Transcriptomic and Metabolomic Analyses of Seedlings of Two Grape Cultivars with Distinct Tolerance Responses to Flooding and Post-Flooding Stress Conditions
Abstract
:1. Introduction
2. Results
2.1. Sequencing Quality Statistics
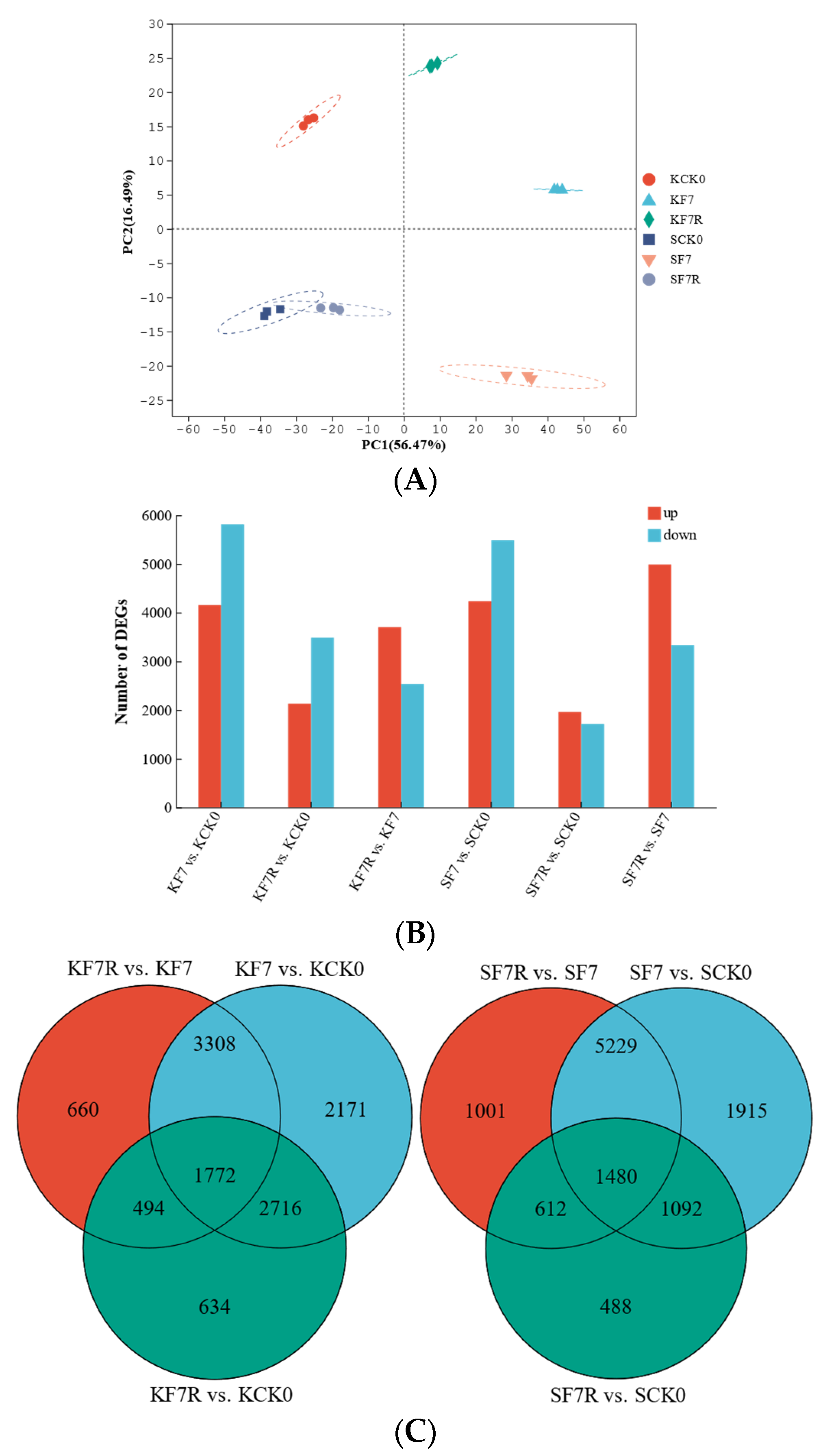
2.2. Relationship between Samples and Differentially Expressed Genes (DEGs)
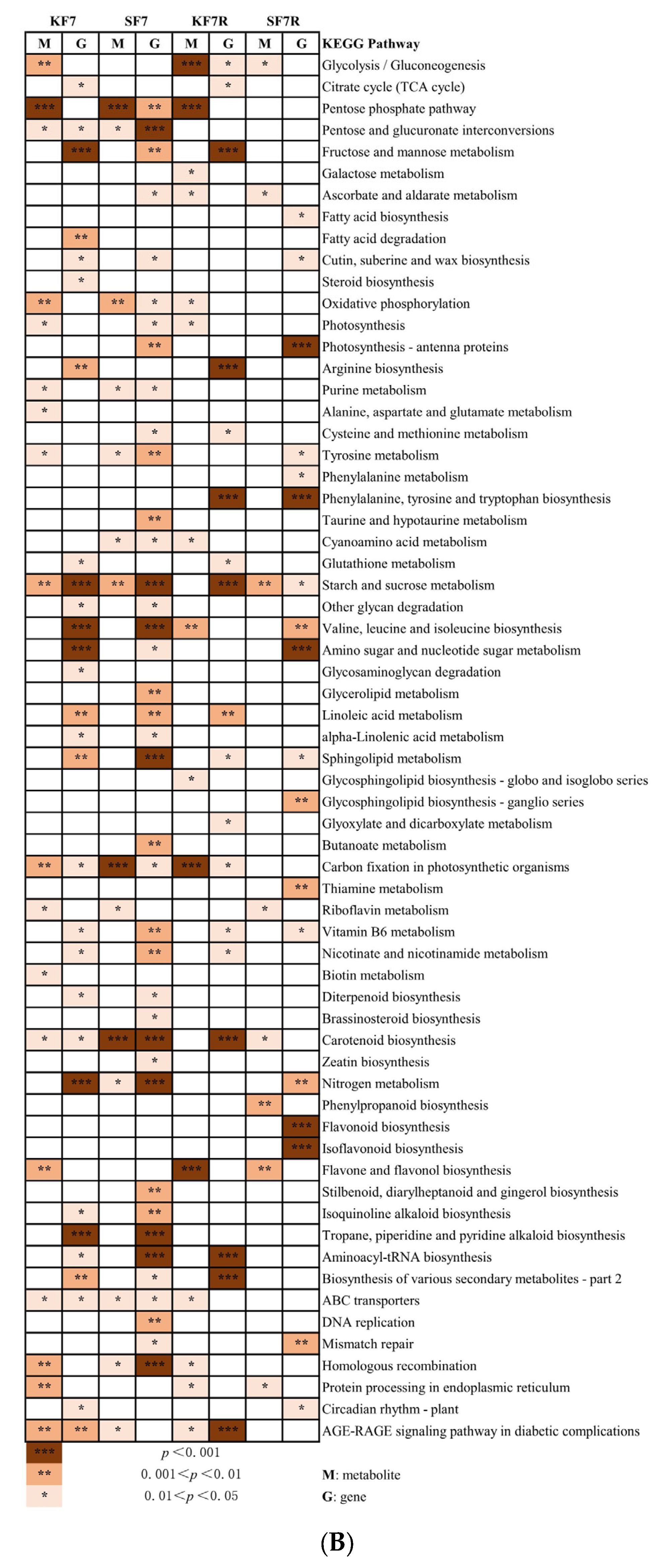
2.3. DEGs with Enriched Gene Ontology (GO) Terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways
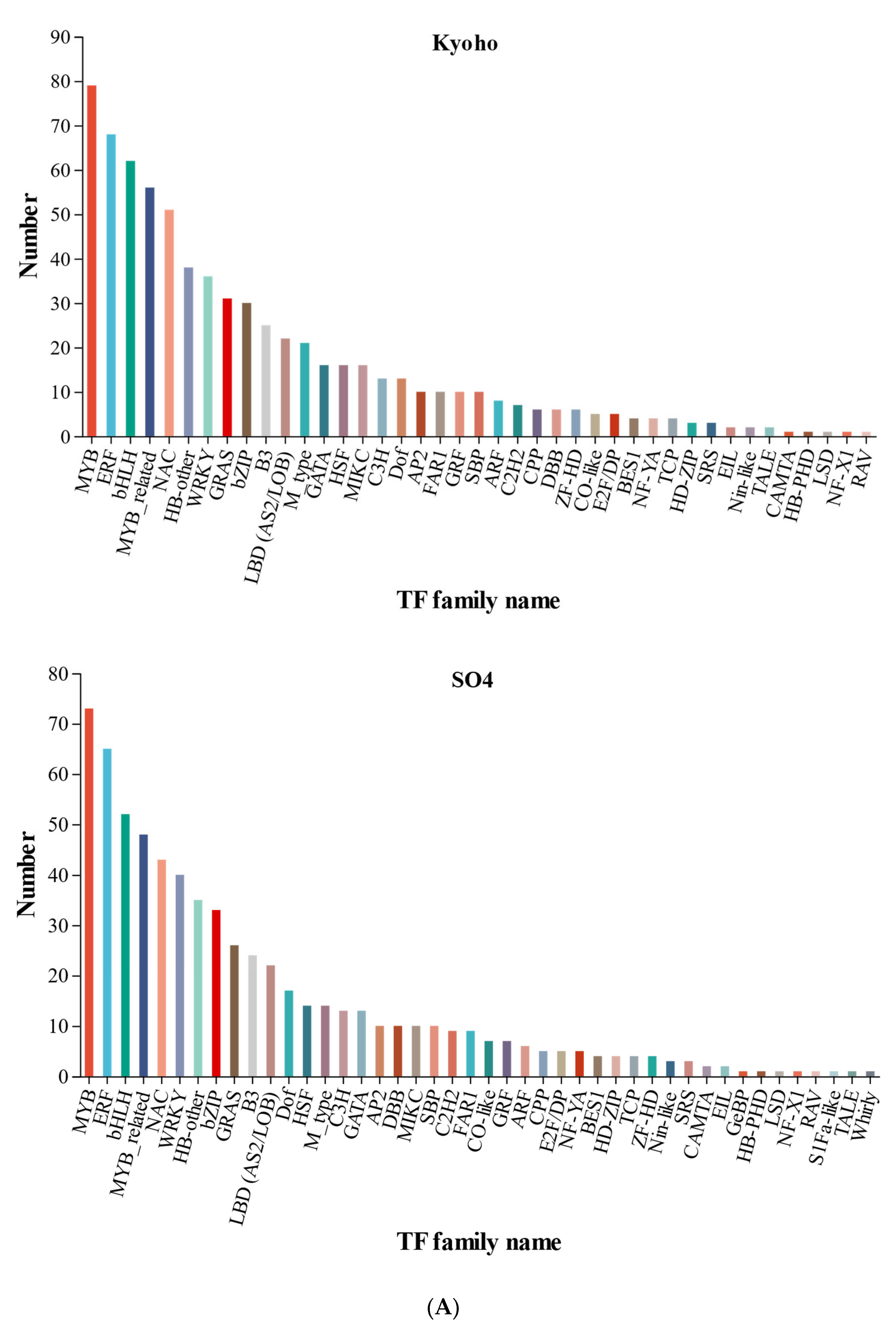
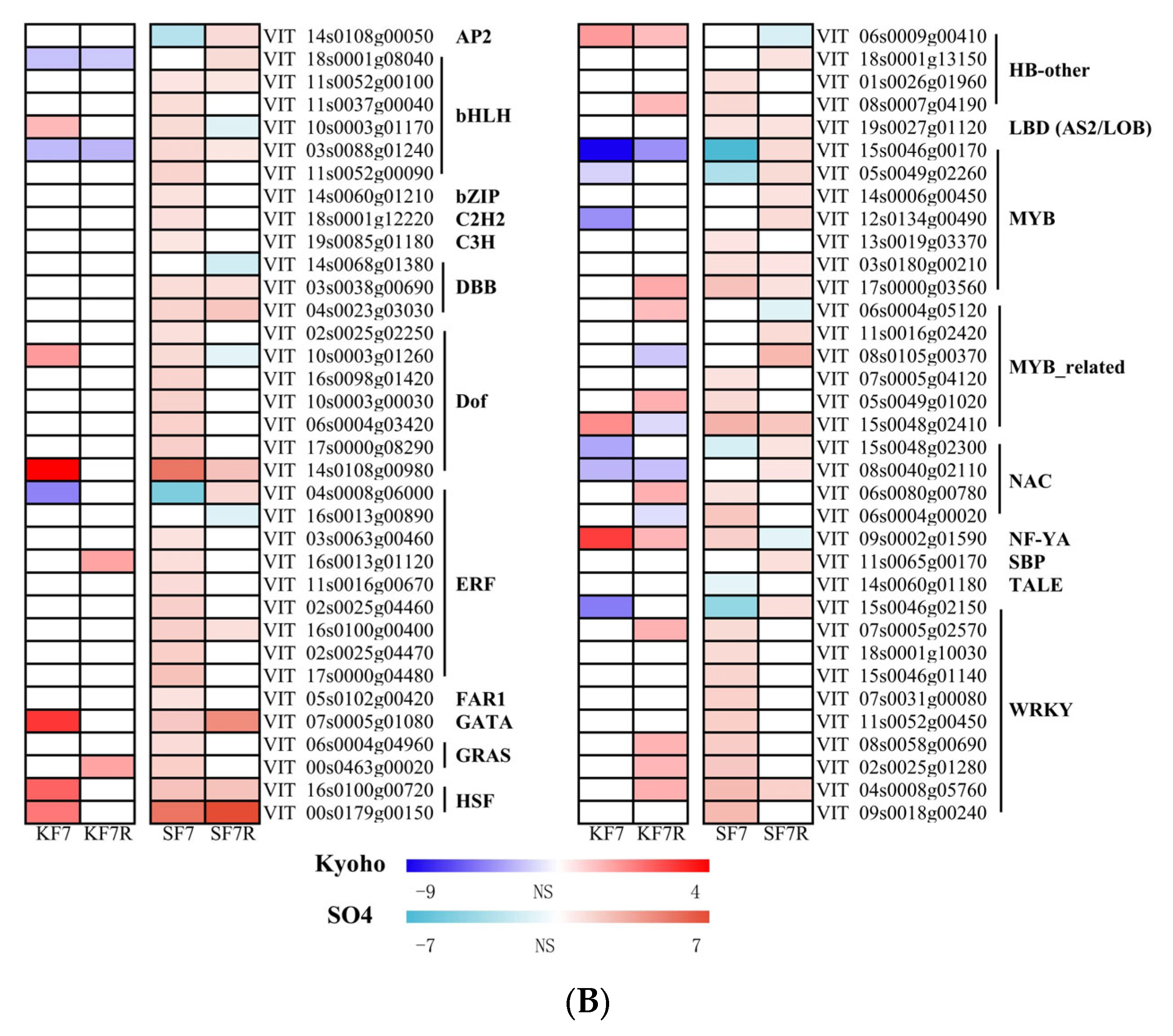
2.4. Differentially Responsive Transcription Factor Genes
2.5. Differential Metabolites in Response to Flooding and Post-Flooding Stress Conditions
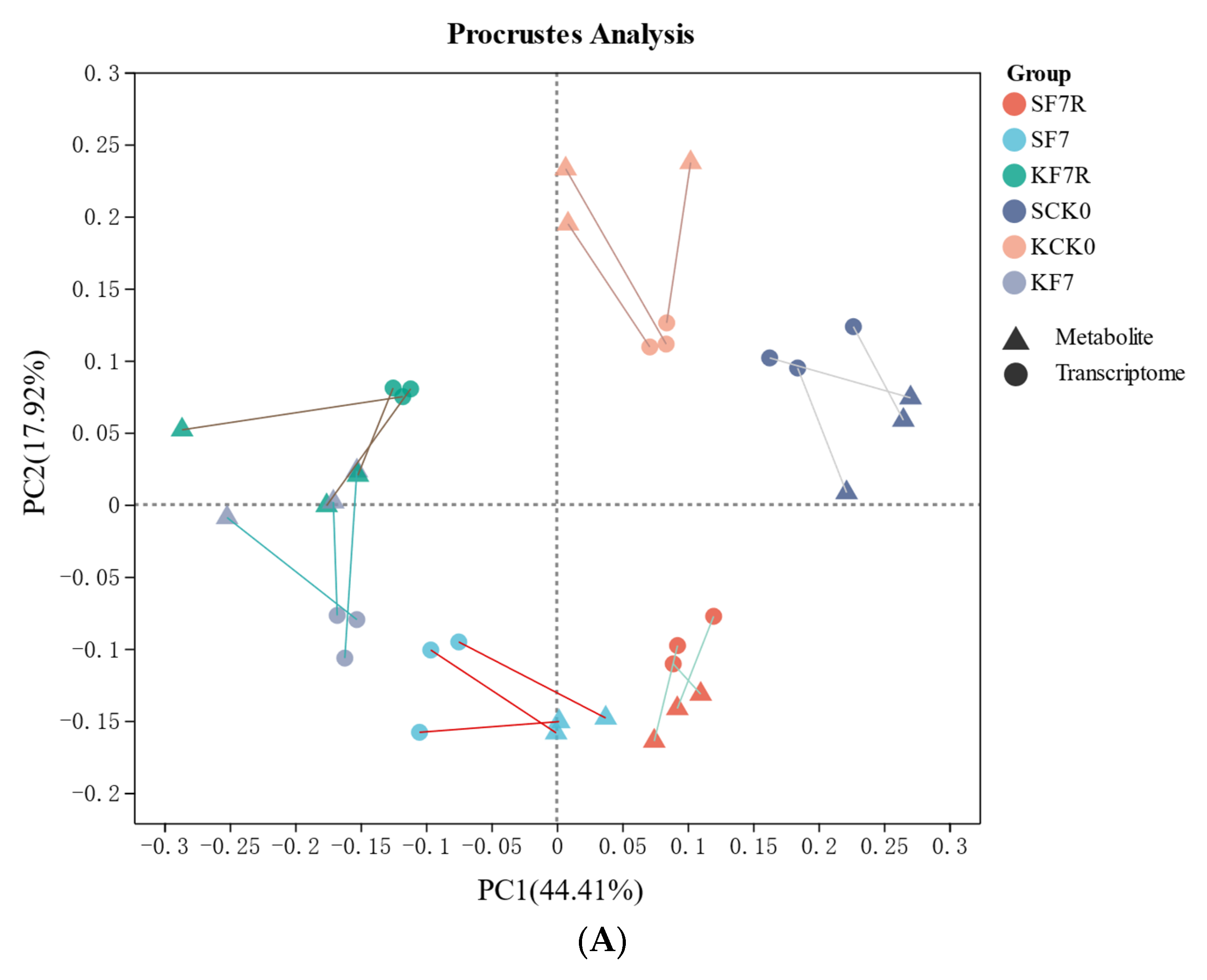
2.6. Integrative Analysis of Transcriptomic and Metabolomic Data
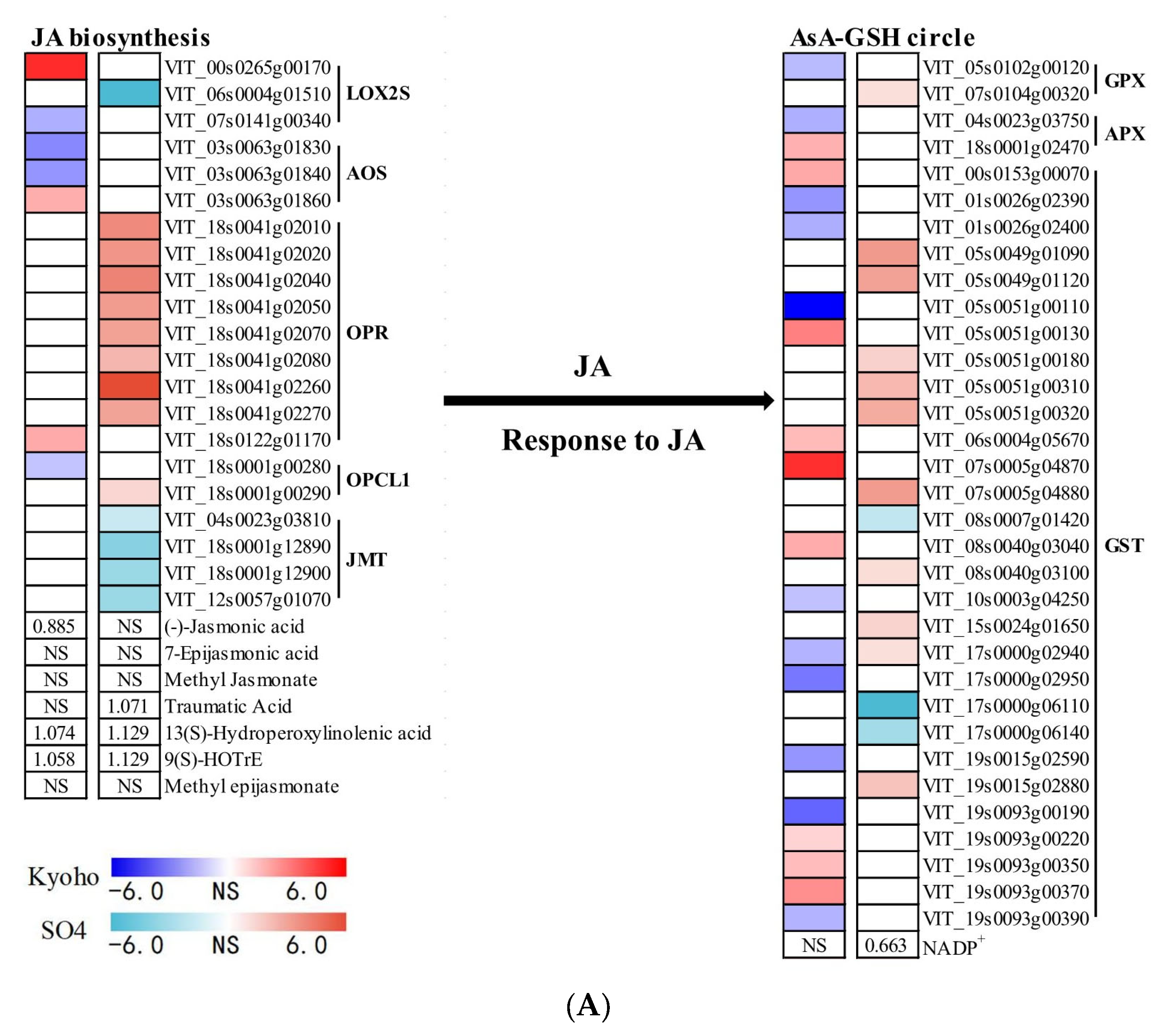
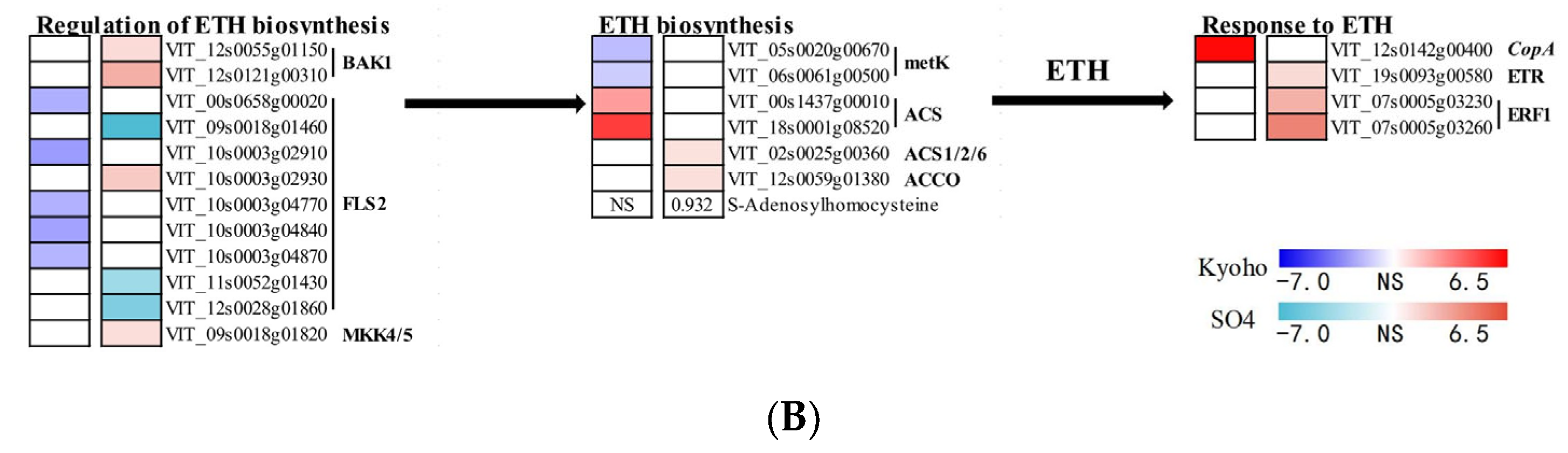
2.7. JA- and ETH-Related Responses of Grapevines to Flooding
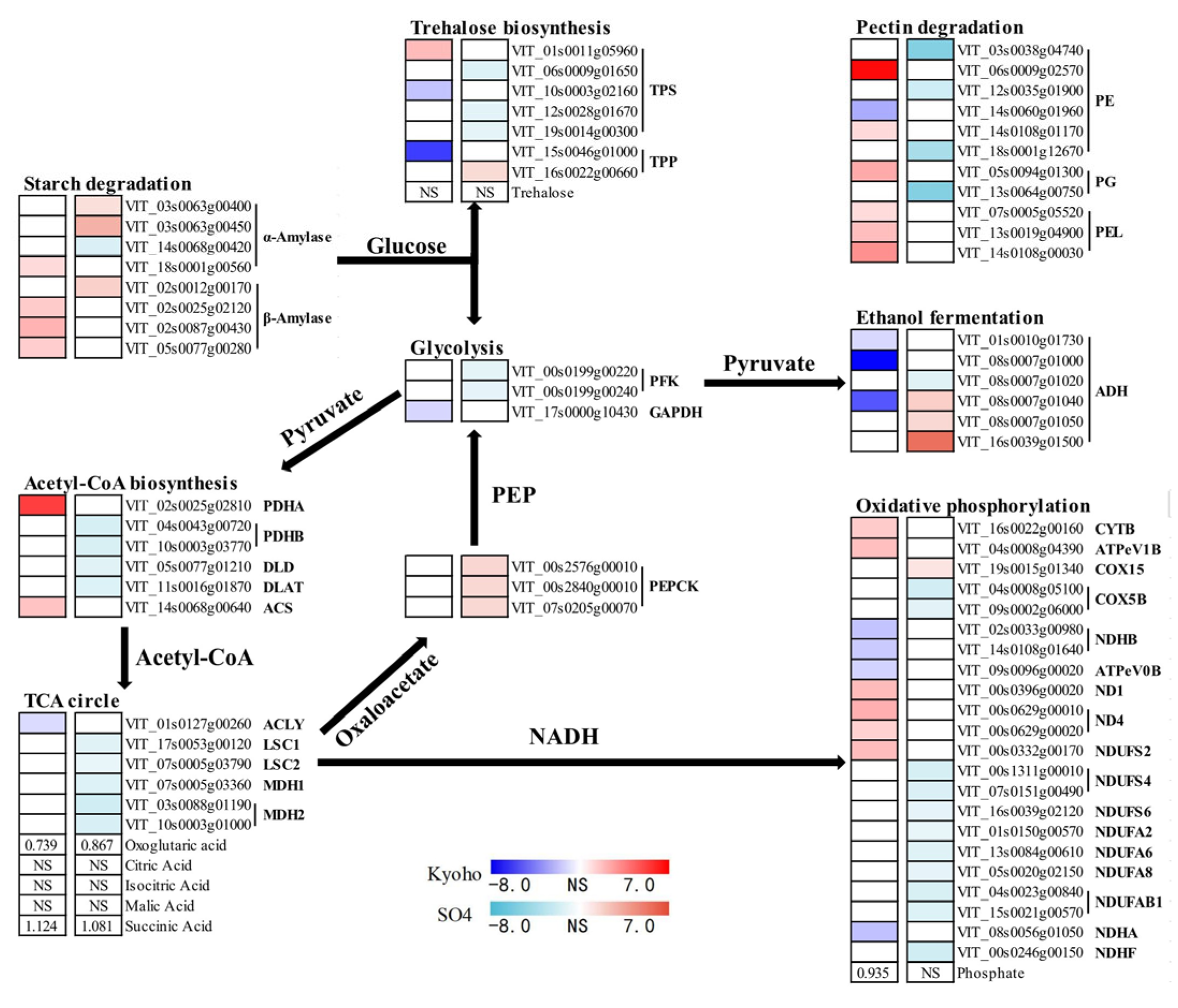
2.8. Carbohydrates Metabolism under Flood Stress
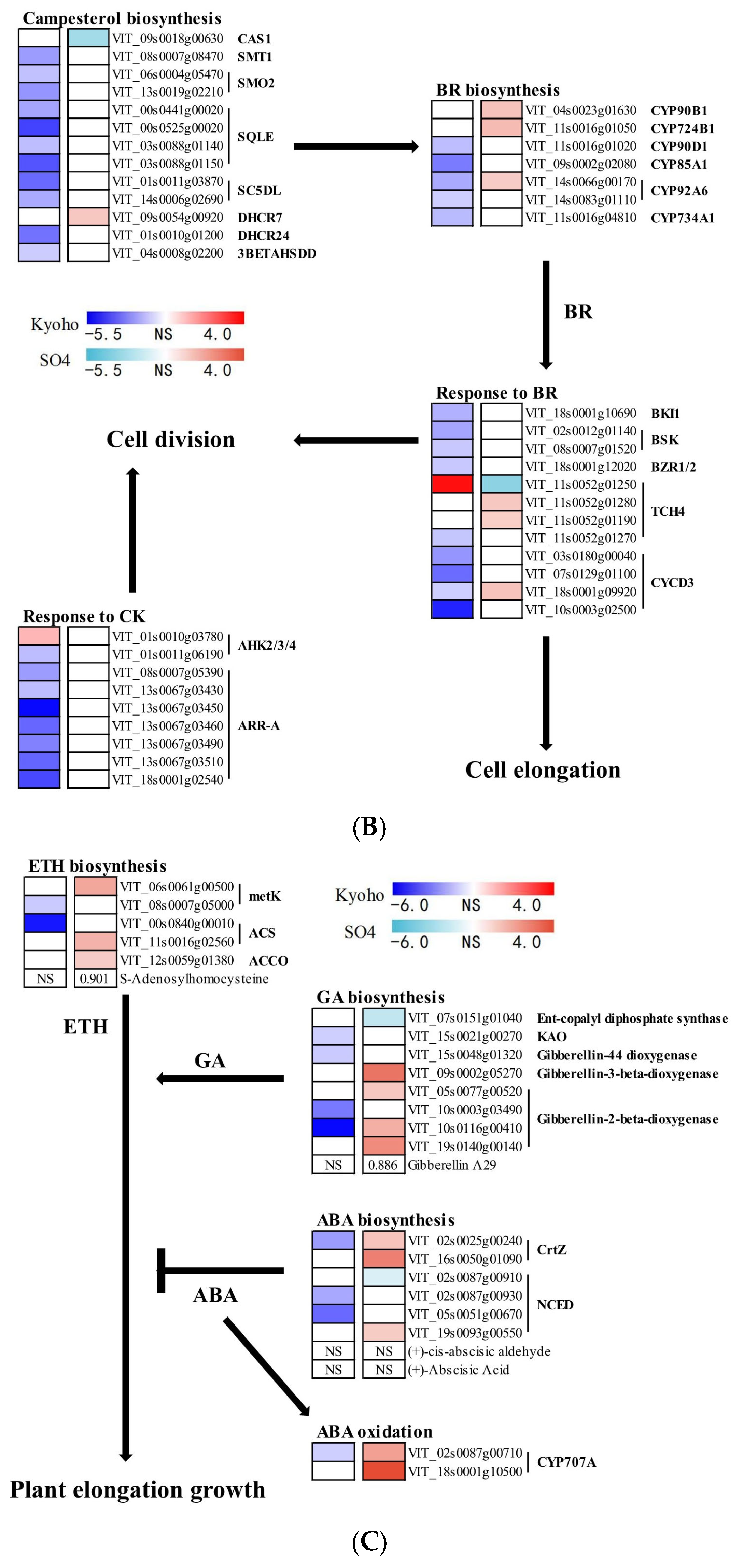
2.9. Phytohormone-Mediated Grapevine Responses during Post-Flooding Recovery
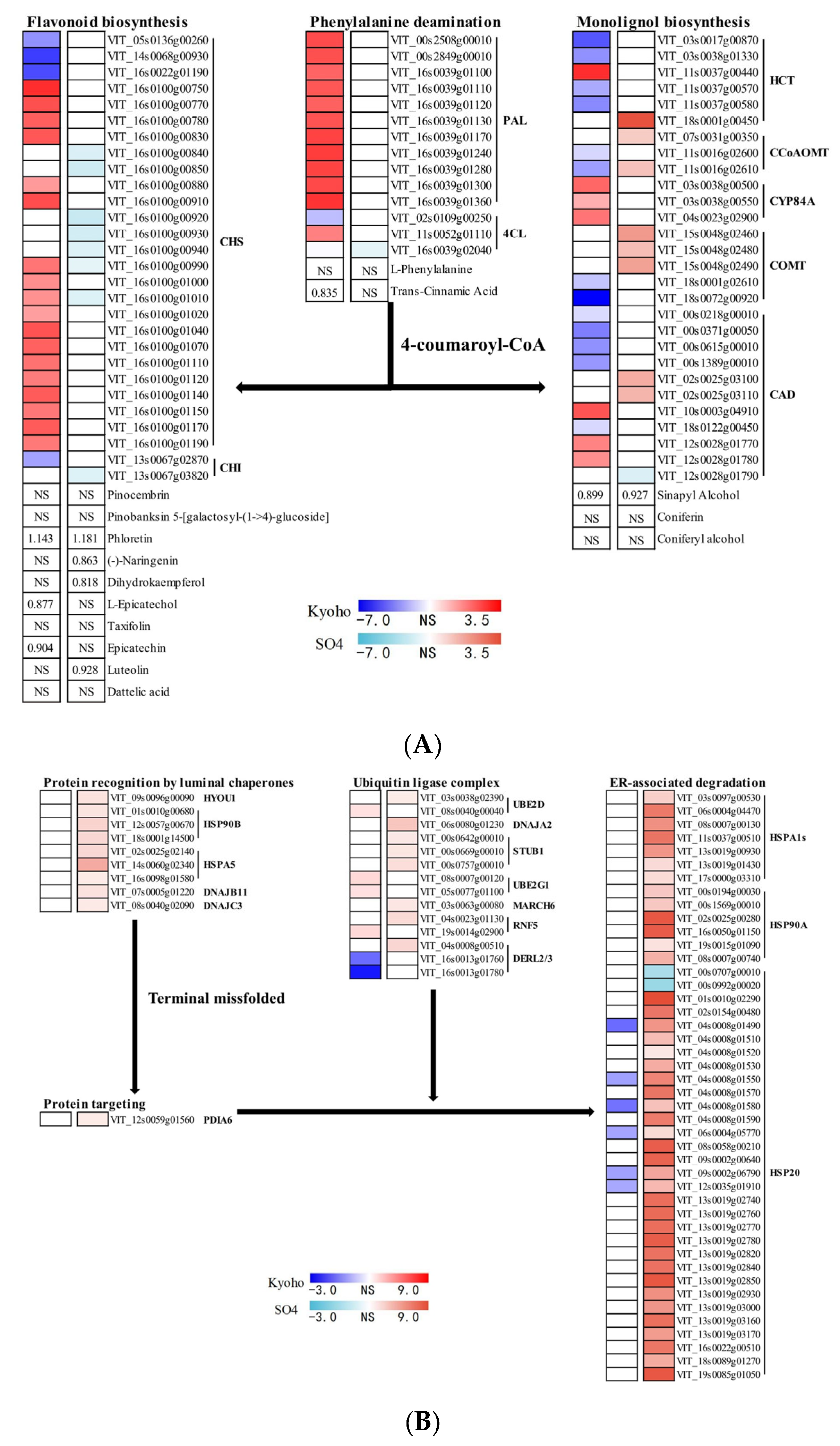
2.10. Flavonoid Biosynthesis, Monolignol Biosynthesis, and Endoplasmic Reticulum Associated Degradation in Post-Flooding Recovery
3. Discussion
3.1. Genotype-Specific TF Genes in Flooding and Post-Flooding Response in SO4
3.2. ETH and JA Regulation under Flooding Situation
3.3. The Shift from Aerobic Respiration to Anaerobic Fermentation under Flooding
3.4. JA Mediated ROS Scavenging System under Post-Flooding
3.5. Phytohormone Mediated Plant Growth in Post-Flooding Stage
3.6. Active ERAD in SO4 under Post-Flooding
4. Materials and Methods
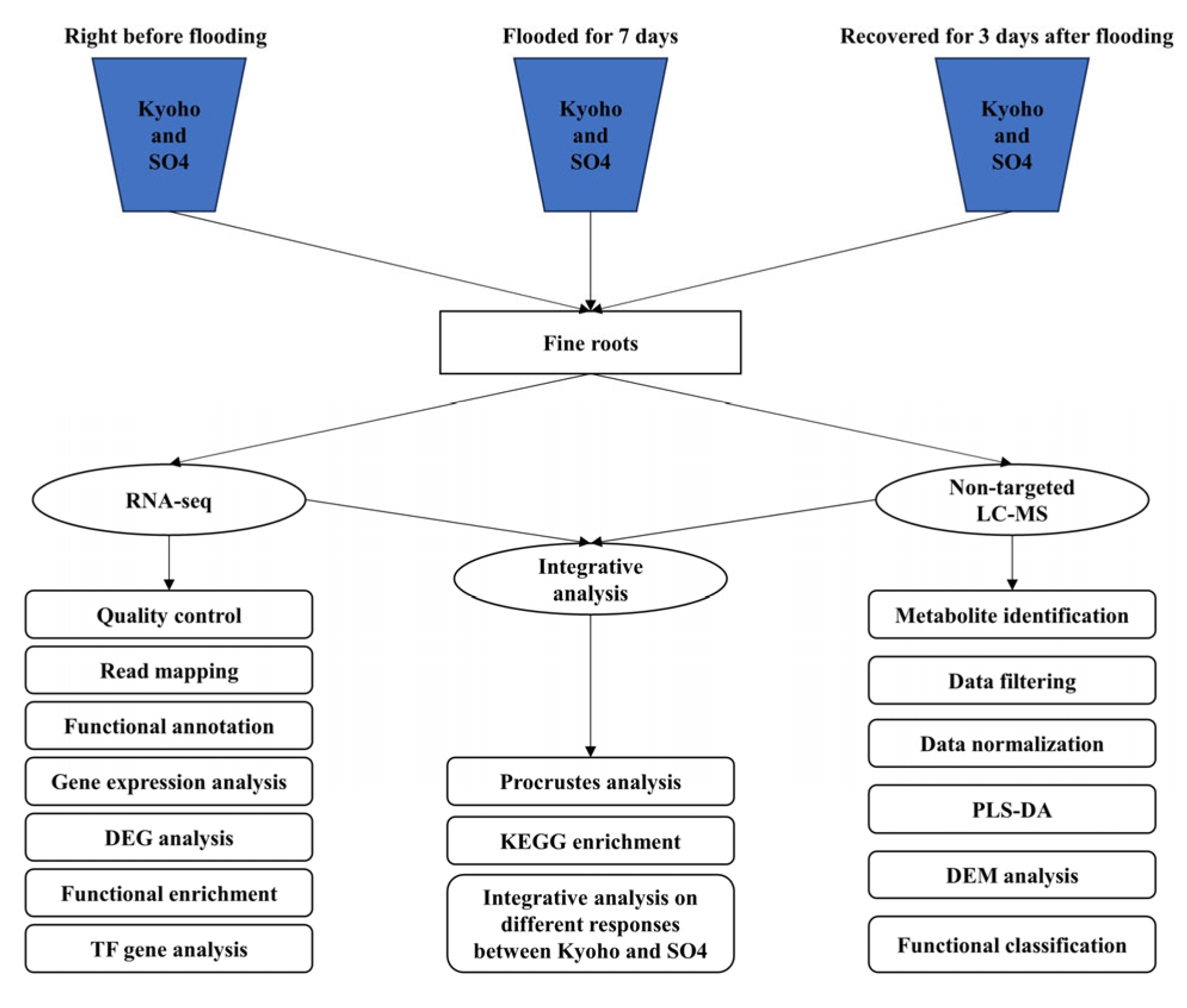
4.1. Plants and Experimental Treatments
4.2. Transcriptomic Analysis
4.3. Metabolomic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pedersen, O.; Perata, P.; Voesenek, L.A.C.J. Flooding and low oxygen responses in plants. Funct. Plant Biol. 2017, 44, iii–vi. [Google Scholar] [CrossRef] [PubMed]
