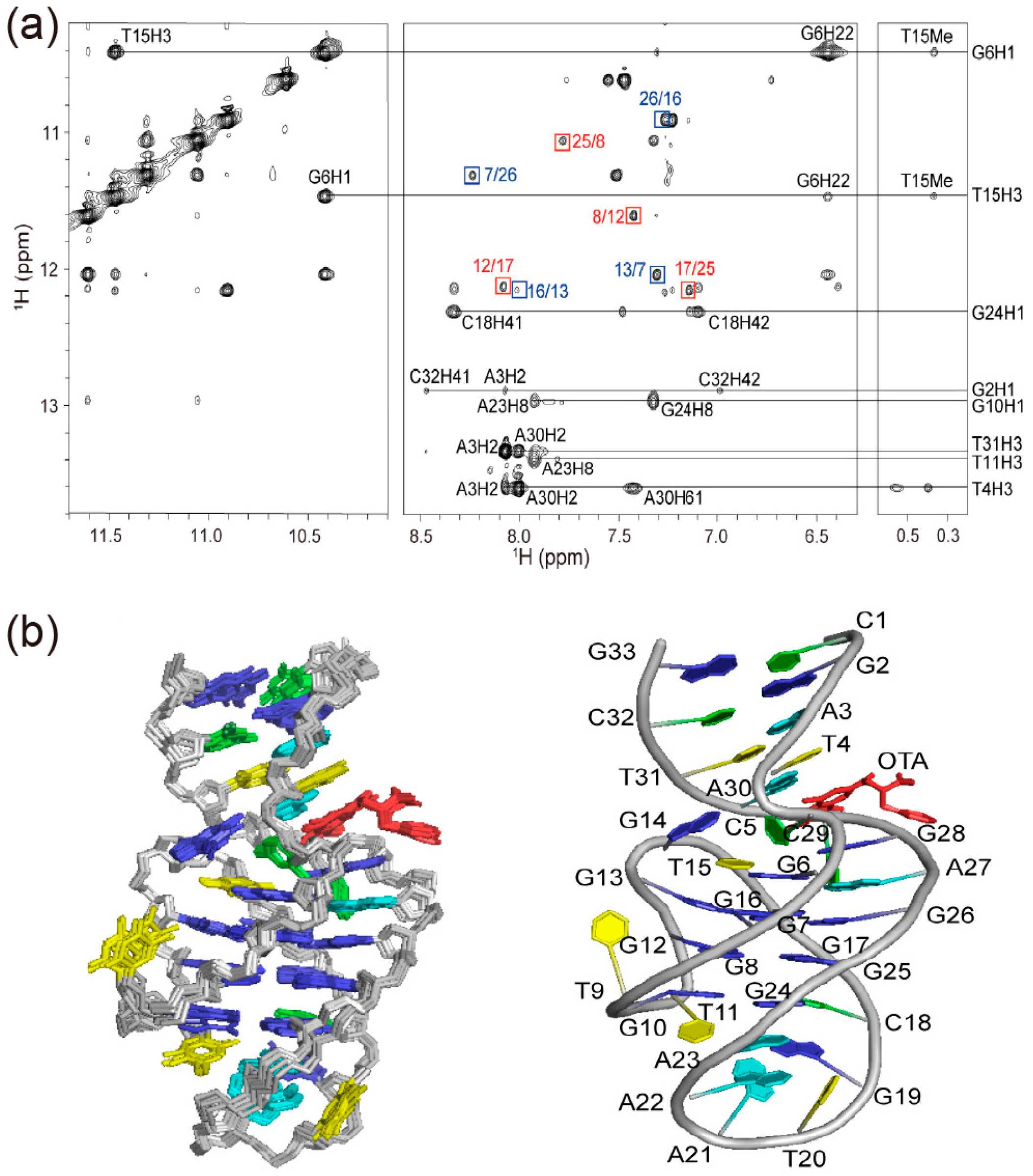
3.2. Ochratoxins
3.3. Fumonisins
3.4. Trichothecenes
3.5. Zearalenone
4. Quantitative Analysis of Mycotoxins Using NMR
5. Challenges and Future Perspectives
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khaneghah, A.M.; Moosavi, M.; Omar, S.S.; Oliveira, C.A.; Karimi-Dehkordi, M.; Fakhri, Y.; Huseyn, E.; Nematollahi, A.; Farahani, M.; Sant’Ana, A.S. The prevalence and concentration of aflatoxin M1 among different types of cheeses: A global systematic review, meta-analysis, and meta-regression. Food Control 2021, 125, 107960. [Google Scholar] [CrossRef]
- Sweeney, M.J.; Dobson, A.D. Mycotoxin production by Aspergillus, Fusarium and Penicillium species. Int. J. Food Microbiol. 1998, 43, 141–158. [Google Scholar] [CrossRef]
- Alshannaq, A.; Yu, J.-H. Occurrence, toxicity, and analysis of major mycotoxins in food. Int. J. Environ. Res. Public Health 2017, 14, 632. [Google Scholar] [CrossRef]
- Tan, H.; Zhou, H.; Guo, T.; Zhou, Y.; Zhang, Q.; Zhang, Y.; Ma, L. Recent advances on formation, transformation, occurrence, and analytical strategy of modified mycotoxins in cereals and their products. Food Chem. 2023, 405, 134752. [Google Scholar] [CrossRef]
- Freire, L.; Sant’Ana, A.S. Modified mycotoxins: An updated review on their formation, detection, occurrence, and toxic effects. Food Chem. Toxicol. 2018, 111, 189–205. [Google Scholar] [CrossRef] [PubMed]
- Tittlemier, S.A.; Cramer, B.; DeRosa, M.C.; Dzuman, Z.; Malone, R.; Maragos, C.; Suman, M.; Sumarah, M.W. Developments in analytical techniques for mycotoxin determination: An update for 2022–23. World Mycotoxin J. 2024, 17, 3–26. