Development, Identification and Validation of a Novel SSR Molecular Marker for Heat Resistance of Grapes Based on miRNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Heat Treatments
2.2. DNA, RNA Extraction and cDNA Synthesis
2.3. Mining of SSRs from Pri-miRNAs and Pre-miRNAs of the Grapevine Genome
2.4. RT-qPCR, PCR, and PCR-Sanger Sequencing Analysis
2.5. About the Naming of miRNA-SSR Molecular Marker Sub
2.6. Prediction of Target Genes for miRNAs and Pathway Analyses
2.7. Statistical Analysis
3. Results
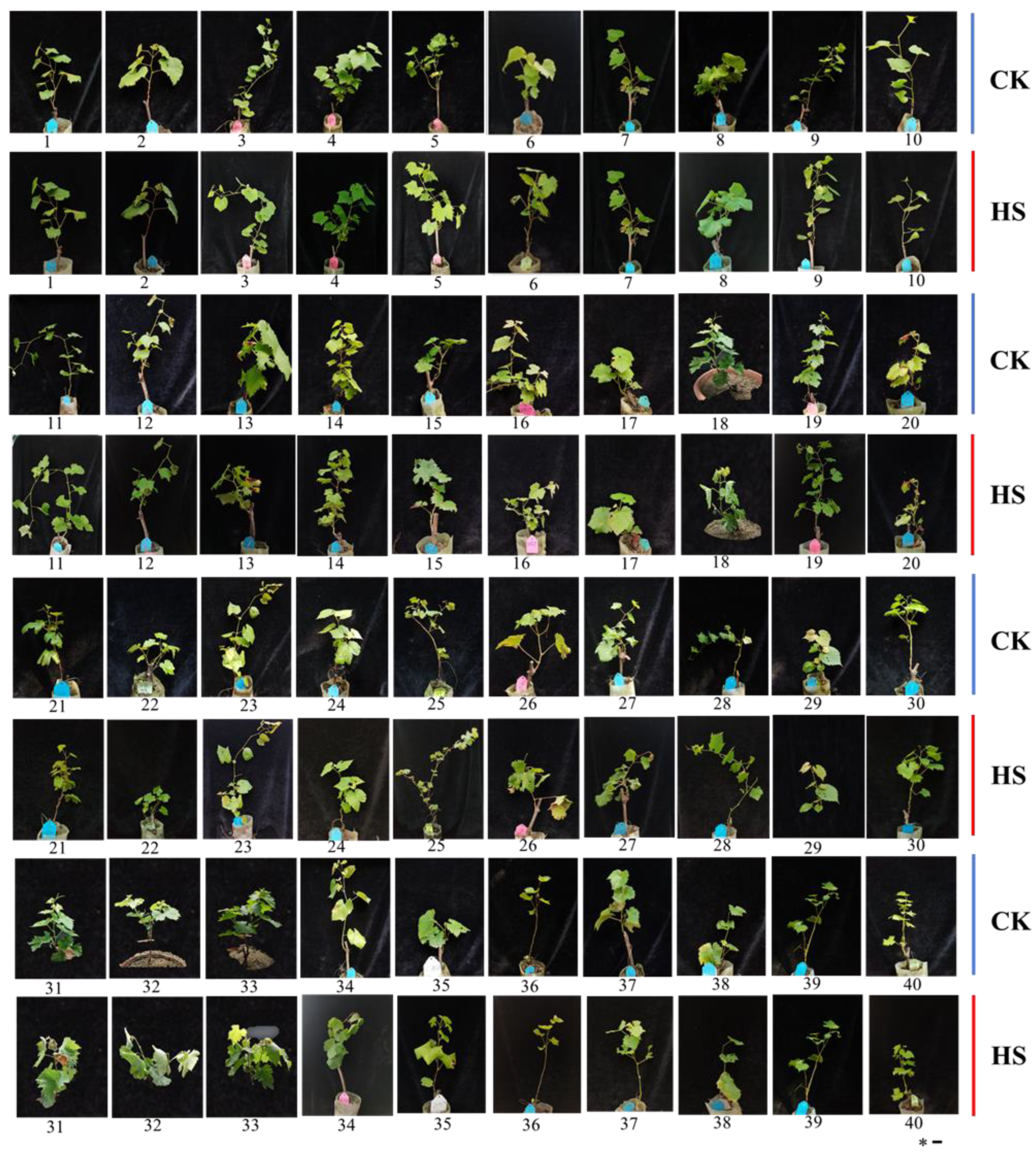
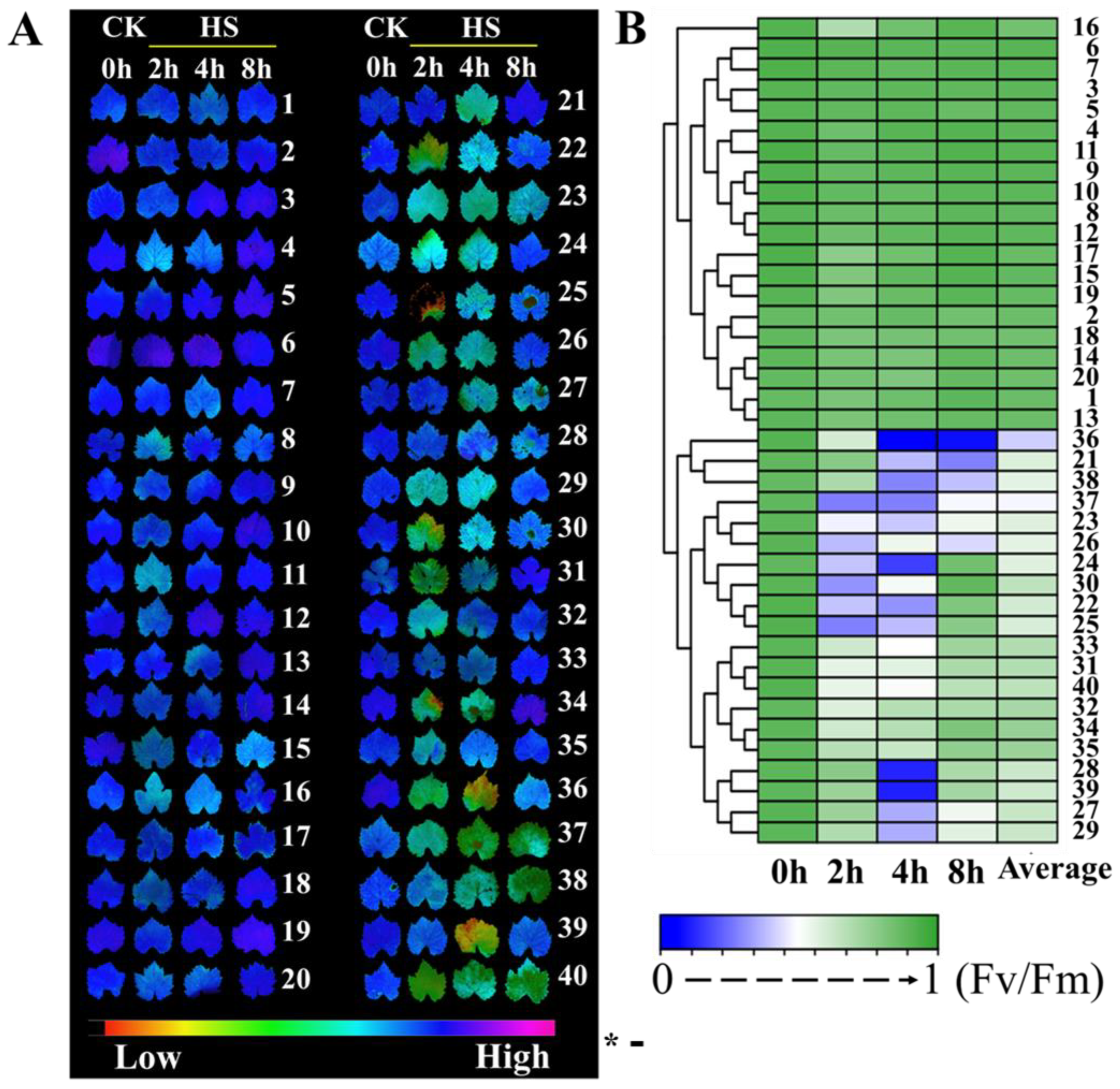
3.1. Heat Injury in 40 Grape Varieties’ Leaves Exposed to High-Temperature Stress
3.2. RT-qPCR Analysis of the Pri-miRNA Response to Heat Stress
3.3. MiRNA and Target Genes Mediated Regulatory Networks
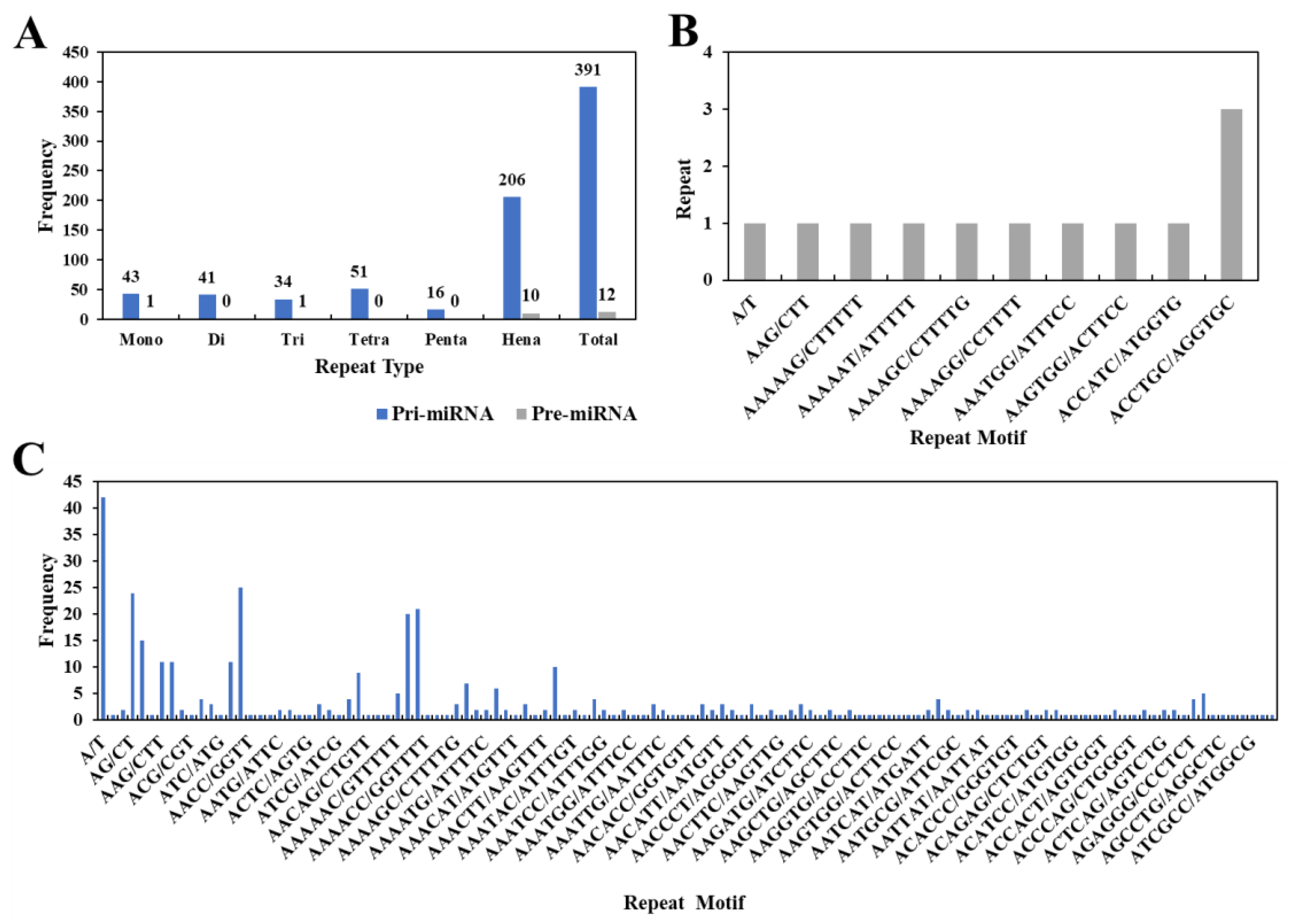
3.4. Identification and Frequency Distribution of miRNA-SSRs in the Grape Genome
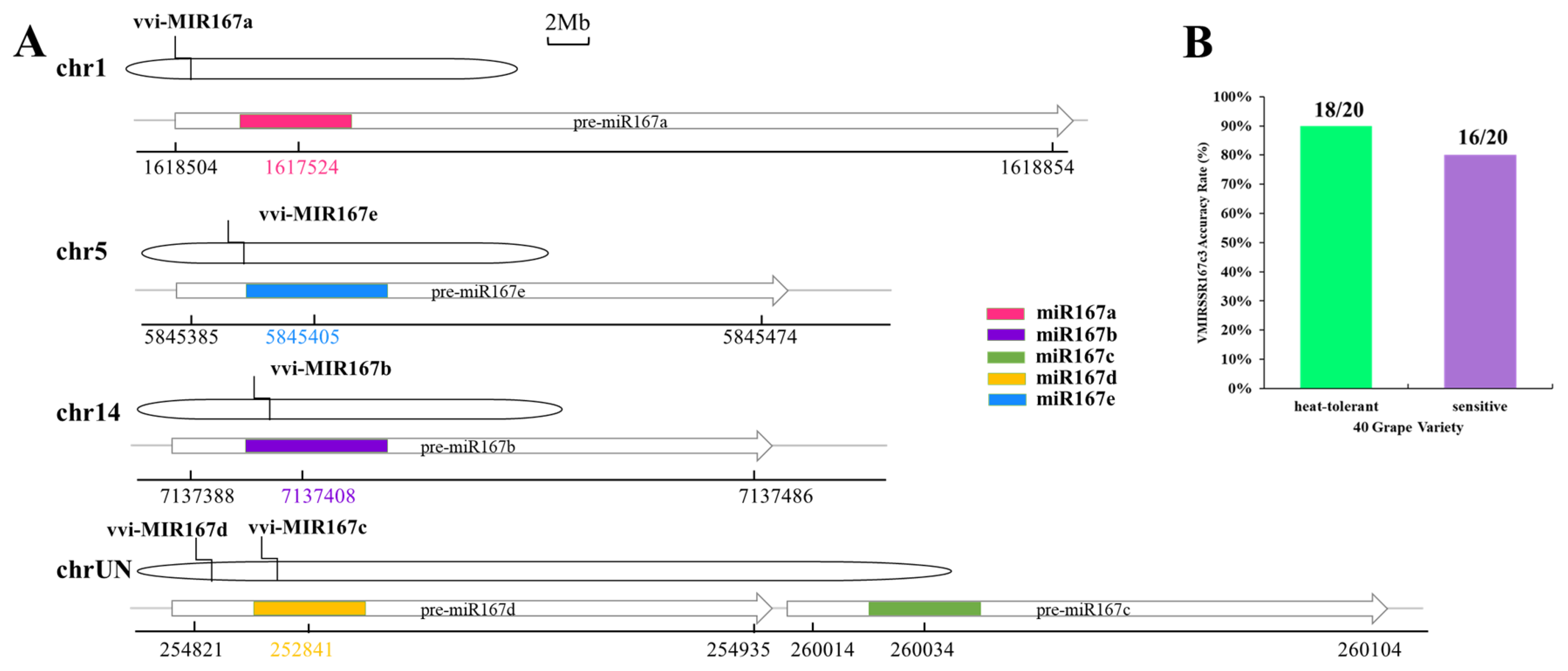
3.5. Distribution of Pri-miRNA-SSR in Grape Chromosomes
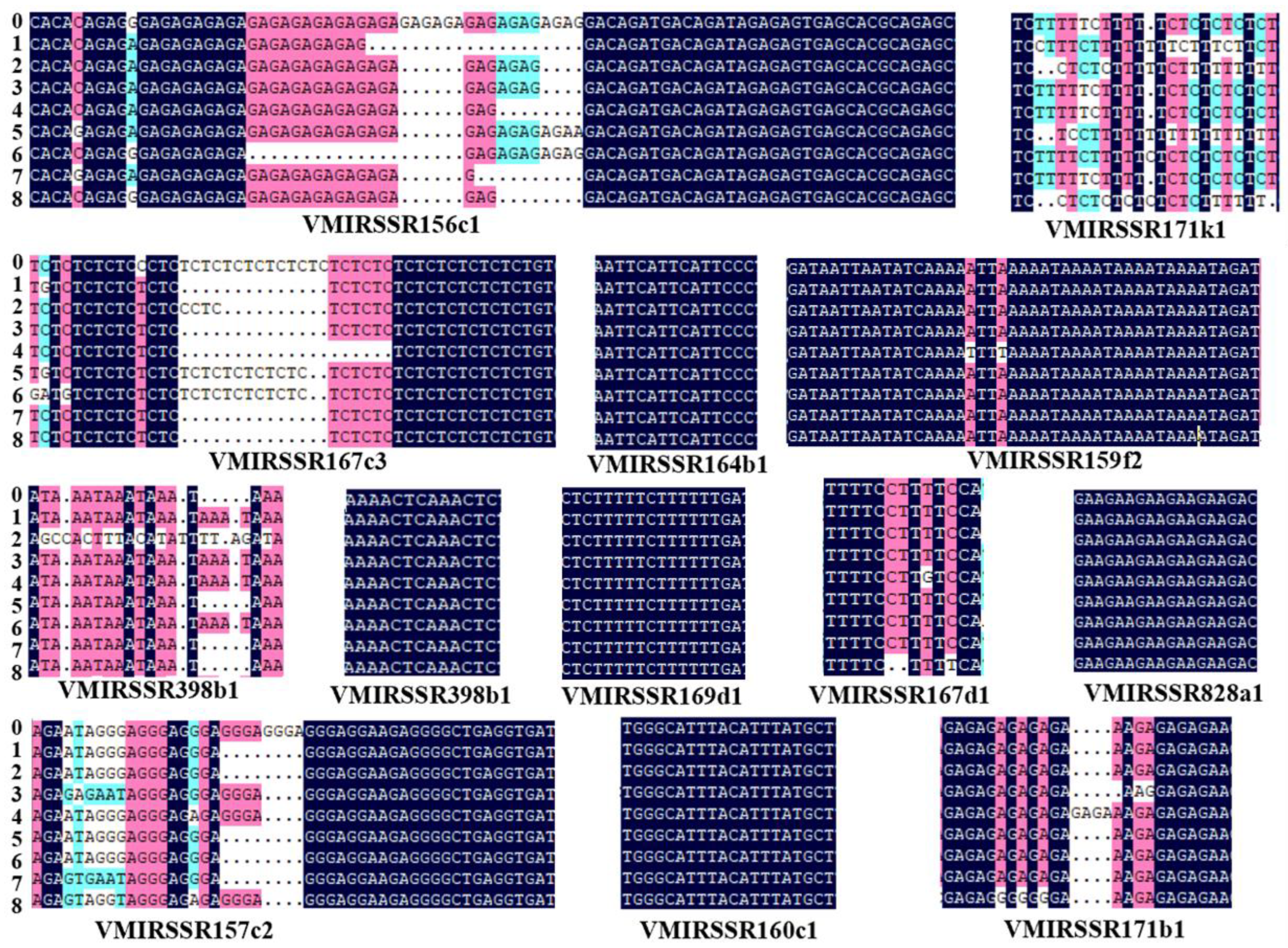
3.6. The Diversity of 13 MIRNA-SSR Loci Was Verified in 8 Grape Germplasm Samples
3.7. Application of VMIRSSRNA Markers in Heat-Resistant and Heat-Sensitive Grape Varieties
3.8. Chromosome Distribution of vvi-miR167 Family Members in Grape
3.9. Phylogenetic Development of the vvi-MIR167s Gene Family of Representative Species
4. Discussion
4.1. Functional Analysis for miRNAs and Their Regulatory Networks
4.2. Development and Application of miRNA-SSR Markers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chen, Q.J.; Zhang, L.P.; Song, S.R.; Wang, L.; Xu, W.P.; Zhang, C.X.; Wang, S.P.; Liu, H.F.; Ma, C. vvi-miPEP172b and vvi-miPEP3635b increase cold tolerance of grapevine by regulating the corresponding MIRNA genes. Plant Sci. 2022, 325, 111450. [Google Scholar] [CrossRef]
- Zha, Q.; Xi, X.; He, Y.; Yin, X.; Jiang, A. Interaction of VvbZIP60s and VvHSP83 in response to high-temperature stress in grapes. Gene 2022, 810, 146053. [Google Scholar] [CrossRef] [PubMed]
- Zha, Q.; Xi, X.; Jiang, A.; Tian, Y. High Temperature Affects Photosynthetic and Molecular Processes in Field-Cultivated Vitis vinifera L. × Vitis labrusca L. Photochem. Photobiol. 2016, 92, 446–454. [Google Scholar] [CrossRef]
