Exploring the Bioprotective Potential of Halophilic Bacteria against Major Postharvest Fungal Pathogens of Citrus Fruit Penicillium digitatum and Penicillium italicum
Abstract
1. Introduction
2. Materials and Methods
2.1. Pathogen Sources
2.2. Fruit Preparation
2.3. Isolating Bacteria from Saline Water and Soil
2.4. Screening for Antagonistic Activity by Direct Confrontation
2.4.1. In Vitro Dual Culture
2.4.2. In Vivo Antagonism Experiment
2.5. Bacterial Identification
2.6. An In-Depth Characterization of Antagonistic Mechanisms
2.6.1. Indirect Antagonist Activity
Volatile Compounds (VOCs) Bioassay
In Vitro Bacterial Cell-Free Filtrates Effect
2.6.2. Bacterial Isolates’ Biochemical Characteristics
Amylase Activity
Protease Activity
Cellulase Activity
Hydrocyanic Acid (HCN) Production
The Antibiotic Biosynthetic Gene Detection
2.7. Semi-Practical Trials
2.8. Statistical Analysis
3. Results
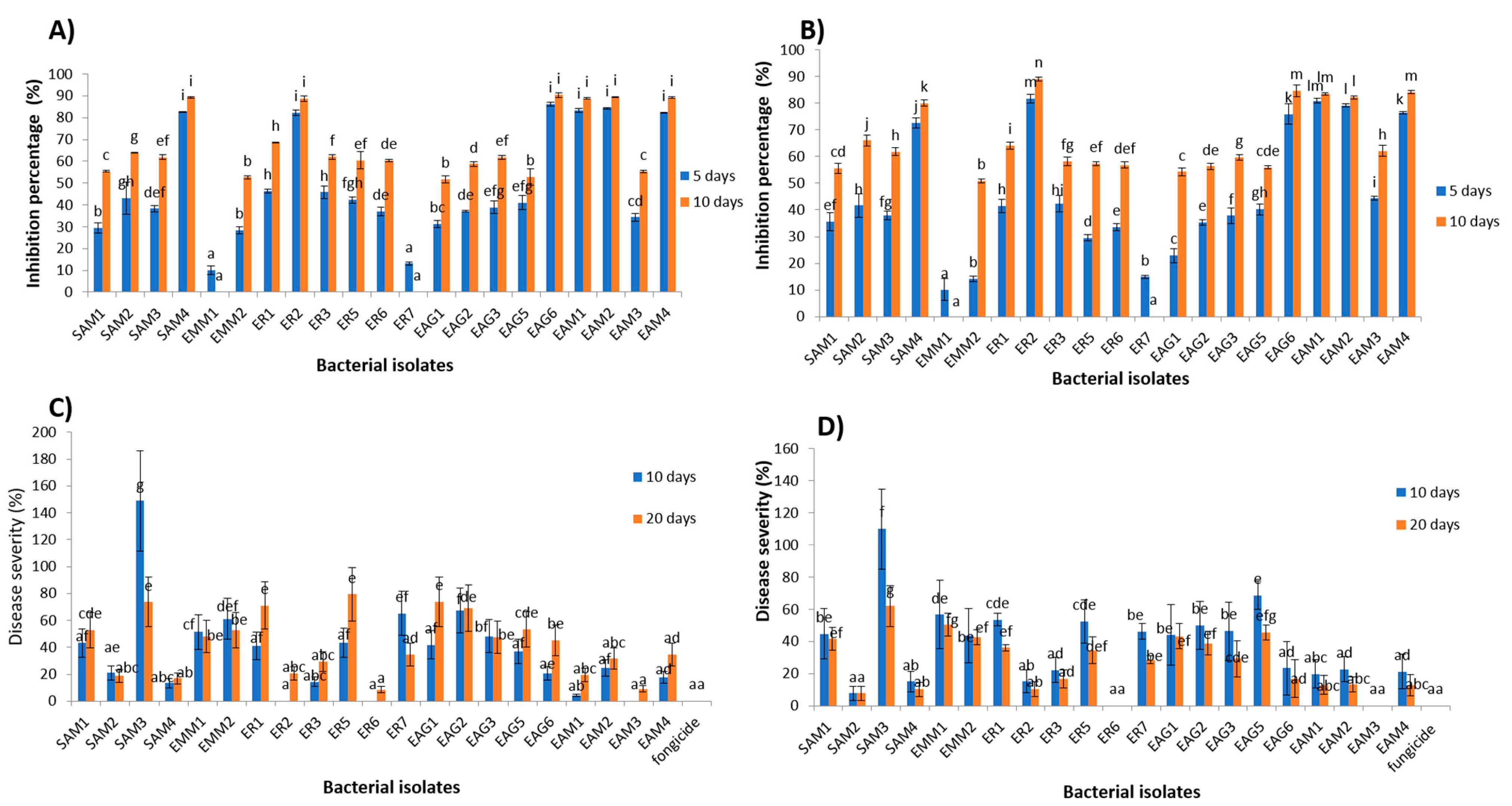
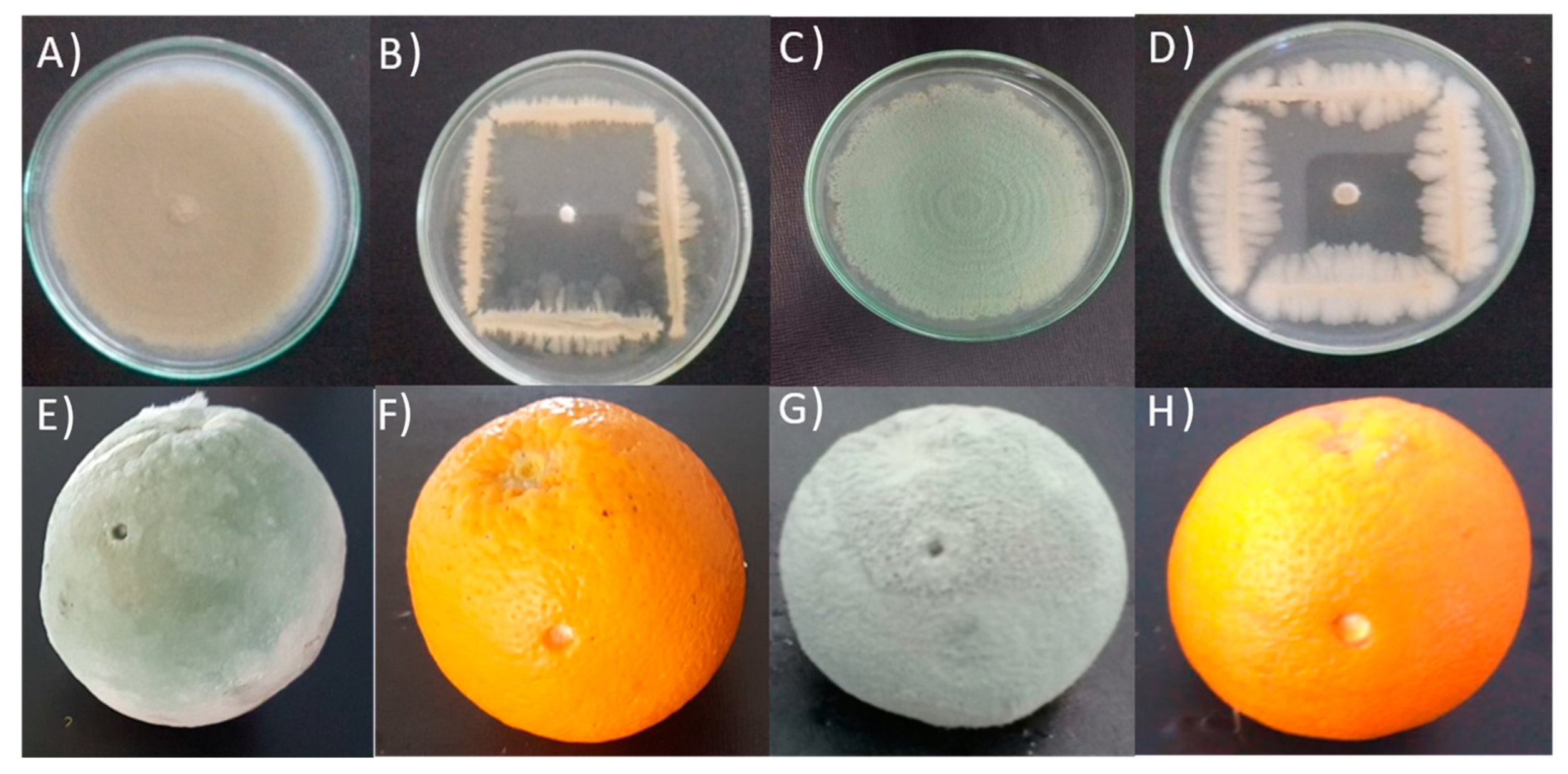
3.1. In Vitro Antagonistic Activity
3.2. In Vivo Antagonism Experiment
3.3. Bacterial Identification
3.4. In Vitro Antagonism via the Bacterial Supernatant
3.5. In Vivo Antagonism via the Bacterial Supernatant
3.6. In Vitro Volatility-Mediated Antagonism
3.7. Biochemical Traits
3.7.1. Amylase Activity
3.7.2. Proteolytic Activity
3.7.3. Cellulose Degradation
3.7.4. Hydrogen Cyanide Production
3.8. Detection of the Antibiotic Biosynthetic Gene
3.9. Semi-Practical Trials
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mditshwa, A.; Magwaza, L.S.; Tesfay, S.Z.; Opara, U.L. Postharvest factors affecting vitamin C content of citrus fruits: A review. Sci. Hortic. 2017, 218, 95–104. [Google Scholar] [CrossRef]
- Chinnici, G.; Zarbà, C.; Hamam, M.; Pecorino, B.; D’Amico, M. A model of circular economy of citrus industry. Int. Multidiscip. Sci. GeoConf. SGEM 2019, 19, 19–26. [Google Scholar]
- Babcock, B.A. Economic Impact of California’s Citrus Industry in 2020. J. Citrus Pathol. 2022, 9, 36–39. [Google Scholar] [CrossRef]
- Ismail, M.; Zhang, J. Post-harvest citrus diseases and their control. Outlooks Pest Manag. 2004, 15, 29. [Google Scholar] [CrossRef]
- Chen, J.; Shen, Y.; Chen, C.; Wan, C. Inhibition of key citrus postharvest fungal strains by plant extracts in vitro and in vivo: A review. Plants 2019, 8, 26. [Google Scholar] [CrossRef]
- Ezrari, S.; Radouane, N.; Tahiri, A.; El Housni, Z.; Mokrini, F.; Özer, G.; Lazraq, A.; Belabess, Z.; Amiri, S.; Lahlali, R. Dry root rot disease, an emerging threat to citrus industry worldwide under climate change: A review. Physiol. Mol. Plant Pathol. 2022, 117, 101753. [Google Scholar] [CrossRef]
- Dwiastuti, M.E.; Soesanto, L.; Aji, T.G.; Devy, N.F. Hardiyanto Biological control strategy for postharvest diseases of citrus, apples, grapes and strawberries fruits and application in Indonesia. Egypt. J. Biol. Pest Control 2021, 31, 141. [Google Scholar] [CrossRef]
