Genome-Wide Identification of Fatty Acyl-CoA Reductase (FAR) Genes in Dendrobium catenatum and Their Response to Drought Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Physicochemical Properties of FAR Genes in D. catenatum
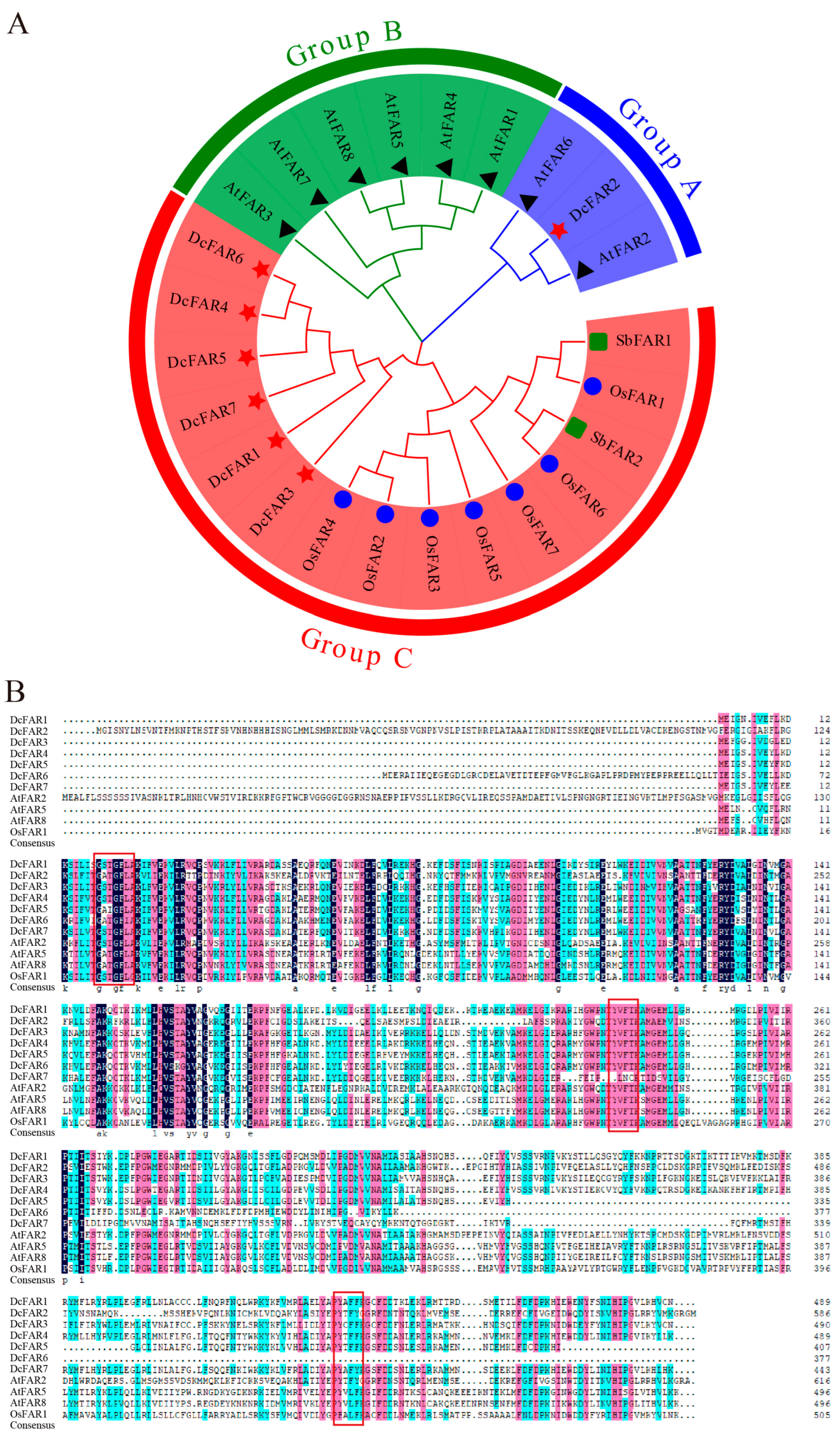
2.2. Phylogenetic Analyses of FAR Family Members
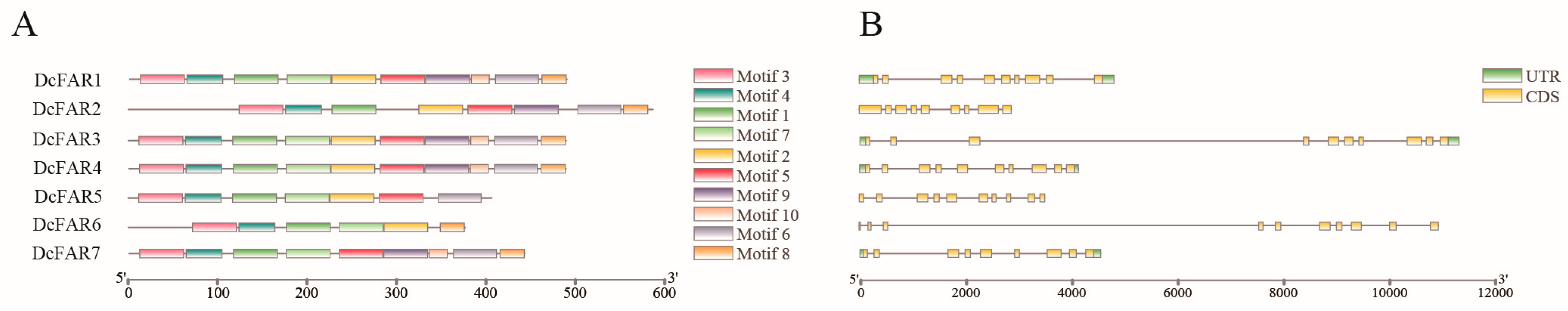
2.3. Conserved Motif and Gene Structure Analyses of DcFARs
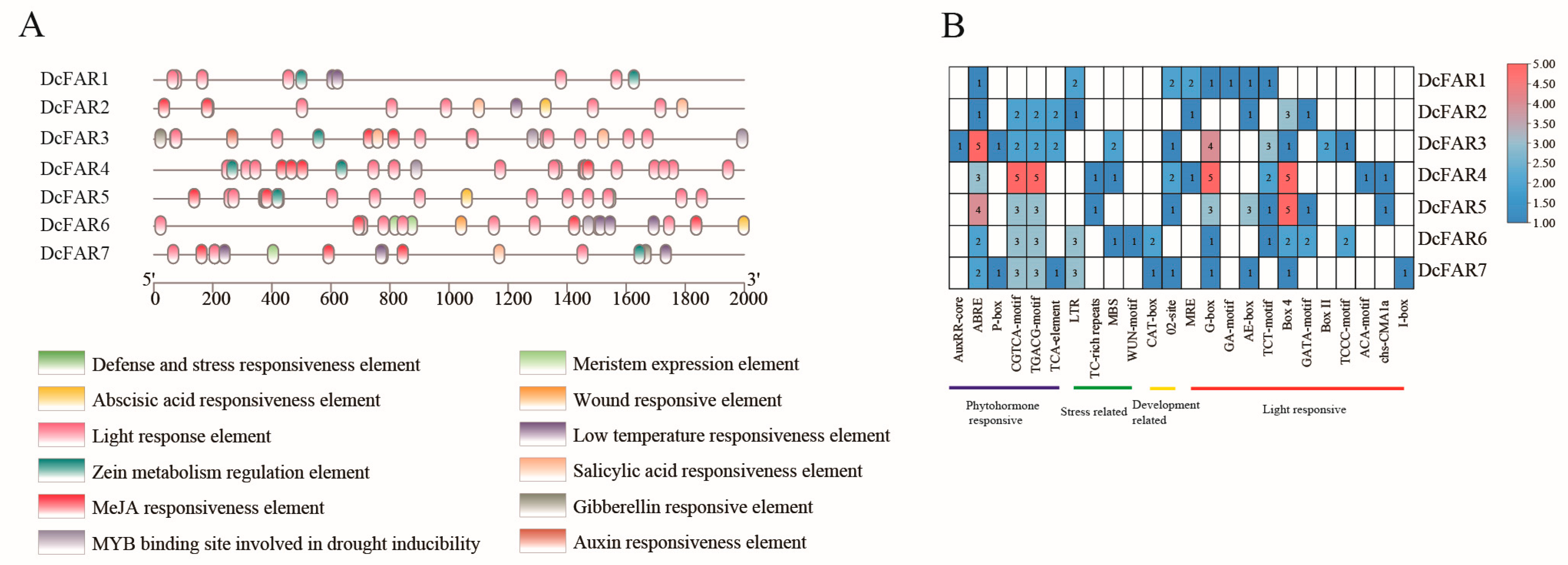
2.4. Prediction of Cis-Regulatory Elements (CREs) for D. catenatum FAR Gene Promoters
2.5. Expression Analysis of DcFAR Genes under Drought Stress
2.6. Expression Analysis of DcFAR Genes in Different Tissues
3. Results
3.1. Identification and Physicochemical Properties of FAR Genes in D. catenatum
3.2. Phylogenetic Analyses of FAR Family Members of D. catenatum and Other Plant Species
3.3. Conserved Motif and Gene Structure Analyses of FAR Family Members
3.4. Prediction of Cis-Regulatory Elements (CREs) for D. catenatum FAR Gene Promoters
3.5. Expression Analysis of DcFAR Genes under Drought Stress
3.6. Expression Analysis of FAR Genes in Different Tissues
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lewandowska, M.; Keyl, A.; Feussner, I. Wax biosynthesis in response to danger: Its regulation upon abiotic and biotic stress. New Phytol. 2020, 227, 698–713. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.B.; Rose, J.K. There’s more than one way to skin a fruit: Formation and functions of fruit cuticles. J. Exp. Bot. 2014, 65, 4639–4651. [Google Scholar] [CrossRef] [PubMed]
- Long, Z.; Tu, M.; Xu, Y.; Pak, H.; Zhu, Y.; Dong, J.; Lu, Y.; Jiang, L. Genome-wide-association study and transcriptome analysis reveal the genetic basis controlling the formation of leaf wax in Brassica napus. J. Exp. Bot. 2023, 74, 2726–2739. [Google Scholar] [CrossRef]
- Buschhaus, C.; Jetter, R. Composition and physiological function of the wax layers coating Arabidopsis leaves: β-amyrin negatively affects the intracuticular water barrier. Plant Physiol. 2012, 160, 1120–1129. [Google Scholar] [CrossRef]
- Muller, Y.; Patwari, P.; Stocker, T.; Zeisler-Diehl, V.; Steiner, U.; Campoli, C.; Grewe, L.; Kuczkowska, M.; Dierig, M.M.; Jose, S.; et al. Isolation and characterization of the gene HvFAR1 encoding acyl-CoA reductase from the cer-za.227 mutant of barley (Hordeum vulgare) and analysis of the cuticular barrier functions. New Phytol. 2023, 239, 1903–1918. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, T.; Griffiths, D.W. The effects of stress on plant cuticular waxes. New Phytol. 2006, 171, 469–499. [Google Scholar] [CrossRef] [PubMed]