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, J.; Dou, J.; Li, T.; Liu, H.; Chang, X.; Qian, S.; Lv, L.; Wu, W.; Sun, C. A novel investigated method for decoupling adsorption and degradation effect on AFB1 based on isotope tracing and NMR analysis. Food Chem. 2023, 405, 134978. [Google Scholar] [CrossRef]
- Solfrizzo, M.; Gambacorta, L.; Bibi, R.; Ciriaci, M.; Paoloni, A.; Pecorelli, I. Multimycotoxin analysis by LC-MS/MS in cereal food and feed: Comparison of different approaches for extraction, purification, and calibration. J. AOAC Int. 2018, 101, 647–657. [Google Scholar] [CrossRef]
- Schothorst, R.C.; Jekel, A.A. Determination of trichothecenes in wheat by capillary gas chromatography with flame ionisation detection. Food Chem. 2001, 73, 111–117. [Google Scholar] [CrossRef]
- Ekwomadu, T.I.; Dada, T.A.; Akinola, S.A.; Nleya, N.; Mwanza, M. Analysis of selected mycotoxins in maize from north-west South Africa using high performance liquid chromatography (HPLC) and other analytical techniques. Separations 2021, 8, 143. [Google Scholar] [CrossRef]
- Xu, G.; Zhao, J.; Yu, H.; Wang, C.; Huang, Y.; Zhao, Q.; Zhou, X.; Li, C.; Liu, M. Structural insights into the mechanism of high-affinity binding of ochratoxin A by a DNA aptamer. J. Am. Chem. Soc. 2022, 144, 7731–7740. [Google Scholar] [CrossRef] [PubMed]
- Yun, S.; Yoon, J.; Kim, B.; Ahn, S.; Choi, K. Purity assessment of Fumonisin B1 by quantitative 1H NMR spectroscopy. Bull. Korean Chem. Soc. 2020, 41, 413–417. [Google Scholar] [CrossRef]
- ISO 24583:2022; Quantitative Nuclear Magnetic Resonance Spectroscopy—Purity Determination of Organic Compounds Used for Foods and Food Products—General Requirements for 1H NMR Internal Standard Method. ISO: Geneva, Switzerland, 2022.