- Zha, Q.; Xi, X.; Jiang, A.; Wang, S.; Tian, Y. Changes in the protective mechanism of photosystem II and molecular regulation in response to high temperature stress in grapevines. Plant Physiol. Biochem. 2016, 101, 43–53. [Google Scholar] [CrossRef]
- Jiang, J.; Liu, X.; Liu, C.; Liu, G.; Li, S.; Wang, L. Integrating Omics and Alternative Splicing Reveals Insights into Grape Response to High Temperature. Plant Physiol. 2017, 173, 1502–1518. [Google Scholar] [CrossRef]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Bokszczanin, K. Perspectives on deciphering mechanisms underlying plant heat stress response and thermotolerance. Front. Plant Sci. 2013, 4, 315. [Google Scholar] [CrossRef]
- Zhang, L.; Fan, D.; Li, H.; Chen, Q.; Zhang, Z.; Liu, M.; Liu, J.; Song, Y.; He, J.; Xu, W.; et al. Characterization and identification of grapevine heat stress-responsive microRNAs revealed the positive regulated function of vvi-miR167 in thermostability. Plant Sci. 2023, 329, 111623. [Google Scholar] [CrossRef] [PubMed]
- Tayade, R.; Nguyen, T.; Oh, S.A.; Hwang, Y.S.; Yoon, I.S.; Deshmuk, R.; Jung, K.-H.; Park, S.K. Effective strategies for enhancing tolerance to high-temperature stress in rice during the reproductive and ripening stages. Plant Breed. Biotechnol. 2018, 6, 1–18. [Google Scholar] [CrossRef]
- Gosal, S.S.; Wani, S.H. Accelerated Plant Breeding; Springer: Berlin/Heidelberg, Germany, 2020; Volume 1. [Google Scholar]
- Russo, G.; Giordano, A. miRNAs: From biogenesis to networks. Protein. Netw. Pathw. Anal. 2009, 563, 303–352. [Google Scholar]
- Li, H.; Gao, Z.; Zahid, M.S.; Li, D.; Javed, H.U.; Wang, L.; Song, S.; Zhao, L.; Xu, W.; Zhang, C.; et al. Small RNA Sequencing Analysis of miRNA Expression Reveals Novel Insihts into Root Formation under Root Restriction Cultivation in Grapevine (Vitis vinifera L.). Int. J. Mol. Sci. 2020, 21, 3513. [Google Scholar] [CrossRef]
- Juarez, M.T.; Kui, J.S.; Thomas, J.; Heller, B.A.; Timmermans, M.C. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 2004, 428, 84–88. [Google Scholar] [CrossRef]
- Liu, W.C.; Song, R.F.; Zheng, S.Q.; Li, T.T.; Zhang, B.L.; Gao, X.; Lu, Y.T. Coordination of plant growth and abiotic stress responses by tryptophan synthase beta subunit 1 through modulation of tryptophan and ABA homeostasis in Arabidopsis. Mol. Plant 2022, 15, 973–990. [Google Scholar] [CrossRef]
