Comparative Transcriptome Analysis Identified Genes Associated with Fruit Size in Pepper (Capsicum annuum L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Fruit Development Investigation
2.3. Histological Sections of the Fruit
2.4. RNA Extraction, Library Preparation, and Sequencing
2.5. Identification and Functional Annotation of Differentially Expressed Genes (DEGs)
2.6. WGCNA Analysis
2.7. Expression Analysis of Quantitative Real-Time PCR
2.8. Statistical Analysis
3. Results
3.1. Physiological Investigation of Fruit Development in Pepper
3.2. Overview of RNA-Sequence Results and Analysis of Differentially Expressed Genes (DEGs)
3.3. GO Enrichment Analysis of DEGs
3.4. KEGG Pathway Analysis of DEGs
3.5. Weighted Gene Co-Expression Network Analysis(WGCNA)
3.6. Expression Analysis of DEGs Associated with Fruit Weight in Pepper (Capsicum annuum)
3.6.1. Genes Associated with Cell Division, Changes in Cell Cycle Control, and Cell Wall
3.6.2. Transcription Factors Associated with Fruit Size Regulation
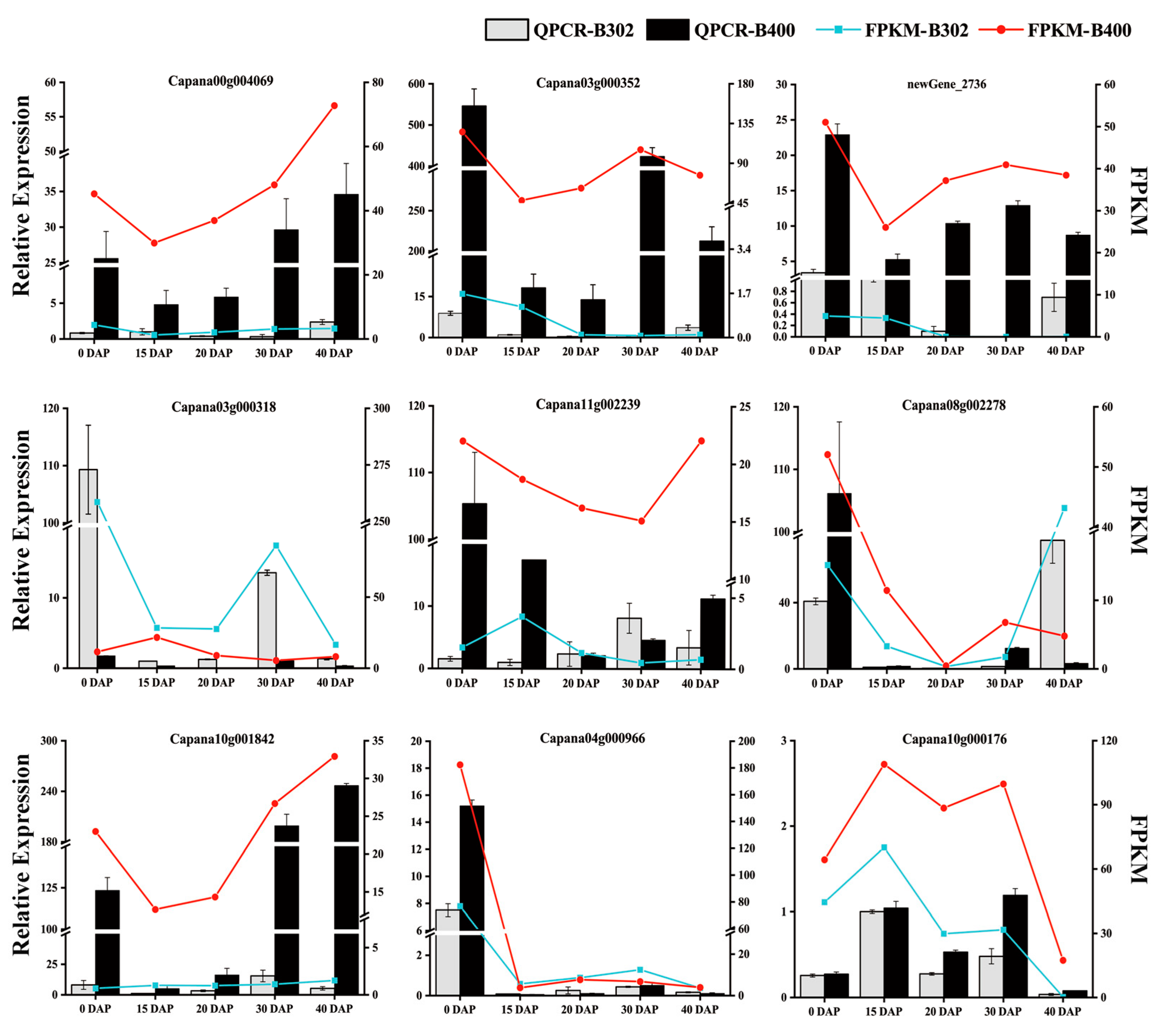
3.7. Verification of Gene Expression Pattern by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, G.; Xu, H.; Liu, Z.; Zhu, F.; Zou, X. The Global Dissemination of Capsicum. Chin. J. Trop. Crops 2023, 44, 1307–1316. [Google Scholar] [CrossRef]
- Lin, Q.; Xin, Z.; Kong, L.; Wang, X.; Yang, X.; He, W. Current situation of pepper industry development and breeding countermeasures in China. J. China Agric. Univ. 2023, 5, 82–95. [Google Scholar] [CrossRef]
- He, H.; Yamamuro, C. Interplays between auxin and GA signaling coordinate early fruit development. Hortic. Res. 2022, 9, uhab078. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, Y.; Hu, L.; Zhang, C.; Wu, W.; Li, W.; Fang, J. Elucidation of the mechanism underlying seedless blueberry formation after GA3 treatment based on the phenotype, physiology, metabolism and transcriptome. Sci. Hortic. 2023, 311, 111781. [Google Scholar] [CrossRef]
- Mu, Q.; Huang, Z.; Chakrabarti, M.; Illa-Berenguer, E.; Liu, X.; Wang, Y.; Ramos, A.; van der Knaap, E. Fruit weight is controlled by Cell Size Regulator encoding a novel protein that is expressed in maturing tomato fruits. PLoS Genet. 2017, 13, e1006930. [Google Scholar] [CrossRef] [PubMed]
- Cheniclet, C.; Rong, W.Y.; Causse, M.; Frangne, N.; Bolling, L.; Carde, J.-P.; Renaudin, J.-P. Cell expansion and endoreduplication show a large genetic variability in pericarp and contribute strongly to tomato fruit growth. Plant Physiol. 2005, 139, 1984–1994. [Google Scholar] [CrossRef] [PubMed]
