A Data Ecosystem for Orchard Research and Early Fruit Traceability
Abstract
:1. Overview
2. Background
2.1. Agriculture Victoria Research
2.2. RDM and FAIR Principles
2.3. Data Standards and Integration
2.4. Traceability
3. Materials and Methods
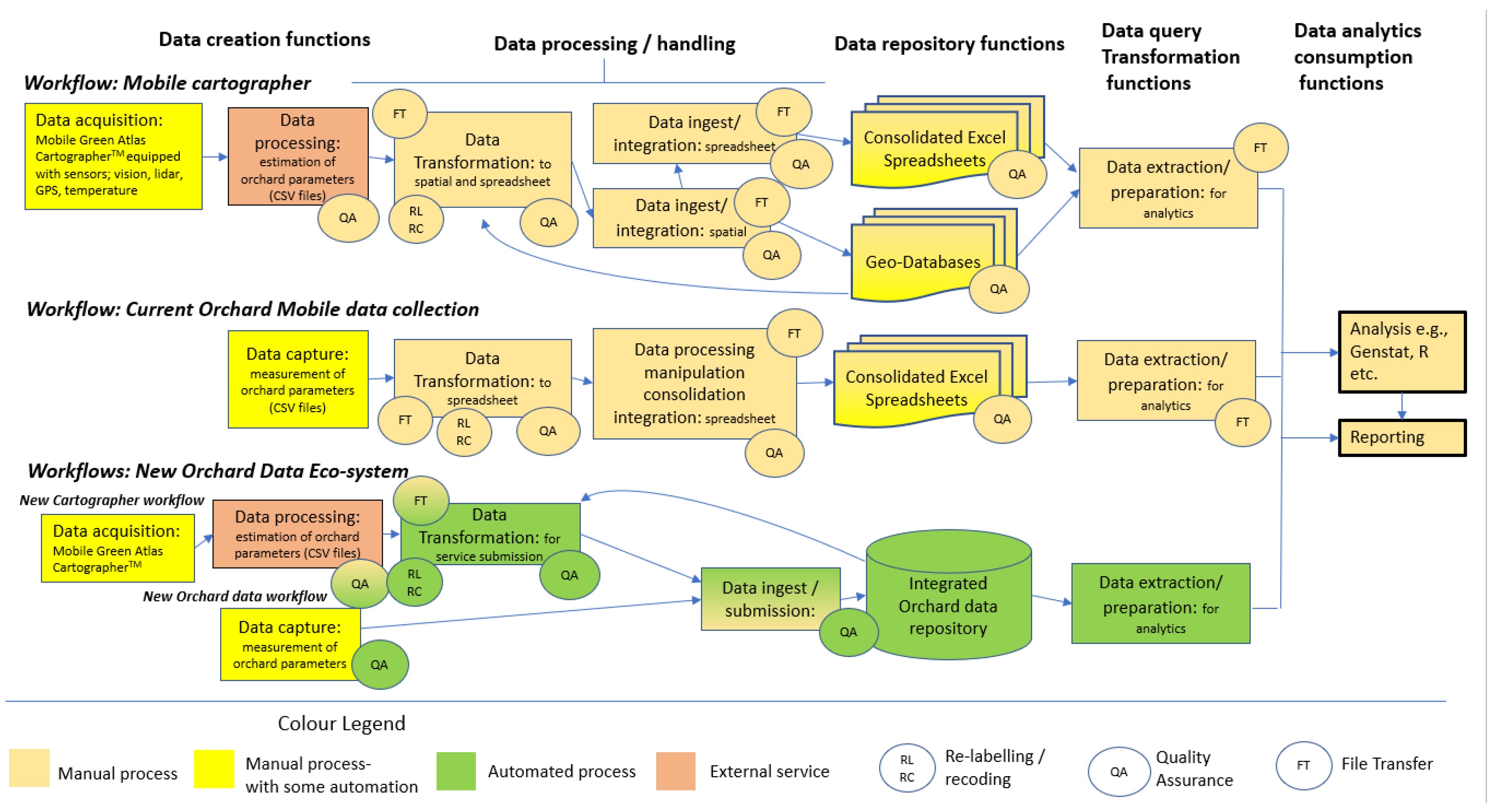
3.1. Requirements Analysis
3.2. Conceptual Information Framework
3.3. Research Information Model and Backend Database Development
3.4. Service Design and Development