- Schwab, R.; Palatnik, J.F.; Riester, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell 2005, 8, 517–527. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, Q.; Liu, J.; Dou, F.; Wang, H.; Song, Y.; Ren, Y.; He, J.; Wang, L.; Zhang, C.; et al. Identification of grape miRNA revealed Vvi-miR164b involved in auxin induced root development. Sci. Hortic. 2022, 295, 110804. [Google Scholar] [CrossRef]
- Gandikota, M.; Birkenbihl, R.P.; Höhmann, S.; Cardon, G.H.; Saedler, H.; Huijser, P. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007, 49, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhao, L.; Malik, S.; Gentile, B.R.; Xiong, V.; Arazi, T.; Owen, H.A.; Friml, J.; Zhao, D. Specification of female germline by microRNA orchestrated auxin signaling in Arabidopsis. Nat. Commun. 2022, 13, 6960. [Google Scholar] [CrossRef] [PubMed]
- Palatnik, J.F.; Wollmann, H.; Schommer, C.; Schwab, R.; Boisbouvier, J.; Rodriguez, R.; Warthmann, N.; Allen, E.; Dezulian, T.; Huson, D.; et al. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev. Cell 2007, 13, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Xia, R.; Zhu, H.; An, Y.-q.; Beers, E.P.; Liu, Z. Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 2012, 13, R47. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Lu, X.; Zeng, H.; Zhang, Y.; Zhu, J. Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 2013, 74, 840–851. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Yang, J.; He, Y. Natural antisense transcripts of MIR398 genes suppress microR398 processing and attenuate plant thermotolerance. Nat. Commun. 2020, 11, 5351. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Zhao, G.; Zhang, S.; Li, Y.; Gu, H.; Li, Y.; Zhao, Q.; Qi, Y. Chloroplast-to-Nucleus Signaling Regulates MicroRNA Biogenesis in Arabidopsis. Dev. Cell 2019, 48, 371–382.e4. [Google Scholar] [CrossRef] [PubMed]
- Stief, A.; Altmann, S.; Hoffmann, K.; Pant, B.D.; Scheible, W.-R.; Bäurle, I. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 2014, 26, 1792–1807. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.S.; Kuo, C.C.; Yang, I.C.; Tsai, W.A.; Shen, Y.H.; Lin, C.C.; Liang, Y.C.; Li, Y.C.; Kuo, Y.W.; King, Y.C.; et al. MicroRNA160 Modulates Plant Development and Heat Shock Protein Gene Expression to Mediate Heat Tolerance in Arabidopsis. Front. Plant Sci. 2018, 9, 68. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Burd, S.; Lers, A. miR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 2015, 84, 169–187. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Chen, Q.; Wang, S.; Lers, A. Downregulation of GeBP-like alpha factor by MiR827 suggests their involvement in senescence and phosphate homeostasis. BMC Biol. 2021, 19, 90. [Google Scholar] [CrossRef]