- Long, L.M.; Patel, H.P.; Cory, W.C.; Stapleton, A.E. The maize epicuticular wax layer provides UV protection. Funct. Plant Biol. 2003, 30, 75–81. [Google Scholar] [CrossRef]
- Sieber, P.; Schorderet, M.; Ryser, U.; Buchala, A.; Kolattukudy, P.; Métraux, J.P.; Nawrath, C. Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell 2000, 12, 721–738. [Google Scholar] [CrossRef]
- Gorb, E.V.; Gorb, S.N. Anti-adhesive effects of plant wax coverage on insect attachment. J. Exp. Bot. 2017, 68, 5323–5337. [Google Scholar] [CrossRef]
- Gaume, L.; Perret, P.; Gorb, E.; Gorb, S.; Labat, J.J.; Rowe, N. How do plant waxes cause flies to slide? Experimental tests of wax-based trapping mechanisms in three pitfall carnivorous plants. Arthropod. Struct. Dev. 2004, 33, 103–111. [Google Scholar] [CrossRef]
- Yang, X.P.; Zhao, H.Y.; Kosma, D.K.; Tomasi, P.; Dyer, J.M.; Li, R.; Liu, X.; Wang, Z.; Parsons, E.P.; Jenks, M.A.; et al. The acyl desaturase CER17 is involved in producing wax unsaturated primary alcohols and cutin monomers. Plant Physiol. 2017, 173, 1109–1124. [Google Scholar] [CrossRef] [PubMed]
- Samuels, L.; Kunst, L.; Jetter, R. Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 2008, 59, 683–707. [Google Scholar] [CrossRef]
- Kunst, L.; Samuels, L. Plant cuticles shine: Advances in wax biosynthesis and export. Curr. Opin. Plant. Biol. 2009, 12, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Chacón, M.G.; Fournier, A.E.; Tran, F.; Dittrich-Domergue, F.; Pulsifer, I.P.; Domergue, F.; Rowland, O. Identification of amino acids conferring chain length substrate specificities on fatty alcohol-forming reductases FAR5 and FAR8 from Arabidopsis thaliana. J. Biol. Chem. 2013, 288, 30345–30355. [Google Scholar] [CrossRef] [PubMed]
- Teerawanichpan, P.; Qiu, X. Fatty acyl-CoA reductase and wax synthase from Euglena gracilis in the biosynthesis of medium-chain wax esters. Lipids 2010, 45, 263–273. [Google Scholar] [CrossRef]
- Metz, J.G.; Pollard, M.R.; Anderson, L.; Hayes, T.R.; Lassner, M.W. Purification of a jojoba embryo fatty acyl-coenzyme A reductase and expression of its cDNA in high erucic acid rapeseed. Plant Physiol. 2000, 122, 635–644. [Google Scholar] [CrossRef]
- Doan, T.T.; Carlsson, A.S.; Hamberg, M.; Bülow, L.; Stymne, S.; Olsson, P. Functional expression of five Arabidopsis fatty acyl-CoA reductase genes in Escherichia coli. J. Plant Physiol. 2009, 166, 787–796. [Google Scholar] [CrossRef]