- Bailey-Serres, J.; Fukao, T.; Gibbs, D.J.; Holdsworth, M.J.; Lee, S.C.; Licausi, F.; Perata, P.; Voesenek, L.A.C.J.; van Dongen, J.T. Making sense of low oxygen sensing. Trends Plant Sci. 2012, 17, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Tewari, S.; Mishra, A. Chapter 18—Flooding Stress in Plants and Approaches to Overcome. In Plant Metabolites and Regulation Under Environmental Stress; Ahmad, P., Ahanger, M.A., Singh, V.P., Tripathi, D.K., Alam, P., Alyemeni, M.N., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 355–366. [Google Scholar]
- Colmer, T.D.; Voesenek, L. Flooding tolerance: Suites of plant traits in variable environments. Funct. Plant Biol. 2009, 36, 665–681. [Google Scholar] [CrossRef] [PubMed]
- Loreti, E.; van Veen, H.; Perata, P. Plant responses to flooding stress. Curr. Opin. Plant Biol. 2016, 33, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Perata, P.; Voesenek, L.A.C.J. Submergence tolerance in rice requires Sub1A, an ethylene-response-factor-like gene. Trends Plant Sci. 2007, 12, 43–46. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhou, Z.; Zhang, Z.; Yu, X.; Zhang, X.; Du, K. Molecular and physiological responses in roots of two full-sib poplars uncover mechanisms that contribute to differences in partial submergence tolerance. Sci. Rep. 2018, 8, 12829. [Google Scholar] [CrossRef]
- Sarwat, M.; Ahmad, A.; Abdin, M.Z.; Ibrahim, M.M. Stress Signaling in Plants: Genomics and Proteomics Perspective; Physiological, Metabolic, and Molecular Responses of Plants to Abiotic Stress; Springer: Berlin/Heidelberg, Germany, 2017; Volume 2, pp. 1–35. [Google Scholar] [CrossRef]
- Sasidharan, R.; Hartman, S.; Liu, Z.; Martopawiro, S.; Sajeev, N.; Veen, H.; Yeung, E.; Voesenek, L. Signal Dynamics and Interactions during Flooding Stress. Plant Physiol. 2017, 176, 1106–1117. [Google Scholar] [CrossRef] [PubMed]
- Lekshmy, S.; Jha, S.K.; Sairam, R.K. Physiological and Molecular Mechanisms of Flooding Tolerance in Plants. In Elucidation of Abiotic Stress Signaling in Plants: Functional Genomics Perspectives; Pandey, G.K., Ed.; Springer: New York, NY, USA, 2015; Volume 2, pp. 227–242. [Google Scholar]