- Westwood, S.; Noordhoek, J.; Lippa, K.; Roos, A.H.; Davies, S.; Nishikida, K.; Schiel, J.E.; Davis, R.; Milton, M.J.T. Methods for the SI-traceable value assignment of the purity of organic compounds (IUPAC Technical Report). Pure Appl. Chem. 2023, 95, 1–77. [Google Scholar] [CrossRef]
- Steyn, P.S. Mycotoxins, general view, chemistry and structure. Toxicol. Lett. 1995, 82–83, 843–851. [Google Scholar] [CrossRef]
- Wang, Y.-D.; Song, C.-G.; Yang, J.; Zhou, T.; Zhao, Y.-Y.; Qin, J.-C.; Guo, L.-P.; Ding, G. Accurate identification of degraded products of aflatoxin B1 under UV irradiation based on UPLC-Q-TOF-MS/MS and NMR analysis. Front. Chem. 2021, 9, 789249. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Wang, C.; Yu, H.; Li, Y.; Zhao, Q.; Zhou, X.; Li, C.; Liu, M. Structural basis for high-affinity recognition of aflatoxin B1 by a DNA aptamer. Nucleic Acids Res. 2023, 51, 7666–7674. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Qin, X.; Tang, Y.; Ma, Q.; Zhang, J.; Zhao, L. CotA laccase, a novel aflatoxin oxidase from Bacillus licheniformis, transforms aflatoxin B1 to aflatoxin Q1 and epi-aflatoxin Q1. Food Chem. 2020, 325, 126877. [Google Scholar] [CrossRef]
- Incze, D.J.; Molnár, Z.; Nagy, G.N.; Leveles, I.; Vértessy, B.G.; Poppe, L.; Bata, Z. Understanding the molecular mechanism of fumonisin esterases by kinetic and structural studies. Food Chem. 2025, 405, 143110. [Google Scholar] [CrossRef]
- Angeli, C.; Nagy, T.M.; Horváth, L.; Varga, M.; Szekeres, A.; Tóth, G.K.; Janáky, T.; Szolomájer, J.; Kovács, M.; Kövér, K.E. Preparation of 3-O-, 5-O- and N-palmitoyl derivatives of fumonisin B1 toxin and their characterisation with NMR and LC-HRMS methods. Food Addit. Contam. Part A 2022, 39, 1759–1771. [Google Scholar] [CrossRef]
- Kumla, D.; Sousa, E.; Marengo, A.; Dethoup, T.; Pereira, J.A.; Gales, L.; Freitas-Silva, J.; Costa, P.M.; Mistry, S.; Silva, A.M. 1,3-Dioxepine and spiropyran derivatives of viomellein and other dimeric naphthopyranones from cultures of Aspergillus elegans KUFA0015 and their antibacterial activity. Phytochemistry 2021, 181, 112575. [Google Scholar] [CrossRef]
- Sueck, F.; Specht, J.; Cramer, B.; Humpf, H.-U. Identification of ochratoxin-N-acetyl-L-cysteine as a new ochratoxin A metabolite and potential biomarker in human urine. Mycotoxin Res. 2020, 36, 1–10. [Google Scholar] [CrossRef]
- Gao, J.; Liu, D.; Nguyen, C.; McCormick, S.P.; Proctor, R.H.; Luo, S.; Zou, Y.; Hai, Y. Biosynthesis of the Central Tricyclic Skeleton of Trichothecene Mycotoxins. J. Am. Chem. Soc. 2025, 147, 8374–8384. [Google Scholar] [CrossRef]
- Shi, Z.-Z.; Liu, X.-H.; Li, X.-N.; Ji, N.-Y. Antifungal and antimicroalgal trichothecene sesquiterpenes from the marine algicolous fungus Trichoderma brevicompactum A-DL-9-2. J. Agric. Food Chem. 2020, 68, 15440–15448. [Google Scholar] [CrossRef]