- Pei, M.; Liu, H.; Wei, T.; Jin, H.; Yu, Y.; Ma, M.; Song, X.; Dai, R.; Guo, D. Identification, characterization, and verification of miR399 target gene in grape. Hortic. Plant J. 2023. [Google Scholar] [CrossRef]
- Joy, N.; Asha, S.; Mallika, V.; Soniya, E.V. De novo transcriptome sequencing reveals a considerable bias in the incidence of simple sequence repeats towards the downstream of ‘pre-miRNAs’ of black pepper. PLoS ONE 2013, 8, e56694. [Google Scholar] [CrossRef]
- Mondal, T.K.; Ganie, S.A. Identification and characterization of salt responsive miRNA-SSR markers in rice (Oryza sativa). Gene 2014, 535, 204–209. [Google Scholar] [CrossRef]
- Ganie, S.A.; Mondal, T.K. Genome-wide development of novel miRNA-based microsatellite markers of rice (Oryza sativa) for genotyping applications. Mol. Breed. 2015, 35, 51. [Google Scholar] [CrossRef]
- Ferrão, L.F.V.; Caixeta, E.T.; Pena, G.; Zambolim, E.M.; Cruz, C.D.; Zambolim, L.; Ferrao, M.A.G.; Sakiyama, N.S. New EST–SSR markers of Coffea arabica: Transferability and application to studies of molecular characterization and genetic mapping. Mol. Breed. 2015, 35, 31. [Google Scholar] [CrossRef][Green Version]
- Joy, N.; Maimoonath Beevi, Y.; Soniya, E. A deeper view into the significance of simple sequence repeats in pre-miRNAs provides clues for its possible roles in determining the function of microRNAs. BMC Genet. 2018, 19, 29. [Google Scholar] [CrossRef] [PubMed]
- de Lima, J.C.; Loss-Morais, G.; Margis, R. MicroRNAs play critical roles during plant development and in response to abiotic stresses. Genet. Mol. Biol. 2012, 35, 1069–1077. [Google Scholar] [CrossRef] [PubMed]
- Patil, P.G.; Singh, N.; Parashuram, S.; Bohra, A.; Mundewadikar, D.M.; Sangnure, V.R.; Babu, K.D.; Sharma, J. Genome wide identification, characterization and validation of novel miRNA-based SSR markers in pomegranate (Punica granatum L.). Physiol. Mol. Biol. Plants 2020, 26, 683–696. [Google Scholar] [CrossRef]
- Kumar, A.; Chauhan, A.; Sharma, M.; Kompelli, S.K.; Gahlaut, V.; Ijaq, J.; Singh, K.P.; Prasad Gajula, M.; Suravajhala, P.; Singh Balyan, H. Genome-wide mining, characterization and development of miRNA-SSRs in Arabidopsis thaliana. BioRxiv 2017, 203851. [Google Scholar] [CrossRef]