- Striker, G.G. Time is on our side: The importance of considering a recovery period when assessing flooding tolerance in plants. Ecol. Res. 2012, 27, 983–987. [Google Scholar] [CrossRef]
- Yeung, E.; Bailey-Serres, J.; Sasidharan, R. After The Deluge: Plant Revival Post-Flooding. Trends Plant Sci. 2019, 24, 443–454. [Google Scholar] [CrossRef]
- Kreuzwieser, J.; Hauberg, J.; Howell, K.; Carroll, A.; Rennenberg, H.; Millar, A.; Whelan, J. Differential Response of Gray Poplar Leaves and Roots Underpins Stress Adaptation during Hypoxia. Plant Physiol. 2008, 149, 461–473. [Google Scholar] [CrossRef]
- Peng, Y.; Dong, Y.; Tu, B.; Zhou, Z.; Zheng, B.; Luo, L.; Shi, C.; Du, K. Roots play a vital role in flood-tolerance of poplar demonstrated by reciprocal grafting. Flora Morphol. Distrib. Funct. Ecol. Plants 2013, 208, 479–487. [Google Scholar] [CrossRef]
- Chen, S.; Yin, C.; Strasser, R.J.; Govindjee; Yang, C.; Qiang, S. Reactive oxygen species from chloroplasts contribute to 3-acetyl-5-isopropyltetramic acid-induced leaf necrosis of Arabidopsis thaliana. Plant Physiol. Biochem. 2012, 52, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Yeung, E.; van Veen, H.; Vashisht, D.; Sobral Paiva, A.L.; Hummel, M.; Rankenberg, T.; Steffens, B.; Steffen-Heins, A.; Sauter, M.; de Vries, M.; et al. A stress recovery signaling network for enhanced flooding tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2018, 115, E6085–E6094. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Cao, P.; Tian, D.; Chen, H.; Zhu, C.; Zhu, B. Postharvest Biology; Technology. Edible coatings inhibit the postharvest berry abscission of table grapes caused by sulfur dioxide during storage. Postharvest Biol. Technol. 2019, 152, 1–8. [Google Scholar] [CrossRef]