- Schruff, M.C.; Spielman, M.; Tiwari, S.; Adams, S.; Fenby, N.; Scott, R.J. The Auxin Response Factor 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 2006, 133, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Deprost, D.; Yao, L.; Sormani, R.; Moreau, M.; Leterreux, G.; Nicolaï, M.; Bedu, M.; Robaglia, C.; Meyer, C. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007, 8, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Krizek, B.A. Making bigger plants: Key regulators of final organ size. Curr. Opin. Plant Biol. 2009, 12, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Horváth, B.M.; Magyar, Z.; Zhang, Y.; Hamburger, A.W.; Bakó, L.; Visser, R.G.F.; Bachem, C.W.B.; Bögre, L. EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J. 2006, 25, 4909–4920. [Google Scholar] [CrossRef] [PubMed]
- Rachappanavar, V.; Padiyal, A.; Sharma, J.K.; Gupta, S.K. Plant hormone-mediated stress regulation responses in fruit crops—A review. Sci. Hortic. 2022, 304, 111302. [Google Scholar] [CrossRef]
- Santner, A.; Calderon-Villalobos, L.I.A.; Estelle, M. Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 2009, 5, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Zong, X.; Ren, P.; Qian, Y.; Fu, A. Basic Helix-Loop-Helix (bHLH) Transcription Factors Regulate a Wide Range of Functions in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 7152. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Hu, G.; Wang, K.; Frasse, P.; Maza, E.; Djari, A.; Deng, W.; Pirrello, J.; Burlat, V.; Pons, C.; et al. Interaction of two MADS-box genes leads to growth phenotype divergence of all-flesh type of tomatoes. Nat. Commun. 2021, 12, 6892. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Theissen, G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 2003, 29, 464–489. [Google Scholar] [CrossRef]
- Shore, P.; Sharrocks, A.D. The MADS-box family of transcription factors. Eur. J. Biochem. 1995, 229, 1–13. [Google Scholar] [CrossRef]
- Guo, X.H.; Li, H.; Yin, L.L.; Liu, W.Y.; Naeem, M.; Guo, M. The Mechanism of MADS-box Gene SlMBP3 Modulating Tomato Fruit Size. Russ. J. Plant Physiol. 2022, 69, 63. [Google Scholar] [CrossRef]
- Mizukami, Y.; Ma, H. Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 1992, 71, 119–131. [Google Scholar] [CrossRef]
- Mizukami, Y.; Ma, H. Determination of Arabidopsis floral meristem identity by AGAMOUS. Plant Cell 1997, 9, 393–408. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhao, Y. A Role for Auxin in Flower Development. J. Integr. Plant Biol. 2007, 49, 99–104. [Google Scholar] [CrossRef]
- Liljegren, S.J.; Ditta, G.S.; Eshed, Y.; Savidge, B.; Bowman, J.L.; Yanofsky, M.F. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 2000, 404, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Ferrándiz, C.; Yanofsky, M.F.; Martienssen, R. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 1998, 125, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Gong, P.; Liu, H.; Zhang, M.; He, C. Multiple and integrated functions of floral C-class MADS-box genes in flower and fruit development of Physalis floridana. Plant Mol. Biol. 2021, 107, 101–116. [Google Scholar] [CrossRef]