- Ravishankar, K.; Anand, L.; Dinesh, M. Assessment of genetic relatedness among mango cultivars of India using RAPD markers. J. Hortic. Sci. Biotechnol. 2000, 75, 198–201. [Google Scholar] [CrossRef]
- Li, M.; Li, X.; Han, Z.H.; Shu, H.; Li, T.Z. Molecular analysis of two Chinese pear (Pyrus bretschneideri Rehd.) spontaneous self-compatible mutants, Yan Zhuang and Jin Zhui. Plant Biol. 2009, 11, 774–783. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ma, C.; Lu, Y.; Bai, S.; Zhang, W.; Duan, X.; Meng, D.; Wang, Z.; Wang, A.; Zhou, Z.; Li, T. Cloning and characterization of miRNAs and their targets, including a novel miRNA-targeted NBS-LRR protein class gene in apple (Golden delicious). Mol. Plant 2014, 7, 218–230. [Google Scholar] [CrossRef]
- Murchie, E.H.; Lawson, T. Chlorophyll fluorescence analysis: A guide to good practice and understanding some new applications. J. Exp. Bot. 2013, 64, 3983–3998. [Google Scholar] [CrossRef]
- Ashraf, M.; Harris, P.J.C. Photosynthesis under stressful environments: An overview. Photosynthetica 2013, 51, 163–190. [Google Scholar] [CrossRef]
- Xu, H.; Liu, G.; Liu, G.; Yan, B.; Duan, W.; Wang, L.; Li, S. Comparison of investigation methods of heat injury in grapevine (Vitis) and assessment to heat tolerance in different cultivars and species. BMC Plant Biol. 2014, 14, 156. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Chen, Q.; Deng, B.; Gao, J.; Zhao, Z.; Chen, Z.; Song, S.; Wang, L.; Zhao, L.; Xu, W.; Zhang, C.; et al. Comparative Analysis of miRNA Abundance Revealed the Function of Vvi-miR828 in Fruit Coloring in Root Restriction Cultivation Grapevine (Vitis vinifera L.). Int. J. Mol. Sci. 2019, 20, 4058. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.J.; Deng, B.H.; Gao, J.; Zhao, Z.Y.; Chen, Z.L.; Song, S.R.; Wang, L.; Zhao, L.P.; Xu, W.P.; Zhang, C.X.; et al. A miRNA-Encoded Small Peptide, vvi-miPEP171d1, Regulates Adventitious Root Formation. Plant Physiol. 2020, 183, 656–670. [Google Scholar] [CrossRef]
- Liu, X.; Liu, S.; Chen, X.; Prasanna, B.M.; Ni, Z.; Li, X.; He, Y.; Fan, Z.; Zhou, T. Maize miR167-ARF3/30-polyamine oxidase 1 module-regulated H2O2 production confers resistance to maize chlorotic mottle virus. Plant Physiol. 2022, 189, 1065–1082. [Google Scholar] [CrossRef]