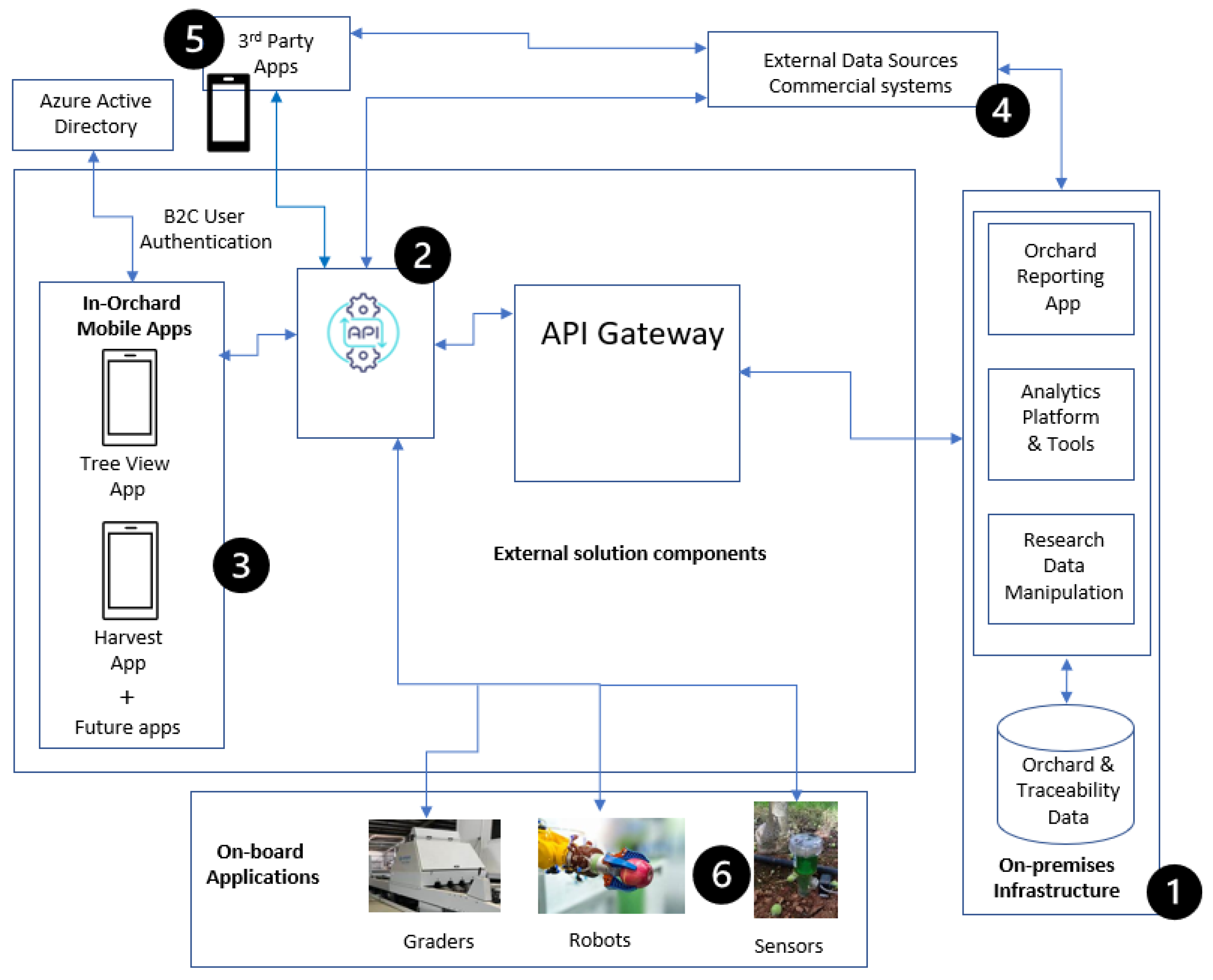
3.5. Solution Architecture and Application Development
- Orchard Reporting App: The database server includes an Orchard Reporting App, which leverages the stored data to generate comprehensive reports on various aspects of orchard research. The app utilises ESRI ArcGIS to visualise and analyse integrated, comprehensive data, allowing users to generate reports for any orchard features.