- Gomes, A.A.M.; Queiroz, M.V.; Pereira, O.L. Mycofumigation for the biological control of post-harvest diseases in fruits and vegetables: A review. Austin J. Biotechnol. Bioeng. 2015, 2, 1–8. [Google Scholar]
- Bradford, K.J.; Dahal, P.; Van Asbrouck, J.; Kunusoth, K.; Bello, P.; Thompson, J.; Wu, F. The dry chain: Reducing postharvest losses and improving food safety in humid climates. In Food Industry Wastes; Elsevier: Amsterdam, The Netherlands, 2020; pp. 375–389. [Google Scholar]
- Sommer, N.F. Postharvest Handling Practices. Plant Dis. 1982, 66, 357. [Google Scholar] [CrossRef]
- Holmes, G.J.; Eckert, J.W. Sensitivity of Penicillium digitatum and P. italicum to postharvest citrus fungicides in California. Phytopathology 1999, 89, 716–721. [Google Scholar] [CrossRef]
- Zacarias, L.; Cronje, P.J.R.; Palou, L. Postharvest technology of citrus fruits. In The Genus Citrus; Elsevier: Amsterdam, The Netherlands, 2020; pp. 421–446. [Google Scholar]
- Macarisin, D.; Cohen, L.; Eick, A.; Rafael, G.; Belausov, E.; Wisniewski, M.; Droby, S. Penicillium digitatum suppresses production of hydrogen peroxide in host tissue during infection of citrus fruit. Phytopathology 2007, 97, 1491–1500. [Google Scholar] [CrossRef]
- Moss, M.O. Fungi, quality and safety issues in fresh fruits and vegetables. J. Appl. Microbiol. 2008, 104, 1239–1243. [Google Scholar] [CrossRef] [PubMed]
- Zamani, M.; Tehrani, A.S.; Ahmadzadeh, M.; Hosseininaveh, V.; Mostofy, Y. Control of Penicillium digitatum on orange fruit combining Pantoea agglomerans with hot sodium bicarbonate dipping. J. Plant Pathol. 2009, 91, 437–442. [Google Scholar]
- Perez, M.F.; Ibarreche, J.P.; Isas, A.S.; Sepulveda, M.; Ramallo, J.; Dib, J.R. Antagonistic yeasts for the biological control of Penicillium digitatum on lemons stored under export conditions. Biol. Control 2017, 115, 135–140. [Google Scholar] [CrossRef]
- Prusky, D.; McEvoy, J.L.; Saftner, R.; Conway, W.S.; Jones, R. Relationship between host acidification and virulence of Penicillium spp. on apple and citrus fruit. Phytopathology 2004, 94, 44–51. [Google Scholar] [CrossRef]
- Mercier, J.; Smilanick, J.L. Control of green mold and sour rot of stored lemon by biofumigation with Muscodor albus. Biol. Control 2005, 32, 401–407. [Google Scholar] [CrossRef]
- Gong, L.; Liu, Y.; Xiong, Y.; Li, T.; Yin, C.; Zhao, J.; Yu, J.; Yin, Q.; Gupta, V.K.; Jiang, Y. New insights into the evolution of host specificity of three Penicillium species and the pathogenicity of P. Italicum involving the infection of Valencia orange (Citrus sinensis). Virulence 2020, 11, 748–768. [Google Scholar] [CrossRef]
- Moussa, H.; El Omari, B.; Chefchaou, H.; Tanghort, M.; Mzabi, A.; Chami, N.; Remmal, A. Action of thymol, carvacrol and eugenol on Penicillium and Geotrichum isolates resistant to commercial fungicides and causing postharvest citrus decay. Can. J. Plant Pathol. 2020, 43, 26–34. [Google Scholar] [CrossRef]