- Mancuso, S.; Marras, A.M. Adaptative Response of Vitis Root to Anoxia. Plant Cell Physiol. 2006, 47, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Mugnai, S.; Marras, A.; Mancuso, S. Effect of Hypoxic Acclimation on Anoxia Tolerance in Vitis Roots: Response of Metabolic Activity and K+ Fluxes. Plant Cell Physiol. 2011, 52, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Ruperti, B.; Botton, A.; Populin, F.; Eccher, G.; Brilli, M.; Quaggiotti, S.; Trevisan, S.; Cainelli, N.; Guarracino, P.; Schievano, E.; et al. Flooding Responses on Grapevine: A Physiological, Transcriptional, and Metabolic Perspective. Front. Plant Sci. 2019, 10, 339. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Chen, F.; Meng, Y.; Chandrasekaran, U.; Luo, X.; Yang, W.; Shu, K. Plant waterlogging/flooding stress responses: From seed germination to maturation. Plant Physiol. Biochem. 2020, 148, 228–236. [Google Scholar] [CrossRef]
- Panat, N.A.; Maurya, D.K.; Ghaskadbi, S.S.; Sandur, S.K. Troxerutin, a plant flavonoid, protects cells against oxidative stress-induced cell death through radical scavenging mechanism. Food Chem. 2016, 194, 32–45. [Google Scholar] [CrossRef]
- Hodaei, M.; Rahimmalek, M.; Arzani, A.; Talebi, M. The effect of water stress on phytochemical accumulation, bioactive compounds and expression of key genes involved in flavonoid biosynthesis in Chrysanthemum morifolium L. Ind. Crops Prod. 2018, 120, 295–304. [Google Scholar] [CrossRef]
- Yang, L.-l.; Yang, L.; Yang, X.; Zhang, T.; Lan, Y.-m.; Zhao, Y.; Han, M.; Yang, L.-m. Drought stress induces biosynthesis of flavonoids in leaves and saikosaponins in roots of Bupleurum chinense DC. Phytochemistry 2020, 177, 112434. [Google Scholar] [CrossRef] [PubMed]
- Banti, V.; Mafessoni, F.; Loreti, E.; Alpi, A.; Perata, P. The Heat-Inducible Transcription Factor HsfA2 Enhances Anoxia Tolerance in Arabidopsis. Plant Physiol. 2010, 152, 1471–1483. [Google Scholar] [CrossRef] [PubMed]
- Rauf, M.; Arif, M.; Fisahn, J.; Xue, G.-P.; Balazadeh, S.; Mueller-Roeber, B. NAC transcription factor SPEEDY HYPONASTIC GROWTH regulates flooding-induced leaf movement in Arabidopsis. Plant Cell 2013, 25, 4941–4955. [Google Scholar] [CrossRef] [PubMed]