- Analytics Platform and Tools: The database server incorporates an analytics platform that enables advanced data analysis and modelling. Leveraging tools such as ArcGIS Spatial Analyst and ArcGIS Geostatistical Analyst, users can perform statistical analysis, geospatial modelling, and predictive analytics on the orchard research and traceability data. The platform can support a range of analytical capabilities.
- Research Data Manipulation: The on-premises database server provides functionalities for the efficient manipulation of research data. Researchers and administrators can perform data transformations, cleaning, and aggregation within the database environment. Advanced data manipulation techniques, such as querying, filtering, and joining, are supported to extract specific subsets of data for further analysis or reporting purposes.
4. Results
4.1. Ecosystem Model Evaluation
4.1.1. In-Orchard Data collection
4.1.2. Postharvest and Traceability Data Collection
5. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed]
- Birkbeck, G.; Nagle, T.; Sammon, D. Challenges in research data management practices: A literature analysis. J. Decis. Syst. 2022, 31 (Suppl. S1), 153–167. [Google Scholar] [CrossRef]
- ISO 19156:2011; Geographic Information—Observations and Measurements. International Organization for Standardization: Geneva, Switzerland, 2011. [CrossRef]
- Botts, A.; Robin, A.; Hirschorn, E. OGC SensorML: Model and XML Encoding Standard; Open Geospatial Consortium Pub.: Arlington, TX, USA, 2020; Available online: http://docs.opengeospatial.org/is/12-000r2/12-000r2.html (accessed on 23 May 2023).
- Pylianidis, C.; Osinga, S.; Ioannis, N.; Athanasiadis, I.N. Introducing digital twins to agriculture. Comput. Electron. Agric. 2021, 184, 105942. [Google Scholar] [CrossRef]
- Moghadam, P.; Lowe, T.; Edwards, E.J. Digital Twin for the Future of Orchard Production Systems. Proceedings 2019, 36, 92. [Google Scholar] [CrossRef]
- Queensland Alliance for Agriculture and Food Innovation: ’Digital Twin’ Concept Boosts Food Production. Available online: https://qaafi.uq.edu.au/article/2021/07/digital-twin-concept-boosts-food-production (accessed on 23 May 2023).
- Ali, B.; Dahlhaus, P. The Role of FAIR Data towards Sustainable Agricultural Performance: A Systematic Literature Review. Agriculture 2022, 12, 309. [Google Scholar] [CrossRef]
- Senft, M.; Stahl, U.; Svoboda, N. Research data management in agricultural sciences in Germany: We are not yet where we want to be. PLoS ONE 2022, 17, e0274677. [Google Scholar] [CrossRef] [PubMed]
- DataCite Metadata Working Group. DataCite Metadata Schema Documentation for the Publication and Citation of Research Data and Other Research Outputs. Version 4.4; DataCite: Hannover, Germany, 2021. [Google Scholar] [CrossRef]
- Pommier, C.; Coppens, F.; Ćwiek-Kupczyńska, H.; Faria, D.; Beier, S.; Miguel, C.; Michotey, C.; D’anna, F.; Owen, S.; Gruden, K. Plant Science Data Integration, from Building Community Standards to Defining a Consistent Data Lifecycle. In Towards Responsible Plant Data Linkage: Data Challenges for Agricultural Research and Development; Williamson, H.F., Leonelli, S., Eds.; Springer: Cham, Switzerland, 2023. [Google Scholar] [CrossRef]