- Chen, D.; Forster, H.; Adaskaveg, J.E. Baseline Sensitivities of Major Citrus, Pome, and Stone Fruits Postharvest Pathogens to Natamycin and Estimation of the Resistance Potential in Penicillium digitatum. Plant Dis. 2021, 105, 2114–2121. [Google Scholar] [CrossRef]
- Tanaka, T.; Suzuki, J.; Inomata, A.; Moriyasu, T. Combined effects of maternal exposure to fungicides on behavioral development in F1-generation mice: 3. Fixed-dose study of imazalil. Birth Defects Res. 2021, 113, 1390–1406. [Google Scholar] [CrossRef]
- Ezrari, S.; Mhidra, O.; Radouane, N.; Tahiri, A.; Polizzi, G.; Lazraq, A.; Lahlali, R. Potential role of rhizobacteria isolated from citrus rhizosphere for biological control of citrus dry root rot. Plants 2021, 10, 872. [Google Scholar] [CrossRef]
- Lahlali, R.; Mchachti, O.; Radouane, N.; Ezrari, S.; Belabess, Z.; Khayi, S.; Mentag, R.; Tahiri, A.; Barka, E.A. The Potential of Novel Bacterial Isolates from Natural Soil for the Control of Brown Rot Disease (Monilinia fructigena) on Apple Fruits. Agronomy 2020, 10, 1814. [Google Scholar] [CrossRef]
- Llop, P.; Bonaterra, A.; Peñalver, J.; López, M.M. Development of a Highly Sensitive Nested-PCR Procedure Using a Single Closed Tube for Detection of Erwinia amylovora in Asymptomatic Plant Material. Appl. Environ. Microbiol. 2000, 66, 2071–2078. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.E.; Lorsch, J. Sanger Dideoxy Sequencing of DNA. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2013; pp. 171–184. [Google Scholar]
- Lamia, L.; Souad, A.-G. Action de Paenibacillus polymyxa SGK2 sur quelques champignons de la fusariose du blé dur (Triticum durum) en Algérie. Alger. J. Nat. Prod. 2014, 2, 35–42. [Google Scholar]
- Trivedi, P.; Pandey, A.; Palni, L.M.S. In vitro evaluation of antagonistic properties of Pseudomonas corrugata. Microbiol. Res. 2008, 163, 329–336. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Lahlali, R.; Aksissou, W.; Lyousfi, N.; Ezrari, S.; Blenzar, A.; Tahiri, A.; Ennahli, S.; Hrustić, J.; MacLean, D.; Amiri, S. Biocontrol activity and putative mechanism of Bacillus amyloliquefaciens (SF14 and SP10), Alcaligenes faecalis ACBC1, and Pantoea agglomerans ACBP1 against brown rot disease of fruit. Microb. Pathog. 2020, 139, 103914. [Google Scholar] [CrossRef]
- Sodhi, H.K.; Sharma, K.; Gupta, J.K.; Soni, S.K. Production of a thermostable α-amylase from Bacillus sp. PS-7 by solid state fermentation and its synergistic use in the hydrolysis of malt starch for alcohol production. Process Biochem. 2005, 40, 525–534. [Google Scholar] [CrossRef]