- Sessa, G.; Carabelli, M.; Possenti, M.; Morelli, G.; Ruberti, I. Multiple Links between HD-Zip Proteins and Hormone Networks. Int. J. Mol. Sci. 2018, 19, 4047. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.C.; Hrmova, M.; Lopato, S.; Langridge, P. Modulation of plant growth by HD-Zip class I and II transcription factors in response to environmental stimuli. New Phytol. 2011, 190, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.E.; Debernardi, J.M.; Palatnik, J.F. Morphogenesis of simple leaves: Regulation of leaf size and shape. Wiley Interdiscip. Rev. Dev. Biol. 2014, 3, 41–57. [Google Scholar] [CrossRef] [PubMed]
- Sablowski, R. Control of patterning, growth, and differentiation by floral organ identity genes. J. Exp. Bot. 2015, 66, 1065–1073. [Google Scholar] [CrossRef]
- Khan, M.; Xu, H.; Hepworth, S.R. BLADE-ON-PETIOLE genes: Setting boundaries in development and defense. Plant Sci. 2014, 215–216, 157–171. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, G.R.; Muños, S.; Anderson, C.; Sim, S.-C.; Michel, A.; Causse, M.; Gardener, B.B.M.; Francis, D.; van der Knaap, E. Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol. 2011, 156, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Xiao, H.; Cabrera, A.; Meulia, T.; van der Knaap, E. SUN regulates vegetative and reproductive organ shape by changing cell division patterns. Plant Physiol. 2011, 157, 1175–1186. [Google Scholar] [CrossRef]
- Chakrabarti, M.; Zhang, N.; Sauvage, C.; Muños, S.; Blanca, J.; Cañizares, J.; Diez, M.J.; Schneider, R.; Mazourek, M.; McClead, J.; et al. A cytochrome P450 regulates a domestication trait in cultivated tomato. Proc. Natl. Acad. Sci. USA 2013, 110, 17125–17130. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Van Eck, J.; Cong, B.; Tanksley, S.D. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. USA 2002, 99, 13302–13306. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; van der Knaap, E. Tomato fruit weight 11.3 maps close to fasciated on the bottom of chromosome 11. Theor. Appl. Genet. 2011, 123, 465–474. [Google Scholar] [CrossRef]
- Chaim, A.; Borovsky, Y.; Rao, G.; Gur, A.; Zamir, D.; Paran, I. Comparative QTL mapping of fruit size and shape in tomato and pepper. Isr. J. Plant Sci. 2006, 54, 191–203. [Google Scholar] [CrossRef]
- Tsaballa, A.; Pasentsis, K.; Darzentas, N.; Tsaftaris, A.S. Multiple evidence for the role of an Ovate-like gene in determining fruit shape in pepper. BMC Plant Biol. 2011, 11, 46. [Google Scholar] [CrossRef] [PubMed]
- Chunthawodtiporn, J.; Hill, T.; Stoffel, K.; Van Deynze, A. Quantitative Trait Loci Controlling Fruit Size and Other Horticultural Traits in Bell Pepper (Capsicum annuum). Plant Genome 2018, 11, 160125. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, X.; Li, Q.; Xu, N.; He, C. A lineage-specific arginine in POS1 is required for fruit size control in Physaleae (Solanaceae) via gene co-option. Plant J. 2022, 111, 183–204. [Google Scholar] [CrossRef] [PubMed]