- Teerawanichpan, P.; Robertson, A.J.; Qiu, X. A fatty acyl-CoA reductase highly expressed in the head of honey bee (Apis mellifera) involves biosynthesis of a wide range of aliphatic fatty alcohols. Insect Biochem. Mol. Biol. 2010, 40, 641–649. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, 225–229. [Google Scholar] [CrossRef]
- Kallberg, Y.; Oppermann, U.; Persson, B. Classification of the short-chain dehydrogenase/reductase superfamily using hidden Markov models. FEBS J. 2010, 277, 2375–2386. [Google Scholar] [CrossRef]
- Aarts, M.G.; Hodge, R.; Kalantidis, K.; Florack, D.; Wilson, Z.A.; Mulligan, B.J.; Stiekema, W.J.; Scott, R.; Pereira, A. The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 1997, 12, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Hooker, T.S.; Millar, A.A.; Kunst, L. Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol. 2002, 129, 1568–1580. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, M.; Sun, Y.; Wang, Y.; Li, T.; Chai, G.; Jiang, W.; Shan, L.; Li, C.; Xiao, E.; et al. FAR5, a fatty acyl-coenzyme A reductase, is involved in primary alcohol biosynthesis of the leaf blade cuticular wax in wheat (Triticum aestivum L.). J. Exp. Bot. 2015, 66, 1165–1178. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, Y.; Wu, H.; Xu, J.; Li, T.; Hegebarth, D.; Jetter, R.; Chen, L.; Wang, Z. Three TaFAR genes function in the biosynthesis of primary alcohols and the response to abiotic stresses in Triticum aestivum. Sci. Rep. 2016, 6, 25008. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, Y.; You, Q.; Luo, W.; Wang, C.; Zhao, S.; Chai, G.; Li, T.; Shi, X.; Li, C.; et al. Three fatty acyl-coenzyme A reductases, BdFAR1, BdFAR2 and BdFAR3, are involved in cuticular wax primary alcohol biosynthesis in brachypodium distachyon. Plant Cell Physiol. 2018, 59, 527–543. [Google Scholar] [CrossRef] [PubMed]
- Domergue, F.; Vishwanath, S.J.; Joubès, J.; Ono, J.; Lee, J.A.; Bourdon, M.; Alhattab, R.; Lowe, C.; Pascal, S.; Lessire, R.; et al. Three Arabidopsis fatty acyl-coenzyme A reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiol. 2010, 153, 1539–1554. [Google Scholar] [CrossRef]
- Denic, V.; Weissman, J.S. A molecular caliper mechanism for determining very long-chain fatty acid length. Cell 2007, 130, 663–677. [Google Scholar] [CrossRef]