- Nisa, R.M.; Irni, M.; Amaryllis, A.; Sugeng, S.; Iman, R. Chitinolytic bacteria isolated from chili rhizosphere: Chitinase characterization and its application as biocontrol for whitefly (Bemisia tabaci Genn.). Am. J. Agric. Biol. Sci. 2010, 5, 430–435. [Google Scholar]
- Cattelan, A.J.; Hartel, P.G.; Fuhrmann, J.J. Screening for plant growth–promoting rhizobacteria to promote early soybean growth. Soil Sci. Soc. Am. J. 1999, 63, 1670–1680. [Google Scholar] [CrossRef]
- Dimkić, I.; Živković, S.; Berić, T.; Ivanović, Ž.; Gavrilović, V.; Stanković, S.; Fira, D. Characterization and evaluation of two Bacillus strains, SS-12.6 and SS-13.1, as potential agents for the control of phytopathogenic bacteria and fungi. Biol. Control 2013, 65, 312–321. [Google Scholar] [CrossRef]
- Ramarathnam, R.; Bo, S.; Chen, Y.; Fernando, W.G.D.; Xuewen, G.; De Kievit, T. Molecular and biochemical detection of fengycin-and bacillomycin D-producing Bacillus spp., antagonistic to fungal pathogens of canola and wheat. Can. J. Microbiol. 2007, 53, 901–911. [Google Scholar] [CrossRef]
- Hsieh, F.C.; Li, M.C.; Lin, T.C.; Kao, S.S. Rapid detection and characterization of surfactin-producing Bacillus subtilis and closely related species based on PCR. Curr. Microbiol. 2004, 49, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Etesami, H.; Glick, B.R. Halotolerant plant growth–promoting bacteria: Prospects for alleviating salinity stress in plants. Environ. Exp. Bot. 2020, 178, 104124. [Google Scholar] [CrossRef]
- Pérez-Inocencio, J.; Iturriaga, G.; Aguirre-Mancilla, C.L.; Ramírez-Pimentel, J.G.; Vásquez-Murrieta, M.S.; Álvarez-Bernal, D. Identification of Halophilic and Halotolerant Bacteria from the Root Soil of the Halophyte Sesuvium verrucosum Raf. Plants 2022, 11, 3355. [Google Scholar] [CrossRef]
- Diba, H.; Cohan, R.A.; Salimian, M.; Mirjani, R.; Soleimani, M.; Khodabakhsh, F. Isolation and characterization of halophilic bacteria with the ability of heavy metal bioremediation and nanoparticle synthesis from Khara salt lake in Iran. Arch. Microbiol. 2021, 203, 3893–3903. [Google Scholar] [CrossRef]
- Essghaier, B.; Fardeau, M.-L.; Cayol, J.-L.; Hajlaoui, M.R.; Boudabous, A.; Jijakli, H.; Sadfi-Zouaoui, N. Biological control of grey mould in strawberry fruits by halophilic bacteria. J. Appl. Microbiol. 2009, 106, 833–846. [Google Scholar] [CrossRef] [PubMed]
- Sadfi-Zouaoui, N.; Hannachi, I.; Andurand, D.; Essghaier, B.; Boudabous, A.; Nicot, P. Biological control of Botrytis cinerea on stem wounds with moderately halophilic bacteria. World J. Microbiol. Biotechnol. 2008, 24, 2871–2877. [Google Scholar] [CrossRef]
- Tabassum, B.; Samuel, A.O.; Bhatti, M.U.; Fatima, N.; Shahid, N.; Nasir, I.A. Bottlenecks in Commercialization and Future Prospects of Beneficial Halotolerant Microorganisms for Saline Soils. In Saline Soil-Based Agriculture by Halotolerant Microorganisms; Springer: Berlin/Heidelberg, Germany, 2019; pp. 187–208. [Google Scholar]