- Davidson, N.M.; Oshlack, A. Corset: Enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 2014, 15, 410. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.C.; Nadeau, K.; Abbasi, M.; Lachance, C.; Nguyen, M.; Fenrich, J. The ultimate qPCR experiment: Producing publication quality, reproducible data the first time. Trends Biotechnol. 2019, 37, 761–774. [Google Scholar] [CrossRef] [PubMed]
- Mestdagh, P.; Van Vlierberghe, P.; De Weer, A.; Muth, D.; Westermann, F.; Speleman, F.; Vandesompele, J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009, 10, R64. [Google Scholar] [CrossRef] [PubMed]
- Singh, A. SPSS—An Overview; Institute of Agricultural Sciences: Zürich. Switzerland. 2015.
- Ariel, F.D.; Manavella, P.A.; Dezar, C.A.; Chan, R.L. The true story of the HD-Zip family. Trends Plant Sci. 2007, 12, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Dell, B.; Huang, L. Physiological response of plants to low boron. Plant Soil. 1997, 193, 103–120. [Google Scholar] [CrossRef]
- Da Silva, M.P.S.; Mendonça Freitas, M.S.; Cesar Santos, P.; De Carvalho, A.J.C.; Jorge, T.S. Capsicum annuum var. annuum under macronutrients and boron deficiencies: Leaf content and visual symptoms. J. Plant Nutr. 2019, 42, 417–427. [Google Scholar] [CrossRef]
- Sarafi, E.; Tsouvaltzis, P.; Chatzissavvidis, C.; Siomos, A.; Therios, I. Melatonin and resveratrol reverse the toxic effect of high boron (B) and modulate biochemical parameters in pepper plants (Capsicum annuum L.). Plant Physiol. Biochem. 2017, 112, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Miwa, K.; Fujiwara, T. Boron transport in plants: Co-ordinated regulation of transporters. Ann. Bot. 2010, 105, 1103–1108. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Wallace, I.S.; Takano, J.; Roberts, D.M.; Fujiwara, T. NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. Plant Cell 2008, 20, 2860–2875. [Google Scholar] [CrossRef] [PubMed]
- Miwa, K.; Takano, J.; Fujiwara, T. Improvement of seed yields under boron-limiting conditions through overexpression of BOR1, a boron transporter for xylem loading, in Arabidopsis thaliana. Plant J. 2006, 46, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Miwa, K.; Wakuta, S.; Takada, S.; Ide, K.; Takano, J.; Naito, S.; Omori, H.; Matsunaga, T.; Fujiwara, T. Roles of BOR2, a boron exporter, in cross linking of rhamnogalacturonan II and root elongation under boron limitation in Arabidopsis. Plant Physiol. 2013, 163, 1699–1709. [Google Scholar] [CrossRef]
- Wang, H.; Tang, S.; Zhi, H.; Xing, L.; Zhang, H.; Tang, C.; Wang, E.; Zhao, M.; Jia, G.; Feng, B.; et al. The boron transporter SiBOR1 functions in cell wall integrity, cellular homeostasis, and panicle development in foxtail millet. Crop J. 2022, 10, 342–353. [Google Scholar] [CrossRef]
- Mauxion, J.-P.; Chevalier, C.; Gonzalez, N. Complex cellular and molecular events determining fruit size. Trends Plant Sci. 2021, 26, 1023–1038. [Google Scholar] [CrossRef]