- Yamazaki, H.; Yagi, A.; Takahashi, O.; Yamaguchi, Y.; Saito, A.; Namikoshi, M.; Uchida, R. Antifungal trichothecene sesquiterpenes obtained from the culture broth of marine-derived Trichoderma cf. brevicompactum and their structure-activity relationship. Bioorg. Med. Chem. Lett. 2020, 30, 127375. [Google Scholar] [CrossRef]
- Yang, H.-X.; Wu, X.; Chi, M.-J.; Li, Z.-H.; Feng, T.; Ai, H.-L.; Liu, J.-K. Structure and cytotoxicity of trichothecenes produced by the potato-associated fungus Trichothecium crotocinigenum. Bioorg. Chem. 2021, 111, 104874. [Google Scholar] [CrossRef]
- Peters, J.; Ash, E.; Gerssen, A.; Van Dam, R.; Franssen, M.C.; Nielen, M.W. Controlled production of zearalenone-glucopyranoside standards with Cunninghamella strains using sulphate-depleted media. Toxins 2021, 13, 366. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.-G.; Wang, Y.-D.; Huang, W.-F.; Liu, J.; Chen, X.-W. Molecular reaction mechanism for elimination of zearalenone during simulated alkali neutralization process of corn oil. Food Chem. 2020, 307, 125546. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Liu, C.; Yang, J.; Tang, Y. Conversion of zearalenone to β-zearalenol and zearalenone-14,16-diglucoside by Candida parapsilosis ATCC 7330. Food Control 2022, 131, 108429. [Google Scholar] [CrossRef]
- Guo, W.; Meng, J.; Wang, X.; Li, Z.; Li, J.; Niu, X.; Zhao, Z.; Han, Z. Preparation and characterization of the aflatoxin B1 purity certified reference material (GBW (E) 100599). Microchem. J. 2024, 199, 109919. [Google Scholar] [CrossRef]
- Paymode, D.J.; Sharma, I. Rhodium-Catalyzed [3+2]-Annulation of Ortho-Diazoquinones with Enol Ethers: Diversity-Oriented Total Synthesis of Aflatoxin B2. Eur. J. Org. Chem. 2021, 2021, 2034–2040. [Google Scholar] [CrossRef]
- Pfohl-Leszkowicz, A.; Manderville, R.A. An update on direct genotoxicity as a molecular mechanism of ochratoxin A carcinogenicity. Chem. Res. Toxicol. 2012, 25, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Bittner, A.; Kümmel, D.; Schulz, M.; Osenberg, M.; Cramer, B.; Humpf, H.U. Structure elucidation and in vitro cytotoxicity of ochratoxin α amide, a new degradation product of ochratoxin A. Mycotoxin Res. 2015, 31, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Savard, M.E. Deoxynivalenol fatty acid and glucoside conjugates. J. Agric. Food Chem. 1991, 39, 570–574. [Google Scholar] [CrossRef]
- Safwan, S.; Wang, S.-W.; Hsiao, G.; Hsiao, S.-W.; Hsu, S.-J.; Lee, T.-H.; Lee, C.-K. New trichothecenes isolated from the marine algicolous fungus Trichoderma brevicompactum. Mar. Drugs 2022, 20, 80. [Google Scholar] [CrossRef]
- Nahle, S.; El Khoury, A.; Atoui, A. Current status on the molecular biology of zearalenone: Its biosynthesis and molecular detection of zearalenone-producing Fusarium species. Eur. J. Plant Pathol. 2021, 159, 247–258. [Google Scholar] [CrossRef]