- Waewthongrak, W.; Pisuchpen, S.; Leelasuphakul, W. Effect of Bacillus subtilis and chitosan applications on green mold (Penicilium digitatum Sacc.) decay in citrus fruit. Postharvest Biol. Technol. 2015, 99, 44–49. [Google Scholar] [CrossRef]
- Hammami, R.; Oueslati, M.; Smiri, M.; Nefzi, S.; Ruissi, M.; Comitini, F.; Romanazzi, G.; Cacciola, S.O.; Sadfi Zouaoui, N. Epiphytic yeasts and bacteria as candidate biocontrol agents of green and blue molds of citrus fruits. J. Fungi 2022, 8, 818. [Google Scholar] [CrossRef]
- Chen, K.; Tian, Z.; He, H.; Long, C.; Jiang, F. Bacillus species as potential biocontrol agents against citrus diseases. Biol. Control 2020, 151, 104419. [Google Scholar] [CrossRef]
- Wang, Z.; Sui, Y.; Li, J.; Tian, X.; Wang, Q. Biological control of postharvest fungal decays in citrus: A review. Crit. Rev. Food Sci. Nutr. 2020, 62, 861–870. [Google Scholar] [CrossRef] [PubMed]
- Lastochkina, O.; Seifikalhor, M.; Aliniaeifard, S.; Baymiev, A.; Pusenkova, L.; Garipova, S.; Kulabuhova, D.; Maksimov, I. Bacillus spp.: Efficient biotic strategy to control postharvest diseases of fruits and vegetables. Plants 2019, 8, 97. [Google Scholar] [CrossRef] [PubMed]
- Arroyave-Toro, J.J.; Mosquera, S.; Villegas-Escobar, V. Biocontrol activity of Bacillus subtilis EA-CB0015 cells and lipopeptides against postharvest fungal pathogens. Biol. Control 2017, 114, 195–200. [Google Scholar] [CrossRef]
- Chen, K.; Tian, Z.; Luo, Y.; Cheng, Y.; Long, C. Antagonistic activity and the mechanism of Bacillus amyloliquefaciens DH-4 against citrus green mold. Phytopathology 2018, 108, 1253–1262. [Google Scholar] [CrossRef]
- Tian, Z.; Chen, C.; Chen, K.; Liu, P.; Fan, Q.; Zhao, J.; Long, C. Biocontrol and the mechanisms of Bacillus sp. w176 against postharvest green mold in citrus. Postharvest Biol. Technol. 2020, 159, 111022. [Google Scholar] [CrossRef]
- Li, Y.; Xia, M.; He, P.; Yang, Q.; Wu, Y.; He, P.; Ahmed, A.; Li, X.; Wang, Y.; Munir, S. Developing Penicillium digitatum Management Strategies on Post-Harvest Citrus Fruits with Metabolic Components and Colonization of Bacillus subtilis L1-21. J. Fungi 2022, 8, 80. [Google Scholar] [CrossRef]
- Hao, W.; Li, H.; Hu, M.; Yang, L.; Rizwan-ul-Haq, M. Integrated control of citrus green and blue mold and sour rot by Bacillus amyloliquefaciens in combination with tea saponin. Postharvest Biol. Technol. 2011, 59, 316–323. [Google Scholar] [CrossRef]
- Calvo, H.; Marco, P.; Blanco, D.; Oria, R.; Venturini, M.E. Potential of a New Strain of Bacillus Amyloliquefaciens BUZ-14 as a Biocontrol Agent of Postharvest Fruit Diseases; Elsevier: Amsterdam, The Netherlands, 2017; Volume 63, ISBN 3497676268. [Google Scholar]
- Jiang, C.H.; Liao, M.J.; Wang, H.K.; Zheng, M.Z.; Xu, J.J.; Guo, J.H. Bacillus velezensis, a potential and efficient biocontrol agent in control of pepper gray mold caused by Botrytis cinerea. Biol. Control 2018, 126, 147–157. [Google Scholar] [CrossRef]