- Bernard, A.; Domergue, F.; Pascal, S.; Jetter, R.; Renne, C.; Faure, J.D.; Haslam, R.P.; Napier, J.A.; Lessire, R.; Joubès, J. Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 2012, 24, 3106–3118. [Google Scholar] [CrossRef]
- Sun, J.; Guo, Y.; Fu, X.; Wang, Y.; Liu, Y.; Huo, B.; Sheng, J.; Hu, X. Dendrobium candidum inhibits MCF-7 cells proliferation by inducing cell cycle arrest at G2/M phase and regulating key biomarkers. OncoTargets Ther. 2015, 9, 21–30. [Google Scholar] [CrossRef]
- Zotz, G.; Winkler, U. Aerial roots of epiphytic orchids: The velamen radicum and its role in water and nutrient uptake. Oecologia 2013, 171, 733–741. [Google Scholar] [CrossRef]
- Zou, L.H.; Wan, X.; Deng, H.; Zheng, B.Q.; Li, B.J.; Wang, Y. Data descriptor: RNA-seq transcriptomic profiling of crassulacean acid metabolism pathway in Dendrobium catenatum. Sci. Data 2018, 5, 180252. [Google Scholar] [CrossRef]
- Ng, T.B.; Liu, J.; Wong, J.H.; Ye, X.; Sze, S.C.W.; Tong, Y.; Zhang, K.Y. Review of research on Dendrobium, a prized folk medicine. Appl. Microbiol. Biotechnol. 2012, 93, 1795–1803. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.T.; Cui, Z.; Li, Y.; Kang, Y.; Song, X.; Wang, J.; Zhou, Y. Genome-wide identification and expression analysis of MYB transcription factor superfamily in Dendrobium catenatum. Front. Genet. 2021, 12, 714696. [Google Scholar] [CrossRef]
- Zhang, T.T.; Xu, Y.; Ding, Y.; Yu, W.; Wang, J.; Lai, H.; Zhou, Y. Identification and expression analysis of WRKY gene family in response to abiotic stress in Dendrobium catenatum. Front. Genet. 2022, 13, 800019. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, T.T.; Xing, W.; Wang, J.; Yu, W.; Zhou, Y. Comprehensive genomic characterization of the NAC transcription factors and their response to drought stress in Dendrobium catenatum. Agronomy 2022, 12, 2753. [Google Scholar] [CrossRef]
- Wang, P.; Li, Y.X.; Zhang, T.T.; Kang, Y.Q.; Li, W.; Wang, J.; Yu, W.; Zhou, Y. Identification of the bZIP gene family and investigation of their response to drought stress in Dendrobium catenatum. Agronomy 2023, 13, 236. [Google Scholar] [CrossRef]
- Yang, T.; Zhang, T.; Li, Y.; Kang, Y.; Wang, P.; Liu, W.; Wang, Y.; Tian, L.; Dai, J.; Zhou, Y. Genome-wide identification and expression analysis of the chalcone synthase (CHS) gene family in Dendrobium catenatum. Agronomy 2023, 13, 1488. [Google Scholar] [CrossRef]
- Zhang, T.T.; Li, Y.X.; Wang, P.; Luo, Q.; Fu, S.; Kang, Y.; Zhou, Y. Characterization of Dendrobium catenatum CBL-CIPK signaling networks and their response to abiotic stress. Int. J. Biol. Macromol. 2023, 236, 124010. [Google Scholar] [CrossRef]