- Raineri, J.; Ribichich, K.; Chan, R. The sunflower transcription factor HaWRKY76 confers drought and flood tolerance to Arabidopsis thaliana plants without yield penalty. Plant Cell Rep. 2015, 34, 2065–2080. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Qiuming, Y.; Patil, G.; Agarwal, G.; Deshmukh, R.; Lin, L.; Wang, B.; Wang, Y.; Prince, S.; Song, L.; et al. Identification and Comparative Analysis of Differential Gene Expression in Soybean Leaf Tissue under Drought and Flooding Stress Revealed by RNA-Seq. Front. Plant Sci. 2016, 7, 1044. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, R.; Voesenek, L.A. Ethylene-Mediated Acclimations to Flooding Stress. Plant Physiol. 2015, 169, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Vidoz, M.L.; Loreti, E.; Mensuali, A.; Alpi, A.; Perata, P. Hormonal interplay during adventitious root formation in flooded tomato plants. Plant J. Cell Mol. Biol. 2010, 63, 551–562. [Google Scholar] [CrossRef]
- Sauter, M. Root responses to flooding. Curr. Opin. Plant Biol. 2013, 16, 282–286. [Google Scholar] [CrossRef]
- Yamauchi, T.; Watanabe, K.; Fukazawa, A.; Mori, H.; Abe, F.; Kawaguchi, K.; Oyanagi, A.; Nakazono, M. Ethylene and reactive oxygen species are involved in root aerenchyma formation and adaptation of wheat seedlings to oxygen-deficient conditions. J. Exp. Bot. 2014, 65, 261–273. [Google Scholar] [CrossRef]
- Khan, M.I.R.; Trivellini, A.; Chhillar, H.; Chopra, P.; Ferrante, A.; Khan, N.A.; Ismail, A.M. The significance and functions of ethylene in flooding stress tolerance in plants. Environ. Exp. Bot. 2020, 179, 104188. [Google Scholar] [CrossRef]
- Steffens, B.; Geske, T.; Sauter, M. Aerenchyma formation in the rice stem and its promotion by H2O2. New Phytol. 2011, 190, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Steffens, B.; Kovalev, A.; Gorb, S.N.; Sauter, M. Emerging roots alter epidermal cell fate through mechanical and reactive oxygen species signaling. Plant Cell 2012, 24, 3296–3306. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhou, Z.; Tong, R.; Hu, X.; Du, K. Anatomy and ultrastructure adaptations to soil flooding of two full-sib poplar clones differing in flood-tolerance. Flora 2017, 233, 90–98. [Google Scholar] [CrossRef]