- Calvo, H.; Mendiara, I.; Arias, E.; Gracia, A.P.; Blanco, D.; Venturini, M.E. Antifungal activity of the volatile organic compounds produced by Bacillus velezensis strains against postharvest fungal pathogens. Postharvest Biol. Technol. 2020, 166, 111208. [Google Scholar] [CrossRef]
- Li, Y.; Wang, R.; Liu, J.; Xu, L.; Ji, P.; Sun, L.; Pan, H.; Jiang, B.; Li, L. Identification of a biocontrol agent Bacillus vallismortis BV23 and assessment of effects of its metabolites on Fusarium graminearum causing corn stalk rot. Biocontrol Sci. Technol. 2019, 29, 263–275. [Google Scholar] [CrossRef]
- Liu, C.; Yin, X.; Wang, Q.; Peng, Y.; Ma, Y.; Liu, P.; Shi, J. Antagonistic activities of volatiles produced by two Bacillus strains against Monilinia fructicola in peach fruit. J. Sci. Food Agric. 2018, 98, 5756–5763. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-W.; Balaraju, K.; Kim, J.-W.; Lee, S.-W.; Park, K. Systemic resistance and growth promotion of chili pepper induced by an antibiotic producing Bacillus vallismortis strain BS07. Biol. Control 2013, 65, 246–257. [Google Scholar] [CrossRef]
- Duan, Y.; Chen, R.; Zhang, R.; Jiang, W.; Chen, X.; Yin, C.; Mao, Z. Isolation and identification of Bacillus vallismortis HSB-2 and its biocontrol potential against apple replant disease. Biol. Control 2022, 170, 104921. [Google Scholar] [CrossRef]
- Zang, C.; Lin, Q.; Xie, J.; Lin, Y.; Zhao, K.; Liang, C. The biological control of the grapevine downy mildew disease using Ochrobactrum sp. Plant Prot. Sci. 2020, 56, 52–61. [Google Scholar] [CrossRef]
- Shoaib, A.; Ali, H.; Javaid, A.; Awan, Z.A. Contending charcoal rot disease of mungbean by employing biocontrol Ochrobactrum ciceri and zinc. Physiol. Mol. Biol. Plants 2020, 26, 1385–1397. [Google Scholar] [CrossRef]
- Erguven, G.O.; Demirci, U. Using Ochrobactrum thiophenivorans and Sphingomonas melonis for bioremediation of Imidacloprid. Environ. Technol. Innov. 2021, 21, 101236. [Google Scholar] [CrossRef]
- You, W.; Ge, C.; Jiang, Z.; Chen, M.; Li, W.; Shao, Y. Screening of a broad-spectrum antagonist-Bacillus siamensis, and its possible mechanisms to control postharvest disease in tropical fruits. Biol. Control 2021, 157, 104584. [Google Scholar] [CrossRef]
- Gao, L.; Ma, J.; Liu, Y.; Huang, Y.; Mohamad, O.A.A.; Jiang, H.; Egamberdieva, D.; Li, W.; Li, L. Diversity and biocontrol potential of cultivable endophytic bacteria associated with halophytes from the west Aral Sea Basin. Microorganisms 2021, 9, 1448. [Google Scholar] [CrossRef]
- Can-Herrera, L.A.; Gutierrez-Canul, C.D.; Dzul-Cervantes, M.A.A.; Pacheco-Salazar, O.F.; Chi-Cortez, J.D.; Carbonell, L.S. Identification by molecular techniques of halophilic bacteria producing important enzymes from pristine area in Campeche, Mexico. Braz. J. Biol. 2021, 83, e246038. [Google Scholar] [CrossRef]