- Zhang, T.T.; Li, Y.; Kang, Y.; Wang, P.; Li, W.; Yu, W.; Wang, J.; Wang, J.; Song, X.; Jiang, X.Y.; et al. The Dendrobium catenatum DcCIPK24 increases drought and salt tolerance of transgenic Arabidopsis. Ind. Crop. Prod. 2022, 187, 115375. [Google Scholar] [CrossRef]
- Zhang, G.Q.; Xu, Q.; Bian, C.; Tsai, W.C.; Yeh, C.M.; Liu, K.W.; Yoshida, K.; Zhang, L.S.; Chang, S.B.; Chen, F.; et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 2016, 6, 19029. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.C.; Jain, P.K.; Bhat, S.R.; Srinivasan, R. Upstream sequence of fatty acyl-CoA reductase (FAR6) of Arabidopsis thaliana drives wound-inducible and stem-specific expression. Plant Cell Rep. 2012, 31, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, M.; Sun, Y.; Hegebarth, D.; Li, T.; Jetter, R.; Wang, Z. Molecular Characterization of TaFAR1 involved in primary alcohol biosynthesis of cuticular wax in hexaploid wheat. Plant Cell Physiol. 2015, 56, 1944–1961. [Google Scholar] [CrossRef]
- Chai, G.; Li, C.; Xu, F.; Li, Y.; Shi, X.; Wang, Y.; Wang, Z. Three endoplasmic reticulum-associated fatty acyl-coenzyme a reductases were involved in the production of primary alcohols in hexaploid wheat (Triticum aestivum L.). BMC Plant Biol. 2018, 18, 41. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, S.; Niu, C.; Liu, D.; Yan, T.; Tian, Y.; Liu, S.; Xie, K.; Li, Z.; Wang, Y.; et al. ZmMs25 encoding a plastid-localized fatty acyl reductase is critical for anther and pollen development in maize. J. Exp. Bot. 2021, 72, 4298–4318. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Cheng, X.; Jia, M.; Zhang, X.; Xue, F.; Li, Y.; Sun, J.; Liu, F. Silencing GhFAR3.1 reduces wax accumulation in cotton leaves and leads to increased susceptibility to drought stress. Plant Direct. 2021, 5, e00313. [Google Scholar] [CrossRef]
- Guan, L.; Xia, D.; Hu, N.; Zhang, H.; Wu, H.; Jiang, Q.; Li, X.; Sun, Y.; Wang, Y.; Wang, Z. OsFAR1 is involved in primary fatty alcohol biosynthesis and promotes drought tolerance in rice. Planta 2023, 258, 24. [Google Scholar] [CrossRef]
- Vioque, J.; Kolattukudy, P.E. Resolution and purification of an aldehyde-generating and an alcohol-generating fatty acyl-CoA reductase from pea leaves (Pisum sativum L.). Arch. Biochem. Biophys. 1997, 340, 64–72. [Google Scholar] [CrossRef]