- Nittoli, A.C.; Costantini, S.; Sorice, A.; Capone, F.; Ciarcia, R.; Marzocco, S.; Budillon, A.; Severino, L. Effects of α-zearalenol on the metabolome of two breast cancer cell lines by 1H-NMR approach. Metabolomics 2018, 14, 1–11. [Google Scholar] [CrossRef]
- Borzekowski, A.; Drewitz, T.; Keller, J.; Pfeifer, D.; Kunte, H.-J.; Koch, M.; Rohn, S.; Maul, R. Biosynthesis and characterization of zearalenone-14-sulfate, zearalenone-14-glucoside and zearalenone-16-glucoside using common fungal strains. Toxins 2018, 10, 104. [Google Scholar] [CrossRef]
- Bates, J.; Bahadoor, A.; Cui, Y.; Meija, J.; Windust, A.; Melanson, J.E. Certification of ochratoxin A reference materials: Calibration solutions OTAN-1 and OTAL-1 and a mycotoxin-contaminated rye flour MYCO-1. Anal. Bioanal. Chem. 2019, 411, 1756–1766. [Google Scholar] [CrossRef]
- Renaud, J.B.; Walsh, J.P.; Sumarah, M.W. Simplified synthesis and stability assessment of aflatoxin B1-Lysine and aflatoxin G1-Lysine. Toxins 2022, 14, 56. [Google Scholar] [CrossRef]
- Wang, S.; Wang, S.; Li, P.; Li, L.; Ye, J. Establishment of SI-traceable purity assessment of Fumonisin B1 using a combination of quantitative 1H NMR and mass balance. Microchem. J. 2023, 185, 108282. [Google Scholar] [CrossRef]
- Schmidt, J.; Lindemann, V.; Olsen, M.; Cramer, B.; Humpf, H.-U. Dried urine spots as sampling technique for multi-mycotoxin analysis in human urine. Mycotoxin Res. 2021, 37, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Bahadoor, A.; Brinkmann, A.; Melanson, J.E. 13C-Satellite decoupling strategies for improving accuracy in quantitative nuclear magnetic resonance. Anal. Chem. 2020, 93, 851–858. [Google Scholar] [CrossRef]
- Li, X.; Liu, S.; Guo, Z.; Li, X.; Li, X.; Jiao, H.; Zhang, Q. Stability of a calibrant as certified reference material for determination of trans-zearalenone by high performance liquid chromatography–diode array detection–triple quadrupole tandem mass spectrometry. Anal. Bioanal. Chem. 2022, 414, 3631–3641. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Li, X.; Li, H. Certified Reference Materials and Metrological Traceability for Mycotoxin Analysis. J. AOAC Int. 2019, 102, 992–998. [Google Scholar] [CrossRef]
- Steiner, D.; Bartók, T.; Sulyok, M.; Szekeres, A.; Varga, M.; Horváth, L.; Rost, H. Global Perspectives on Mycotoxin Reference Materials (Part I): Insights from Multi-Supplier Comparison Study Including Aflatoxin B1, Deoxynivalenol and Zearalenone. Toxins 2024, 16, 397. [Google Scholar] [CrossRef]
- Choi, K.; Myoung, S.; Seo, Y.; Ahn, S. Quantitative NMR as a Versatile Tool for the Reference Material Preparation. Magnetochemistry 2021, 7, 15. [Google Scholar] [CrossRef]
- Habler, K.; Frank, O.; Rychlik, M. Chemical synthesis of deoxynivalenol-3-β-D-[13C6]-glucoside and application in stable isotope dilution assays. Molecules 2016, 21, 838. [Google Scholar] [CrossRef] [PubMed]