- Dunne, C.; Crowley, J.J.; Moënne-Loccoz, Y.; Dowling, D.N.; O’Gara, F. Biological control of Pythium ultimum by Stenotrophomonas maltophilia W81 is mediated by an extracellular proteolytic activity. Microbiology 1997, 143, 3921–3931. [Google Scholar] [CrossRef]
- Schmoll, M.; Schuster, A. Biology and biotechnology of Trichoderma. Appl. Microbiol. Biotechnol. 2010, 87, 787–799. [Google Scholar]
- Bibi, F.; Strobel, G.A.; Naseer, M.I.; Yasir, M.; Khalaf Al-Ghamdi, A.A.; Azhar, E.I. Microbial Flora Associated with the Halophyte–Salsola imbricate and Its Biotechnical Potential. Front. Microbiol. 2018, 9, 65. [Google Scholar] [CrossRef] [PubMed]
- Bano, N.; Musarrat, J. Characterization of a New Pseudomonas aeruginosa Strain NJ-15 as a Potential Biocontrol Agent. Curr. Microbiol. 2003, 46, 324–328. [Google Scholar] [CrossRef]
- Fira, D.; Dimkić, I.; Berić, T.; Lozo, J.; Stanković, S. Biological control of plant pathogens by Bacillus species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef]
- Cao, Y.; Pi, H.; Chandrangsu, P.; Li, Y.; Wang, Y.; Zhou, H.; Xiong, H.; Helmann, J.D.; Cai, Y. Antagonism of Two Plant-Growth Promoting Bacillus velezensis Isolates Against Ralstonia solanacearum and Fusarium oxysporum. Sci. Rep. 2018, 8, 4360. [Google Scholar] [CrossRef]
- Arrebola, E.; Jacobs, R.; Korsten, L. Iturin A is the principal inhibitor in the biocontrol activity of Bacillus amyloliquefaciens PPCB004 against postharvest fungal pathogens. J. Appl. Microbiol. 2010, 108, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Arrebola, E.; Sivakumar, D.; Korsten, L. Effect of volatile compounds produced by Bacillus strains on postharvest decay in citrus. Biol. Control 2010, 53, 122–128. [Google Scholar] [CrossRef]
- Rodríguez-Chávez, J.L.; Juárez-Campusano, Y.S.; Delgado, G.; Aguilar, J.R.P. Identification of lipopeptides from Bacillus strain Q11 with ability to inhibit the germination of Penicillium expansum, the etiological agent of postharvest blue mold disease. Postharvest Biol. Technol. 2019, 155, 72–79. [Google Scholar] [CrossRef]
- Leelasuphakul, W.; Hemmanee, P.; Chuenchitt, S. Growth inhibitory properties of Bacillus subtilis strains and their metabolites against the green mold pathogen (Penicillium digitatum Sacc.) of citrus fruit. Postharvest Biol. Technol. 2008, 48, 113–121. [Google Scholar] [CrossRef]

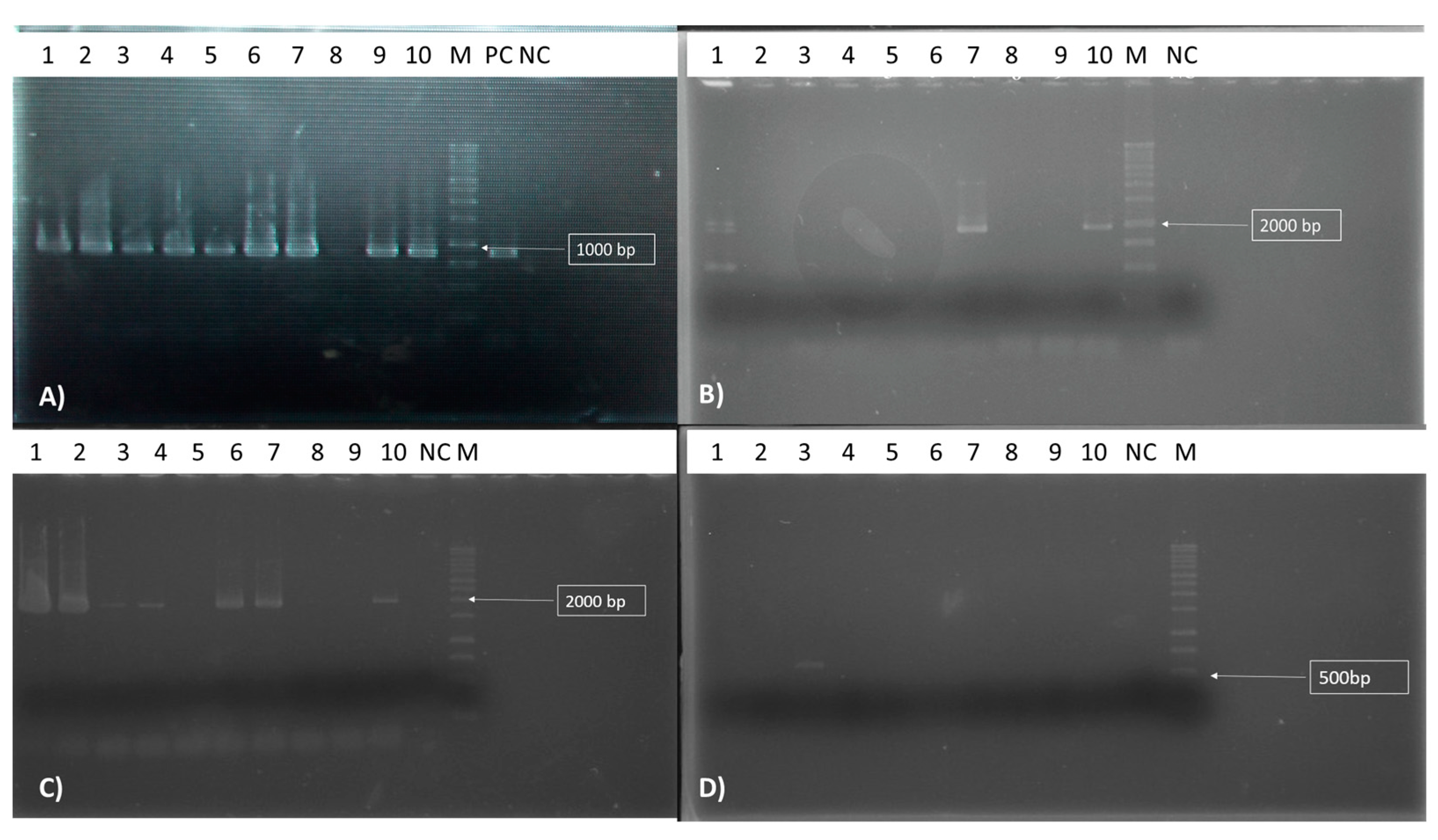

| Lipopeptides | Primer Pair | Primer Sequence | Product Length (bp) | Annealing T° | References |
|---|---|---|---|---|---|
| Bacillomycin | BACC1F /BACC1R | GAAGGACACGGCAGAGAGTC/ CGCTGATGACTGTTCATGCT | 875 bp | 60 °C | [35] |
| Fengycin | FEND1F/ FEND1R | TTTGGCAGCAGGAGAAGTT/ GCTGTCCGTTCTGCTTTTTC | 964 bp | 62 °C | [35] |
| Iturin | ITUP1F/ ITUP2R | AGCTTAGGGAACAATTGTCATCGGGGCTTC/ TCAGATAGGCCGCCATATCGGAATGATTCG | 2 kb | 45 °C | [34] |