- Caverzan, A.; Passaia, G.; Barcellos Rosa, S.; Ribeiro, C.; Lazzarotto, F.; Margis-Pinheiro, M. Plant responses to stresses: Role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 2012, 35, 1011–1019. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.-B.; Dai, Y.-S.; Xie, L.-J.; Yu, L.-J.; Zhou, Y.; Lai, Y.-X.; Yang, Y.-C.; Xu, L.; Chen, Q.-F.; Xiao, S. Jasmonate Regulates Plant Responses to Postsubmergence Reoxygenation through Transcriptional Activation of Antioxidant Synthesis. Plant Physiol. 2017, 173, 1864–1880. [Google Scholar] [CrossRef]
- Farhangi-Abriz, S.; Ghassemi-Golezani, K. Jasmonates: Mechanisms and functions in abiotic stress tolerance of plants. Biocatal. Agric. Biotechnol. 2019, 20, 101210. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Voesenek, L.A. Flooding stress: Acclimations and genetic diversity. Annu. Rev. Plant Biol. 2008, 59, 313–339. [Google Scholar] [CrossRef]
- Petra, R.; Planta, M.J. Flooding tolerance of Carex species. I. Root structure. Planta 1998, 207, 189–198. [Google Scholar]
- Crawford, R.M.M.; Walton, J.C.; Wollenweber-Ratzer, B. Similarities between post-ischaemic injury to animal tissues and post-anoxic injury in plants. Proc. R. Soc. Edinb. Sect. B Biol. Sci. 1994, 102, 325–332. [Google Scholar] [CrossRef]
- Chen, J.; Nolan, T.M.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yin, Y. Arabidopsis WRKY46, WRKY54, and WRKY70 Transcription Factors Are Involved in Brassinosteroid-Regulated Plant Growth and Drought Responses. Plant Cell 2017, 29, 1425–1439. [Google Scholar] [CrossRef]
- Nitsch, L.M.C.; Oplaat, C.; Feron, R.; Ma, Q.; Wolters-Arts, M.; Hedden, P.; Mariani, C.; Vriezen, W.H. Abscisic acid levels in tomato ovaries are regulated by LeNCED1 and SlCYP707A1. Planta 2009, 229, 1335–1346. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-X.; Howell, S.H. Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol. 2016, 211, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Xia, R.; Xie, Q.; Wu, Y. Endoplasmic reticulum-related E3 ubiquitin ligases: Key regulators of plant growth and stress responses. Plant Commun. 2021, 2, 100186. [Google Scholar] [CrossRef] [PubMed]