- Sura, A.; Muna, A.; Ali, E.H. Meta-Analysis for Agricultural Research (Review). Mesop. J. Agric. 2020, 48, 21–32. [Google Scholar]
- Dundore-Arias, J.P.; Eloe-Fadrosh, E.; Schriml, L.; Beattie, G.; Brennan, F.; Busby, P.; Calderon, R.; Castle, S.; Emerson, J.; Everhart, S.; et al. Community-Driven Metadata Standards for Agricultural Microbiome Research. Phytobiomes J. 2020, 4, 115–121. [Google Scholar] [CrossRef]
- Jacobsen, A.; de Miranda Azevedo, R.; Juty, N.; Batista, D.; Coles, S.; Cornet, R.; Courtot, M.; Crosas, M.; Dumontier, M.; Evelo, C.T.; et al. FAIR Principles: Interpretations and Implementation Considerations. Data Intell. 2020, 2, 10–29. [Google Scholar] [CrossRef]
- Rawlings, R.; Castells, N.; Glendining, M. Linking Legacies: Realising the Potential of the Rothamsted Long-Term Agricultural Experiments. In Towards Responsible Plant Data Linkage; Springer: Cham, Switzerland; University of Exeter: Exeter, UK, 2023; pp. 107–123. [Google Scholar]
- Rawlings, C.J.; Davey, R.P. From Farm to FAIR: The Trials of Linking and Sharing Wheat Research Data. In Towards Responsible Plant Data Linkage: Data Challenges for Agricultural Research and Development; Williamson, H.F., Leonelli, S., Eds.; Springer: Cham, Switzerland, 2023. [Google Scholar] [CrossRef]
- Radick, G. Theory-Ladenness as a Problem for Plant Data Linkage. In Towards Responsible Plant Data Linkage; Springer: Cham, Switzerland; University of Exeter: Exeter, UK, 2023; pp. 27–36. [Google Scholar]
- Leach, P.; Mealling, M.; Salz, R. A Universally Unique IDentifier (UUID) URN Namespace. Available online: https://www.researchgate.net/publication/215758035_A_Universally_Unique_IDentifier_UUID_URN_Namespace (accessed on 12 June 2023).
- Baker, T.; Whitehead, B.; Musker, R.; Keizer, J. Global agricultural concept space: Lightweight semantics for pragmatic interoperability. NPJ Sci. Food 2019, 3, 16. [Google Scholar] [CrossRef] [PubMed]
- GS1 Global Location Number. Available online: https://www.gs1au.org/what-we-do/standards/global-location-number-glna. (accessed on 4 June 2023).
- Chalmers, D.; van den Ende, B.; van Heek, L. Productivity and Mechanization of the Tatura Trellis Orchard1. HortScience 1978, 13, 517–521. [Google Scholar] [CrossRef]
- Ionic Framework. Available online: https://ionicframework.com/ (accessed on 12 June 2023).
- Islam, M.S.; Scalisi, A.; O’Connell, M.G.; Morton, P.; Scheding, S.; Underwood, J.; Goodwin, I. A Ground-based Platform for Reliable Estimates of Fruit Number, Size, and Color in Stone Fruit Orchards. HortTechnology 2022, 32, 510–522. [Google Scholar] [CrossRef]
- Scalisi, A.; Pelliccia, D.; O’Connell, M.G. Maturity Prediction in Yellow Peach (Prunus persica L.) Cultivars Using a Fluorescence Spectrometer. Sensors 2020, 20, 6555. [Google Scholar] [CrossRef] [PubMed]
- EPCIS and CBV Implementation Guideline Release 2.0. Available online: https://ref.gs1.org/guidelines/epcis-cbv/2.0.0/ (accessed on 14 June 2023).
- Global Trade Item Number (GTIN). Available online: https://www.gs1.org/standards/id-keys/gtin (accessed on 14 June 2023).
- Sileshi, G.W. Analytic transparency is key for reproducibility of agricultural research. CABI Agric. Biosci. 2023, 4, 2. [Google Scholar] [CrossRef]
- Protocolexchange. Available online: https://protocolexchange.researchsquare.com/ (accessed on 25 June 2023).