- Schwacke, R.; Schneider, A.; van der Graaff, E.; Fischer, K.; Catoni, E.; Desimone, M.; Frommer, W.B.; Flügge, U.I.; Kunze, R. ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 2003, 131, 16–26. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Y.; Ayaz, A.; Zhao, H.; Lv, S. The plant fatty acyl reductases. Int. J. Mol. Sci. 2022, 23, 16156. [Google Scholar] [CrossRef]
- Chen, W.; Yu, X.H.; Zhang, K.; Shi, J.; De Oliveira, S.; Schreiber, L.; Shanklin, J.; Zhang, D. Male Sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol. 2011, 157, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Dobritsa, A.A.; Shrestha, J.; Morant, M.; Pinot, F.; Matsuno, M.; Swanson, R.; Møller, B.L.; Preuss, D. CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol. 2009, 151, 574–589. [Google Scholar] [CrossRef] [PubMed]
- Doan, T.T.; Domergue, F.; Fournier, A.E.; Vishwanath, S.J.; Rowland, O.; Moreau, P.; Wood, C.C.; Carlsson, A.S.; Hamberg, M.; Hofvander, P. Biochemical characterization of a chloroplast localized fatty acid reductase from Arabidopsis thaliana. Biochim. Biophys. Acta 2012, 1821, 1244–1255. [Google Scholar] [CrossRef]
- Shi, J.; Tan, H.; Yu, X.H.; Liu, Y.; Liang, W.; Ranathunge, K.; Franke, R.B.; Schreiber, L.; Wang, Y.; Kai, G.; et al. Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 2011, 23, 2225–2246. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Rowland, O.; Zheng, H.; Hepworth, S.R.; Lam, P.; Jetter, R.; Kunst, L. CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 2006, 142, 866–877. [Google Scholar] [CrossRef]
- Ma, J.; Deng, S.; Chen, L.; Jia, Z.; Sang, Z.; Zhu, Z.; Ma, L.; Chen, F. Gene duplication led to divergence of expression patterns, protein-protein interaction patterns and floral development functions of AGL6-like genes in the basal angiosperm Magnolia wufengensis (Magnoliaceae). Tree Physiol. 2019, 39, 861–876. [Google Scholar] [CrossRef]
- Sornaraj, P.; Luang, S.; Lopato, S.; Hrmova, M. Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: A molecular model of a wheat bZIP factor and implications of its structure in function. Biochim. Biophys. 2016, 1860, 46–56. [Google Scholar] [CrossRef]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant. 2013, 147, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Marquis, V.; Smirnova, E.; Graindorge, S.; Delcros, P.; Villette, C.; Zumsteg, J.; Heintz, D.; Heitz, T. Broad-spectrum stress tolerance conferred by suppressing jasmonate signaling attenuation in Arabidopsis JASMONIC ACID OXIDASE mutants. Plant J. 2022, 109, 856–872. [Google Scholar] [CrossRef]