- Sarikas, A.; Hartmann, T.; Pan, Z.-Q. The Cullin Protein Family. Genome Biol. 2011, 12, 220. [Google Scholar] [CrossRef]
- Zhou, Y.-H.; Xia, J.; Xu, W.-H.; Zhu, X.; Wu, X.-H.; Hua, D.; Xing, C. Cullin-1 promotes cell proliferation in human breast cancer and is related to diabetes. Int. J. Biol. Markers 2016, 31, e375–e381. [Google Scholar] [CrossRef]
- Citterio, S.; Sgorbati, S.; Scippa, S.; Sparvoli, E. Ascorbic acid effect on the onset of cell proliferation in pea root. Physiol. Plant 1994, 92, 601–607. [Google Scholar] [CrossRef]
- Chatzopoulou, F.; Sanmartin, M.; Mellidou, I.; Pateraki, I.; Koukounaras, A.; Tanou, G.; Kalamaki, M.S.; Veljović-Jovanović, S.; Antić, T.C.; Kostas, S.; et al. Silencing of ascorbate oxidase results in reduced growth, altered ascorbic acid levels and ripening pattern in melon fruit. Plant Physiol. Biochem. 2020, 156, 291–303. [Google Scholar] [CrossRef]
- Feng, L.; Li, G.; He, Z.; Han, W.; Sun, J.; Huang, F.; Di, J.; Chen, Y. The ARF, GH3, and Aux/IAA gene families in castor bean (Ricinus communis L.): Genome-wide identification and expression profiles in high-stalk and dwarf strains. Ind. Crops Prod. 2019, 141, 111804. [Google Scholar] [CrossRef]
- Leyser, O. Auxin Signaling. Plant Physiol. 2018, 176, 465–479. [Google Scholar] [CrossRef]
- Devos, S.; Laukens, K.; Deckers, P.; Van Der Straeten, D.; Beeckman, T.; Inzé, D.; Van Onckelen, H.; Witters, E.; Prinsen, E. A Hormone and Proteome Approach to Picturing the Initial Metabolic Events During Plasmodiophora brassicae Infection on Arabidopsis. MPMI 2006, 19, 1431–1443. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. Auxin Biosynthesis: A Simple Two-Step Pathway Converts Tryptophan to Indole-3-Acetic Acid in Plants. Mol. Plant 2012, 5, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Calderon-Villalobos, L.I.A.; Sharon, M.; Zheng, C.; Robinson, C.V.; Estelle, M.; Zheng, N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 2007, 446, 640–645. [Google Scholar] [CrossRef]
- Kepinski, S.; Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Marchant, A.; Kargul, J.; May, S.T.; Muller, P.; Delbarre, A.; Perrot-Rechenmann, C.; Bennett, M.J. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 1999, 18, 2066–2073. [Google Scholar] [CrossRef] [PubMed]
- Fendrych, M.; Akhmanova, M.; Merrin, J.; Glanc, M.; Hagihara, S.; Takahashi, K.; Uchida, N.; Torii, K.U.; Friml, J. Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nat. Plants 2018, 4, 453–459. [Google Scholar] [CrossRef]
- Spartz, A.K.; Lee, S.H.; Wenger, J.P.; Gonzalez, N.; Itoh, H.; Inzé, D.; Peer, W.A.; Murphy, A.S.; Overvoorde, P.J.; Gray, W.M. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012, 70, 978–990. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Hu, Z.; Guo, X.; Tian, S.; Chen, G. Genome-Wide Analysis of the MADS-Box Transcription Factor Family in Solanum lycopersicum. Int. J. Mol. Sci. 2019, 20, 2961. [Google Scholar] [CrossRef]