- Westwood, S.; Josephs, R.; Martos, G.; Choteau, T.; Gao, Y.; Un, I.; Gokcen, T.; Santos, L. Purity Evaluation Guideline: Patulin; Bureau International des Poids et Mesures (BIPM): Sèvres, France, 2022; Rapport BIPM-2022/05. [Google Scholar]
- Westwood, S.; Josephs, R.; Martos, G.; Choteau, T.; Li, X.; Un, I.; Santos, L. Purity Evaluation Guideline: Deoxynivalenol; Bureau International des Poids et Mesures (BIPM): Sèvres, France, 2022; Rapport BIPM-2022/04. [Google Scholar]
- Westwood, S.; Josephs, R.; Martos, G.; Choteau, T.; Li, X.; Guo, X.; Li, X.; Garrido, B.; Un, I. Purity Evaluation Guideline: Aflatoxin B1; Bureau International des Poids et Mesures (BIPM): Sèvres, France, 2021; Rapport BIPM-2021/01. [Google Scholar]
- Madelou, N.A.; Melliou, E.; Magiatis, P. Quantitation of Lupinus spp. Quinolizidine Alkaloids by qNMR and Accelerated Debittering with a Resin-Based Protocol. Molecules 2024, 29, 582. [Google Scholar] [CrossRef]
- Ali, S.; Freire, L.G.D.; Rezende, V.T.; Noman, M.; Ullah, S.; Abdullah; Badshah, G.; Afridi, M.S.; Tonin, F.G.; de Oliveira, C.A.F. Occurrence of mycotoxins in foods: Unraveling the knowledge gaps on their persistence in food production systems. Foods 2023, 12, 4314. [Google Scholar] [CrossRef]
- Scheibenzuber, S.; Hoffmann, T.; Effenberger, I.; Schwab, W.; Asam, S.; Rychlik, M. Enzymatic Synthesis of Modified Alternaria Mycotoxins Using a Whole-Cell Biotransformation System. Toxins 2020, 12, 264. [Google Scholar] [CrossRef]
- Wang, Q.; Zou, X.; Chen, Y.; Zhu, Z.; Yan, C.; Shan, P.; Wang, S.; Fu, Y. XGBoost algorithm assisted multi-component quantitative analysis with Raman spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2024, 305, 124917. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Le, D.; Zhang, T.; Lai, Q.; Zhang, J.; Li, B.; Song, Y.; Chen, N. Detection of apple moldy core disease by fusing vibration and Vis/NIR spectroscopy data with dual-input MLP-Transformer. J. Food Eng. 2024, 382, 112219. [Google Scholar] [CrossRef]
- Carbas, B.; Sampaio, P.; Barros, S.C.; Freitas, A.; Silva, A.S.; Brites, C. Rapid screening of fumonisins in maize using near-infrared spectroscopy (NIRS) and machine learning algorithms. Food Chem. X 2025, 27, 102351. [Google Scholar] [CrossRef] [PubMed]
- Bertani, F.; Businaro, L.; Gambacorta, L.; Mencattini, A.; Brenda, D.; Di Giuseppe, D.; De Ninno, A.; Solfrizzo, M.; Martinelli, E.; Gerardino, A. Optical detection of aflatoxins B in grained almonds using fluorescence spectroscopy and machine learning algorithms. Food Control 2020, 112, 107073. [Google Scholar] [CrossRef]
- Cao, R.; Li, J.; Ding, H.; Zhao, T.; Guo, Z.; Li, Y.; Sun, X.; Wang, F.; Qiu, J. Synergistic approaches of AI and NMR in enhancing food component analysis: A comprehensive review. Trends Food Sci. Technol. 2025, 139, 104852. [Google Scholar] [CrossRef]