- Lü, S.; Zhao, H.; Des Marais, D.L.; Parsons, E.P.; Wen, X.; Xu, X.; Bangarusamy, D.K.; Wang, G.; Rowland, O.; Juenger, T.; et al. Arabidopsis ECERIFERUM9 Involvement in Cuticle Formation and Maintenance of Plant Water Status. Plant Physiol. 2012, 159, 930–944. [Google Scholar] [CrossRef] [PubMed]
- Doblas, V.G.; Amorim-Silva, V.; Posé, D.; Rosado, A.; Esteban, A.; Arró, M.; Azevedo, H.; Bombarely, A.; Borsani, O.; Valpuesta, V.; et al. The SUD1 Gene Encodes a Putative E3 Ubiquitin Ligase and Is a Positive Regulator of 3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase Activity in Arabidopsis. Plant Cell 2013, 25, 728–743. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhang, H.; Cui, P.; Ding, F.; Wang, G.; Li, R.; Jenks, M.A.; Lü, S.; Xiong, L. The Putative E3 Ubiquitin Ligase ECERIFERUM9 Regulates Abscisic Acid Biosynthesis and Response during Seed Germination and Postgermination Growth in Arabidopsis. Plant Physiol. 2014, 165, 1255–1268. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-M.; Lü, S.-Y.; Li, R.-J. The Arabidopsis endoplasmic reticulum associated degradation pathways are involved in the regulation of heat stress response. Biochem. Biophys. Res. Commun. 2017, 487, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Gu, X.; Zhou, Q.; Huang, J.; Liu, Z.; Zhou, Y.; Zheng, Y. Molecular and physiologic mechanisms of advanced ripening by trunk girdling at early veraison of ‘Summer Black’ grape. Front. Plant Sci. 2022, 13, 1012741. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sun, G.; Feng, T.; Zhang, J.; Huang, X.; Wang, T.; Xie, Z.; Chu, X.; Yang, J.; Wang, H.; et al. Sodium oligomannate therapeutically remodels gut microbiota and suppresses gut bacterial amino acids-shaped neuroinflammation to inhibit Alzheimer’s disease progression. Cell Res. 2019, 29, 787–803. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Chen, W.; Lai, X.; Dai, H.; Sun, H.; Zhou, X.; Chen, T. Metabolic responses and their correlations with phytochelatins in Amaranthus hypochondriacus under cadmium stress. Environ. Pollut. 2019, 252, 1791–1800. [Google Scholar] [CrossRef]