- Theissen, G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001, 4, 75–85. [Google Scholar] [CrossRef]
- Khew, C.Y.; Harikrishna, J.A.; Wee, W.Y.; Lau, E.T.; Hwang, S.S. Transcriptional Sequencing and Gene Expression Analysis of Various Genes in Fruit Development of Three Different Black Pepper (Piper Nigrum L.) Varieties. Int. J. Genom. 2020, 2020, 1540915. [Google Scholar] [CrossRef]
- Ribone, P.A.; Capella, M.; Chan, R.L. Functional Characterization of the Homeodomain Leucine Zipper I Transcription Factor AtHB13 Reveals a Crucial Role in Arabidopsis Development. J. Exp. Bot. 2015, 66, 5929–5943. [Google Scholar] [CrossRef] [PubMed]
- Mora, C.C.; Perotti, M.F.; González-Grandío, E.; Ribone, P.A.; Cubas, P.; Chan, R.L. AtHB40 Modulates Primary Root Length and Gravitropism Involving CYCLINB and Auxin Transporters. Plant Sci. 2022, 324, 111421. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Deng, W.; Hu, G.; Hu, N.; Li, Z. Transcriptome Profiling Reveals the Regulatory Mechanism Underlying Pollination Dependent and Parthenocarpic Fruit Set Mainly Mediated by Auxin and Gibberellin. PLoS ONE 2015, 10, e0125355. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Henriksson, E.; Söderman, E.; Henriksson, K.N.; Sundberg, E.; Engström, P. The Arabidopsis Homeobox Gene, ATHB16, Regulates Leaf Development and the Sensitivity to Photoperiod in Arabidopsis. Dev. Biol. 2003, 264, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Richter, R.; Rossmann, S.; Gabriel, D.; Töpfer, R.; Theres, K.; Zyprian, E. Differential expression of transcription factor- and further growth-related genes correlates with contrasting cluster architecture in Vitis vinifera “Pinot Noir” and Vitis spp. genotypes. Theor. Appl. Genet. 2020, 133, 3249–3272. [Google Scholar] [CrossRef]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Ma, Q.; Mao, L.; Wu, Z.; Liu, Z.; Zou, X.; Yang, B. Comparative Transcriptome Analysis Identified Genes Associated with Fruit Size in Pepper (Capsicum annuum L.). Horticulturae 2023, 9, 1009. https://doi.org/10.3390/horticulturae9091009
Zheng Y, Ma Q, Mao L, Wu Z, Liu Z, Zou X, Yang B. Comparative Transcriptome Analysis Identified Genes Associated with Fruit Size in Pepper (Capsicum annuum L.). Horticulturae. 2023; 9(9):1009. https://doi.org/10.3390/horticulturae9091009
Chicago/Turabian StyleZheng, Yuxin, Qilong Ma, Lianzhen Mao, Zhuoxuan Wu, Zhoubin Liu, Xuexiao Zou, and Bozhi Yang. 2023. "Comparative Transcriptome Analysis Identified Genes Associated with Fruit Size in Pepper (Capsicum annuum L.)" Horticulturae 9, no. 9: 1009. https://doi.org/10.3390/horticulturae9091009
APA StyleZheng, Y., Ma, Q., Mao, L., Wu, Z., Liu, Z., Zou, X., & Yang, B. (2023). Comparative Transcriptome Analysis Identified Genes Associated with Fruit Size in Pepper (Capsicum annuum L.). Horticulturae, 9(9), 1009. https://doi.org/10.3390/horticulturae9091009