- Yuan, M.; Shu, G.; Zhou, J.; He, P.; Xiang, L.; Yang, C.; Chen, M.; Liao, Z.; Zhang, F. AabHLH113 integrates jasmonic acid and abscisic acid signaling to positively regulate artemisinin biosynthesis in Artemisia annua. New Phytol. 2023, 237, 885–899. [Google Scholar] [CrossRef] [PubMed]
- An, C.; Mou, Z. Salicylic acid and its function in plant immunity. J. Integr. Plant. Biol. 2011, 53, 412–428. [Google Scholar] [CrossRef] [PubMed]
- Kidd, B.N.; Edgar, C.I.; Kumar, K.K.; Aitken, E.A.; Schenk, P.M.; Manners, J.M.; Kazan, K. The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 2009, 21, 2237–2252. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene ID | Number of AA | Molecular Weight (kDa) | Isoelectric Point | Subcellular Localization |
|---|---|---|---|---|---|
| DcFAR1 | LOC110092925 | 489 | 56.05 | 9.02 | Golgi apparatus |
| DcFAR2 | LOC110103179 | 587 | 66.15 | 9.02 | Chloroplast |
| DcFAR3 | LOC110112054 | 490 | 56.44 | 8.83 | Cytoplasm |
| DcFAR4 | LOC110114508 | 489 | 56.62 | 6.96 | Unidentified |
| DcFAR5 | LOC110114509 | 407 | 46.43 | 5.81 | Chloroplast, Golgi apparatus |
| DcFAR6 | LOC110114627 | 377 | 43.41 | 5.55 | Chloroplast |
| DcFAR7 | LOC110114642 | 443 | 51.14 | 8.49 | Mitochondrion |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, Y.; Wang, P.; Zhang, T.; Liu, W.; Wang, Y.; Dai, J.; Zhou, Y. Genome-Wide Identification of Fatty Acyl-CoA Reductase (FAR) Genes in Dendrobium catenatum and Their Response to Drought Stress. Horticulturae 2023, 9, 982. https://doi.org/10.3390/horticulturae9090982
Ren Y, Wang P, Zhang T, Liu W, Wang Y, Dai J, Zhou Y. Genome-Wide Identification of Fatty Acyl-CoA Reductase (FAR) Genes in Dendrobium catenatum and Their Response to Drought Stress. Horticulturae. 2023; 9(9):982. https://doi.org/10.3390/horticulturae9090982
Chicago/Turabian StyleRen, Yutong, Peng Wang, Tingting Zhang, Wen Liu, Yujuan Wang, Jun Dai, and Yang Zhou. 2023. "Genome-Wide Identification of Fatty Acyl-CoA Reductase (FAR) Genes in Dendrobium catenatum and Their Response to Drought Stress" Horticulturae 9, no. 9: 982. https://doi.org/10.3390/horticulturae9090982
APA StyleRen, Y., Wang, P., Zhang, T., Liu, W., Wang, Y., Dai, J., & Zhou, Y. (2023). Genome-Wide Identification of Fatty Acyl-CoA Reductase (FAR) Genes in Dendrobium catenatum and Their Response to Drought Stress. Horticulturae, 9(9), 982. https://doi.org/10.3390/horticulturae9090982