- Li, J.; Cao, Y.; Zhang, J.; Zhu, C.; Tang, G.; Yan, J. The miR165/166-PHABULOSA module promotes thermotolerance by transcriptionally and post-translationally regulating HSFA1. Plant Cell 2023, 35, 2952–2971. [Google Scholar] [CrossRef]
- Song, X.; Yang, Q.; Bai, Y.; Gong, K.; Wu, T.; Yu, T.; Pei, Q.; Duan, W.; Huang, Z.; Wang, Z.; et al. Comprehensive analysis of SSRs and database construction using all complete gene-coding sequences in major horticultural and representative plants. Hortic. Res. 2021, 8, 122. [Google Scholar] [CrossRef]
- Bowers, J.E.; Dangl, G.S.; Meredith, C.P. Development and characterization of additional microsatellite DNA markers for grape. Am. J. Enol. Vitic. 1999, 50, 243–246. [Google Scholar] [CrossRef]
- Sefc, K.M.; Regner, F.; Turetschek, E.; Glössl, J.; Steinkellner, H. Identification of microsatellite sequences in Vitis riparia and their applicability for genotyping of different Vitis species. Genome 1999, 42, 367–373. [Google Scholar] [CrossRef]
- Thomas, M.; Scott, N. Microsatellite repeats in grapevine reveal DNA polymorphisms when analysed as sequence-tagged sites (STSs). Theor. Appl. Genet. 1993, 86, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Stavrakaki, M.; Biniari, K. Study of the genetic variability of grapevine cultivar Liatiko (Vitis vinifera L.) using the ampelographic description and molecular method SSR. Vegetos 2017, 30, 73–79. [Google Scholar] [CrossRef]
- Dunlevy, J.; Blackmore, D.; Watkins, J.; Edwards, E.; Walker, R.; Walker, A. Short Sequence Repeat (SSR) Genotyping and Sodium Exclusion Phenotyping of a Vitis Hybrid Population (‘K51-40’בSchwarzmann’). In Proceedings of the XII International Conference on Grapevine Breeding and Genetics 1248, Bordeaux, France, 15–20 July 2018; pp. 513–520. [Google Scholar] [CrossRef]
- Zhong, H.; Zhang, F.; Zhou, X.; Pan, M.; Xu, J.; Hao, J.; Han, S.; Mei, C.; Xian, H.; Wang, M. Genome-wide identification of sequence variations and SSR marker development in the Munake Grape cultivar. Front. Ecol. Evol. 2021, 190. [Google Scholar] [CrossRef]
- Sun, X.; Du, Z.; Ren, J.; Amombo, E.; Hu, T.; Fu, J. Association of SSR markers with functional traits from heat stress in diverse tall fescue accessions. BMC Plant Biol. 2015, 15, 116. [Google Scholar] [CrossRef] [PubMed]