- Yun, B.H.; Yu, H.-Y.; Kim, H.; Myoung, S.; Yeo, N.; Choi, J.; Chun, H.S.; Kim, H.; Ahn, S. Geographical discrimination of Asian red pepper powders using ¹H NMR spectroscopy and deep learning-based convolution neural networks. Food Chem. 2023, 428, 138082. [Google Scholar] [CrossRef]
| Toxin Type | NMR Method | Information | Reference | |
|---|---|---|---|---|
| Aflatoxins | AFB1 | 1H NMR, 13C NMR, COSY, HMBC, NOESY | Structural elucidation of UV-degraded products of AFB1 using MS and 2D NMR | [16] |
| AFB1-AF26 aptamer | 1H NMR, TOCSY, DQF-COSY, NOESY, HMBC, 31P–1H COSY, SOFAST-HMQC | Structure determination of AFB1–AF26 aptamer complex using 2D NMR, revealing binding pocket and recognition mechanism | [17] | |
| AFB1 | 1H NMR, 13C NMR, COSY, HSQC, HMBC, ROESY | Structural elucidation of AFB1 degradation products (aflatoxicol and epi-aflatoxicol) using isotope tracing and NMR | [7] | |
| AFQ1 | 1H NMR, 13C NMR, COSY, HMBC | Identification of two stereoisomers of AFQ1 (oxidized products of AFB1) using 1D and 2D NMR | [18] | |
| Fumonisins | FB1 | 1H NMR | Structural analysis of degraded products of FB1 | [19] |
| FB1 derivatives | 1H NMR, 13C NMR, HSQC-CLIP-COSY, HMBC | Structural elucidation of site-specific acylated FB1 derivatives using 2D NMR and LC-HRMS | [20] | |
| Ochratoxins | Ochratoxin A (OTA)–DNA aptamer | 1H NMR, 2D NOESY, TOCSY, DQF-COSY, JR-HMBC, 31P–1H COSY, HSQC | Structural studies of OTA-bound DNA aptamer using 2D NMR; binding mechanism via halogen bonding, π stacking, and hydrophobic interactions | [11] |
| OTA, OTA methyl ester, OTB, ochratoxin β | 1H NMR, 13C NMR, COSY, HSQC, HMBC, NOESY | Structural elucidation of ochratoxins, including ochratoxin β, using 1D and 2D NMR techniques | [21] | |
| OTB-GSH, OTB-NAC | 1H NMR, 13C NMR, COSY, HSQC, HMBC | Structural confirmation of synthesized OTB-GSH and OTB-NAC using 2D NMR with HPLC-MS/MS | [22] | |
| Trichothecenes | EPT skeleton (e.g., DON, T-2 toxin) | 1H NMR, 13C NMR, HSQC, HMBC | Elucidation of the biosynthetic mechanism of the EPT tricyclic core, identifying Tri3 (acetyltransferase) and Tri14 (cyclase) as essential enzymes | [23] |
| Trichodermarins G–N (1–8), Trichoderm | 1H NMR, 13C NMR, COSY, HSQC, HMBC, NOESY | Structural elucidation of 8 new trichothecenes and 2 cuparene derivatives from marine Trichoderma using 2D NMR | [24] | |
| Trichobreols A, D, E | 1H NMR, 13C NMR, COSY, HMBC, NOESY | Elucidation of stereochemistry and structural variations of the isolated trichobreol D and E from marine Trichoderma using 2D NMR | [25] | |
| Trichothecrotocins M–S (1–7) | 1H NMR, 13C NMR, COSY, HMBC, HSQC, ROESY | Structural elucidation of 7 new trichothecenes from Trichothecium crotocinigenum and cytotoxicity evaluation using 2D NMR | [26] | |
| Zearalenone | ZEN glycosides (Z14G, Z16G) | 1H NMR, 13C NMR, DEPT, COSY, HSQC, HMBC | Structure confirmation of Z14G and Z16G using 1D and 2D NMR after biotransformation using Cunninghamella strains | [27] |