- Cadwallader, L.; Hrynaszkiewicz, I. A survey of researchers’ code sharing and code reuse practices, and assessment of interactive notebook prototypes. PeerJ 2022, 10, e13933. [Google Scholar] [CrossRef] [PubMed]
- Ehrlinger, L.; Schrott, J.; Melichar, M.; Kirchmayr, N.; Wöß, W. Data Catalogs: A Systematic Literature Review and Guidelines to Implementation. In Database and Expert Systems Applications—DEXA 2021 Workshops. DEXA 2021; Kotsis, G., Tjoa, A.M., Khalil, I., Moser, B., Mashkoor, A., Sametinger, J., Fensel, A., Martinez-Gil, J., Fischer, L., Czech, G., et al., Eds.; Communications in Computer and Information Science; Springer: Cham, Switzerland, 2021; Volume 1479. [Google Scholar] [CrossRef]
- Research Data Australia. Available online: https://researchdata.edu.au/ (accessed on 25 June 2023).
- CGIAR. Ontologies Most Used in Agriculture. Available online: https://bigdata.cgiar.org/ontologies-for-agriculture/ (accessed on 25 June 2023).
- Arnaud, E.; Laporte, M.A.; Kim, S.; Aubert, C.; Leonelli, S.; Miro, B.; Cooper, L.; Jaiswal, P.; Kruseman, G.; Shrestha, R.; et al. The Ontologies Community of Practice: A CGIAR Initiative for Big Data in Agrifood Systems. Patterns 2020, 1, 100105. [Google Scholar] [CrossRef] [PubMed]
- Roussey, C.; Soulignac, V.; Champomier, J.C.; Abt, V.; Chanet, J.-P. Ontologies in Agriculture. In Proceedings of the AgEng 2010, International Conference on Agricultural Engineering, Clermont-Ferrand, France, 6–8 September 2010. [Google Scholar]
- Aydin, S.; Aydin, M.N. Ontology-based data acquisition model development for agricultural open data platforms and implementation of OWL2MVC tool. Comput. Electron. Agric. 2020, 175, 105589. [Google Scholar] [CrossRef]
- ESRI GeoEvent Server. Available online: https://enterprise.arcgis.com/en/geoevent/ (accessed on 25 June 2023).





| API Service | Brief Description | Functional Group |
|---|---|---|
| getUserInformation | Lookup user details | User management/governance |
| createUserInformation | Create new user | User management/governance |
| getOrchardsRegisteredToUser | Lookup experiments, orchards or locations registered to a user | Data access/governance |
| getOrchardFeatureInformation | Lookup information for a feature | Orchard information |
| geFeaturetInformationNearby | Spatial-enabled lookup of feature information | Orchard Information |
| createFeatureInformation | Add new feature information | Orchard Information |
| getFeatureMeasurementInformation | Get measurements/observations taken on a feature | Orchard observations & measurements |
| createFeatureMeasurementInformation | Submit new measurements/observations for a feature | Orchard observations & measurements |
| getMeasurementsMetadata | Get metadata for registered measurements | Orchard measurements |
| getDeviceData | Get metadata for registered measurement devices | Measurement devices |
| getTraceableObject | Lookup traceable object | Traceability |
| createTraceabilityEvent | Submit a traceability data | Traceability |
| getMetadata | Describes the services | API services |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Williams, S.R.; Agrahari Baniya, A.; Islam, M.S.; Murphy, K. A Data Ecosystem for Orchard Research and Early Fruit Traceability. Horticulturae 2023, 9, 1013. https://doi.org/10.3390/horticulturae9091013
Williams SR, Agrahari Baniya A, Islam MS, Murphy K. A Data Ecosystem for Orchard Research and Early Fruit Traceability. Horticulturae. 2023; 9(9):1013. https://doi.org/10.3390/horticulturae9091013
Chicago/Turabian StyleWilliams, Stephen Ross, Arbind Agrahari Baniya, Muhammad Sirajul Islam, and Kieran Murphy. 2023. "A Data Ecosystem for Orchard Research and Early Fruit Traceability" Horticulturae 9, no. 9: 1013. https://doi.org/10.3390/horticulturae9091013
APA StyleWilliams, S. R., Agrahari Baniya, A., Islam, M. S., & Murphy, K. (2023). A Data Ecosystem for Orchard Research and Early Fruit Traceability. Horticulturae, 9(9), 1013. https://doi.org/10.3390/horticulturae9091013