- Ramya, K.; Jain, N.; Gandhi, N.; Arora, A.; Singh, P.; Singh, A.M.; Singh, G.P.; Prabhu, K. Assessing heat stress tolerance and genetic diversity among exotic and Indian wheat genotypes using simple sequence repeats and agro-physiological traits. Plant Genet. Resour. 2017, 15, 208–220. [Google Scholar] [CrossRef]
- Jha, U.C.; Jha, R.; Bohra, A.; Parida, S.K.; Kole, P.C.; Thakro, V.; Singh, D.; Singh, N.P. Population structure and association analysis of heat stress relevant traits in chickpea (Cicer arietinum L.). 3 Biotech 2018, 8, 43. [Google Scholar] [CrossRef]


| Varieties | VMIRSSRNA-Marker | ||||
|---|---|---|---|---|---|
| NO. | Thermostability | VMIRSSR167c3 (CT)n | VMIRSSR167d1 (TTTTCC)n | VMIRSSR157c2 (AGGG)n | VMIRSSR398b1 (ATAA)n |
| 1 | Niagara Rosada | (CT)20 | (TTTTCC)2 | (AGGG)4 | (ATAA)5 |
| 2 | Benizuiho | (CT)19 | (TTTTCC)2 | (AGGG)3, 1(AGAG) | (ATAA)5 |
| 3 | Vitis × Champion | (CT)18, 1(CC) | (TTTTCC)2 | (AGGG)4 | (ATAA)5 |
| 4 | Cuihong | (CT)17, 1(CC) | (TTTTCC)2 | (AGGG)4, 1(AGAG) | (ATAA)5 |
| 5 | Seibel Blanc | (CT)19 | (TTTTCC)2 | (AGGG)4 | 0 |
| 6 | Lot Sand | (CT)20, 1(CC) | (TTTTCC)2 | (AGGG)4 | (ATAA)5 |
| 7 | Damubo | (CT)23 | (TTTTCC)2 | (AGGG)4 | (ATAA)5 |
| 8 | Aromatic Rachachi | (CT)24 | (TTTTCC)2 | (AGGG)3, 1(AGAG) | (ATAA)5 |
| 9 | Pollux | (CT)22, 1(CC) | (TTTTCC)2 | (AGGG)2 | (ATAA)5 |
| 10 | Anisky | (CT)18, 1(CC) | (TTTTCC)2 | (AGGG)4 | (ATAA)5 |
| 11 | Green beads | (CT)19 | (TTTTCC)2 | (AGGG)4 | (ATAA)5 |
| 12 | Volgarton | (CT)16 | (TTTTCC)2 | (AGGG)3,1(AGAG) | (ATAA)5 |
| 13 | Augusta | (CT)17, 1(CC) | (TTTTCC)2 | (AGGG)4 | (ATAA)5 |
| 14 | Jifeng | (CT)18, 1(CC) | (TTTTCC)2 | (AGGG)3, 1(AGAG) | (ATAA)5 |
| 15 | Grand Noir | (CT)23, 1(CC) | (TTTTCC)2 | (AGGG)3, 1(AGAG) | 0 |
| 16 | Shenmei | (CT)18, 1(CC) | (TTTTCC)2 | (AGGG)4 | 0 |
| 17 | Jiangsu v. azuLene | (CT)24, 1(CC) | (TTTTCC)2 | (AGGG)4 | (ATAA)5 |
| 18 | Shenyue | (CT)18, 1(CC) | (TTTTCC)2 | (AGGG)4 | (ATAA)5 |
| 19 | Cabernet Sauvignon | (CT)17 | (TTTTCC)2 | (AGGG)3, 1(GAAT) | (ATAA)4 |
| 20 | Shenhua | (CT)18, 1(CC) | TTTTCCTTGTCC | (AGGG)4, 1(AGAG) | (ATAA)5 |
| Varieties | VMIRSSRNA-Marker | ||||
|---|---|---|---|---|---|
| NO. | Heat-Sensitive | VMIRSSR167c3 (CT) | VMIRSSR167d1 (TTTTCC) | VMIRSSR157c2 (AGGG) | VMIRSSR398b1 (ATAA) |
| 21 | Ruiduxiangyu | (CT)24, 1(CC) | (TTTTCC)2 | (GA)3, 1(TA) | (ATAA)4 |
| 22 | Shenbao | (CT)19 | (TTTTCC)2 | (AGGG)4 | (ATAA)4 |
| 23 | Hakuho | (CT)16, 1(GT) | (TTTTCC)2 | (AGGG)4 | (ATAA)5 |
| 24 | Xinya | (CT)17 | (TTTTCC)2 | (AGGG)4 | (ATAA)5 |