| ZEN | 1H NMR, 13C NMR, COSY, HSQC | Elucidating the degradation product of ZEN under alkali treatment using 1D and 2D NMR, confirming ring opening and decarboxylation | [28] | |
| ZEN-diglucoside | 1H NMR, 13C NMR | Structural characterization of ZEN-14,16-diglucoside using NMR analysis | [29] | |
| Toxin | NMR Method | Information | Internal Standard | References | |
|---|---|---|---|---|---|
| Aflatoxin | AFB1, AFG1 | 1H qNMR | Quantification of AFB1-Lys and AFG1-Lys by qNMR using maleic acid as the internal standard | Maleic acid | [40] |
| AFB1 | 1H NMR, 13C NMR, 1H qNMR | Purity quantification of AFB1 using 1H qNMR and uncertainty evaluation procedure | Benzoic acid | [30] | |
| Fumonisin | FB1 | 1H qNMR | Optimized qNMR method for FB1 using benzoic acid (internal standard) and DMSO-d6/TFA-d (solvent) | Benzoic acid | [41] |
| FB1 | 1H qNMR | Reliable purity assessment of FB1 using qNMR without interference from impurities | Benzoic acid | [12] | |
| FB1 | 1H qNMR | Determination of the exact concentrations of FB1 using qNMR | Thymol | [42] | |
| Ochratoxins | OTA | qH{13C}NMR | Accurate quantification of OTA using qNMR with 13C decoupling | Maleic acid | [43] |
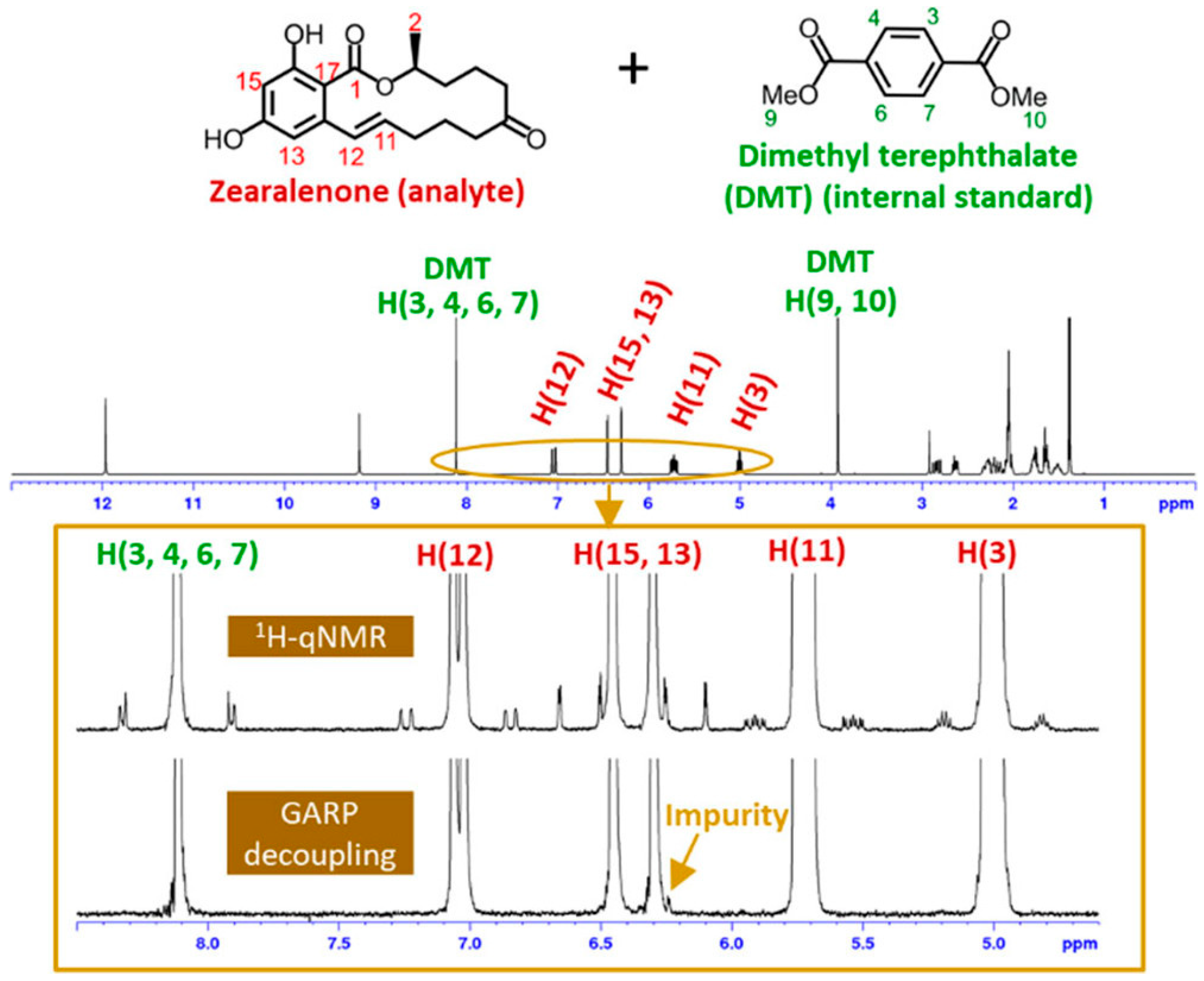
| Trichothecenes | DON | 1H qNMR, qH{13C}NMR, HSQC | Purity determination of DON considering tautomeric and conformational isomers | Dimethyl terephthalate | [43] |
| DON | 1H qNMR | Determination of the exact concentrations of DON using qNMR | Thymol | [42] | |
| Zearalenone | ZEN | 1H qNMR | Purity evaluation of trans-ZEN reference material using qNMR | Benzoic acid | [44] |
| ZEN | 1H qNMR, qH{13C}NMR | Purity determination and detection of low-level impurities using 13C decoupling | Dimethyl terephthalate | [43] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.H.; Lee, S.Y.; Kim, J.Y.; Cho, H.; Chun, H.S.; Ahn, S. Nuclear Magnetic Resonance-Based Approaches for the Structural and Quantitative Analysis of Mycotoxins. Magnetochemistry 2025, 11, 47. https://doi.org/10.3390/magnetochemistry11060047
Kim YH, Lee SY, Kim JY, Cho H, Chun HS, Ahn S. Nuclear Magnetic Resonance-Based Approaches for the Structural and Quantitative Analysis of Mycotoxins. Magnetochemistry. 2025; 11(6):47. https://doi.org/10.3390/magnetochemistry11060047
Chicago/Turabian StyleKim, Yun Hwan, Seon Yeong Lee, Jin Young Kim, Hyojin Cho, Hyang Sook Chun, and Sangdoo Ahn. 2025. "Nuclear Magnetic Resonance-Based Approaches for the Structural and Quantitative Analysis of Mycotoxins" Magnetochemistry 11, no. 6: 47. https://doi.org/10.3390/magnetochemistry11060047
APA StyleKim, Y. H., Lee, S. Y., Kim, J. Y., Cho, H., Chun, H. S., & Ahn, S. (2025). Nuclear Magnetic Resonance-Based Approaches for the Structural and Quantitative Analysis of Mycotoxins. Magnetochemistry, 11(6), 47. https://doi.org/10.3390/magnetochemistry11060047