| Sample Group | Cultivar | Treatment |
|---|---|---|
| KCK0 | Kyoho | Seedlings sampled before flooding treatment, serving as the control |
| KF7 | Kyoho | Seedlings were flooded for 7 days |
| KF7R | Kyoho | Seedlings recovered for 3 days after a 7-day flooding treatment |
| SCK0 | SO4 | Seedlings sampled before flooding treatment, serving as the control |
| SF7 | SO4 | Seedlings were flooded for 7 days |
| SF7R | SO4 | Seedlings recovered for 3 days after a 7-day flooding treatment |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Y.; Chen, J.; Long, W.; He, P.; Zhou, Q.; Hu, X.; Zhou, Y.; Zheng, Y. Transcriptomic and Metabolomic Analyses of Seedlings of Two Grape Cultivars with Distinct Tolerance Responses to Flooding and Post-Flooding Stress Conditions. Horticulturae 2023, 9, 980. https://doi.org/10.3390/horticulturae9090980
Peng Y, Chen J, Long W, He P, Zhou Q, Hu X, Zhou Y, Zheng Y. Transcriptomic and Metabolomic Analyses of Seedlings of Two Grape Cultivars with Distinct Tolerance Responses to Flooding and Post-Flooding Stress Conditions. Horticulturae. 2023; 9(9):980. https://doi.org/10.3390/horticulturae9090980
Chicago/Turabian StylePeng, Yanjie, Jinli Chen, Wenjie Long, Pan He, Qi Zhou, Xia Hu, Yong Zhou, and Ying Zheng. 2023. "Transcriptomic and Metabolomic Analyses of Seedlings of Two Grape Cultivars with Distinct Tolerance Responses to Flooding and Post-Flooding Stress Conditions" Horticulturae 9, no. 9: 980. https://doi.org/10.3390/horticulturae9090980
APA StylePeng, Y., Chen, J., Long, W., He, P., Zhou, Q., Hu, X., Zhou, Y., & Zheng, Y. (2023). Transcriptomic and Metabolomic Analyses of Seedlings of Two Grape Cultivars with Distinct Tolerance Responses to Flooding and Post-Flooding Stress Conditions. Horticulturae, 9(9), 980. https://doi.org/10.3390/horticulturae9090980