| 25 | Beta | (CT)21 | (TTTTCC)2 | (AGGG)4 | (ATAA)4 |
| 26 | Beichun | (CT)13, 1(CC) | (TTTTCC)2 | (AGGG)4 | (ATAA)4 |
| 27 | Molixiang | (CT)16, 1(CC) | TTTTCCTTTTTC | (AGGG)4 | (ATAA)5 |
| 28 | Yan 74 | (CT)12, 1(CC) | (TTTTCC)2 | (AGGG)4 | (ATAA)5 |
| 29 | Μgni Blanc | (CT)19 | (TTTTCC)2 | (AGGG)4, 1(AGAG) | (ATAA)5 |
| 30 | Shine Muscat | (CT)14 | (TTTTCC)2 | (AGGG)4, 1(AGAG) | (ATAA)5 |
| 31 | Fujiminori | (CT)14 | (TTTTCC)2 | (AGGG)3, 1(AGAG) | (ATAA)5 |
| 32 | Italy grape | (CT)17 | (TTTTCC)2 | (AGGG)3, 1(AGAG) | (ATAA)5 |
| 33 | SO4 | (CT)14 | (TTTTCC)2 | (AGGG)4 | 3(ATAA), 1(ATTA) |
| 34 | Wuhecuibao | (CT)14 | (TTTTCC)2 | (AGGG)3, 1(AGAG) | (ATAA)5 |
| 35 | Jumeigui | (CT)14 | (TTTTCC)2 | (AGGG)4 | (ATAA)5 |
| 36 | Kyoho | (CT)17 | (TTTTCC)2 | (AGGG)4 | (ATAA)5 |
| 37 | MuscatHamburgh | (CT)17 | (TTTTCC)2 | (AGGG)3, 1(AGAG) | (ATAA)5 |
| 38 | Thompson Seedless | (CT)17 | (TTTTCC)2 | (AGGG)3, 1(AGAG) | (ATAA)4 |
| 39 | Shuanghong | (CT)15 | TTTTCCTTGTCC | (AGGG)4, 1(AGAG) | (ATAA)5 |
| 40 | 1103P | (CT)14 | (TTTTCC)2 | (AGGG)4 | 3(ATAA), 1(ATTA) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Song, Y.; Li, J.; Liu, J.; Zhang, Z.; Xu, Y.; Fan, D.; Liu, M.; Ren, Y.; Xi, X.; et al. Development, Identification and Validation of a Novel SSR Molecular Marker for Heat Resistance of Grapes Based on miRNA. Horticulturae 2023, 9, 931. https://doi.org/10.3390/horticulturae9080931
Zhang L, Song Y, Li J, Liu J, Zhang Z, Xu Y, Fan D, Liu M, Ren Y, Xi X, et al. Development, Identification and Validation of a Novel SSR Molecular Marker for Heat Resistance of Grapes Based on miRNA. Horticulturae. 2023; 9(8):931. https://doi.org/10.3390/horticulturae9080931
Chicago/Turabian StyleZhang, Lipeng, Yue Song, Junpeng Li, Jingjing Liu, Zhen Zhang, Yuanyuan Xu, Dongying Fan, Mingying Liu, Yi Ren, Xiaojun Xi, and et al. 2023. "Development, Identification and Validation of a Novel SSR Molecular Marker for Heat Resistance of Grapes Based on miRNA" Horticulturae 9, no. 8: 931. https://doi.org/10.3390/horticulturae9080931
APA StyleZhang, L., Song, Y., Li, J., Liu, J., Zhang, Z., Xu, Y., Fan, D., Liu, M., Ren, Y., Xi, X., Chen, Q., He, J., Xu, W., Song, S., Liu, H., & Ma, C. (2023). Development, Identification and Validation of a Novel SSR Molecular Marker for Heat Resistance of Grapes Based on miRNA. Horticulturae, 9(8), 931. https://doi.org/10.3390/horticulturae9080931