| Surfactin | P17/ P18 | ATGAAGATTTACGGAATTTA/ TTATAAAAGCTCTTCGTACG | 675 bp | 53 °C | [36] |
| Isolates | Species | Accession Number |
|---|---|---|
| EAM1 | B. amyloliquefaciens | ON375996 |
| EAM2 | B. subtilis | MW646942 |
| EAM3 | O. thiophenivorans | MW644683 |
| EAM4 | B. amyloliquefaciens | MW644681 |
| SAM2 | B. amyloliquefaciens | MW644680 |
| SAM4 | B. velezensis | MW644682 |
| ER2 | B. amyloliquefaciens | ON376334 |
| ER3 | B. subtilis | ON376748 |
| ER6 | B. vallismortis | MW644685 |
| EAG6 | B. subtilis | MW644684 |
| Isolates | Amylase Production | Protease Production | Cellulase Production | HCN Production |
|---|---|---|---|---|
| SAM2 | 0.00 ± 0.00 a (−) | 0.00 ± 0.00 a (−) | 0.00 ± 0.00 a (−) | − |
| SAM4 | 1.11 ± 0.04 b (+) | 1.62 ± 0.00 c (+) | 1.33 ± 0.03 b (+) | − |
| EAM1 | 1.81 ± 0.03 c (+) | 3.24 ± 0.25 e (+) | 1.40 ± 0.04 b (+) | + |
| EAM2 | 2.28 ± 0.23 d (+) | 3.68 ± 0,32 f (+) | 1.23 ± 0.05 b (+) | − |
| EAM3 | 1.11+ 0.04 b (+) | 1.34 ± 0.06 b (+) | 0.00 ± 0.00 a (−) | + |
| EAM4 | 1.89 ± 0.08 b (+) | 4.24 ± 0.06 g (+) | 0.00 ± 0.00 a (−) | − |
| ER2 | 1.11 b ± 0.03 (+) | 1.60 ± 0.02 bc (+) | 0.00 ± 0.00 a (−) | − |
| ER3 | 1.16 b ± 0.08 (+) | 2.28 ± 0.16 d (+) | 1.75 ± 0.22 c (+) | − |
| ER6 | 0.0 ± 0.00 a (−) | 2.20 ± 0.12 d (+) | 1.30 ± 0.19 b (+) | − |
| EAG6 | 1.18 ± 0.06 b (+) | 4.30 ± 0.01 g (+) | 0.00 ± 0.00 a (−) | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radouane, N.; Adadi, H.; Ezrari, S.; Kenfaoui, J.; Belabess, Z.; Mokrini, F.; Barka, E.A.; Lahlali, R. Exploring the Bioprotective Potential of Halophilic Bacteria against Major Postharvest Fungal Pathogens of Citrus Fruit Penicillium digitatum and Penicillium italicum. Horticulturae 2023, 9, 922. https://doi.org/10.3390/horticulturae9080922
Radouane N, Adadi H, Ezrari S, Kenfaoui J, Belabess Z, Mokrini F, Barka EA, Lahlali R. Exploring the Bioprotective Potential of Halophilic Bacteria against Major Postharvest Fungal Pathogens of Citrus Fruit Penicillium digitatum and Penicillium italicum. Horticulturae. 2023; 9(8):922. https://doi.org/10.3390/horticulturae9080922
Chicago/Turabian StyleRadouane, Nabil, Hasnae Adadi, Said Ezrari, Jihane Kenfaoui, Zineb Belabess, Fouad Mokrini, Essaid Ait Barka, and Rachid Lahlali. 2023. "Exploring the Bioprotective Potential of Halophilic Bacteria against Major Postharvest Fungal Pathogens of Citrus Fruit Penicillium digitatum and Penicillium italicum" Horticulturae 9, no. 8: 922. https://doi.org/10.3390/horticulturae9080922
APA StyleRadouane, N., Adadi, H., Ezrari, S., Kenfaoui, J., Belabess, Z., Mokrini, F., Barka, E. A., & Lahlali, R. (2023). Exploring the Bioprotective Potential of Halophilic Bacteria against Major Postharvest Fungal Pathogens of Citrus Fruit Penicillium digitatum and Penicillium italicum. Horticulturae, 9(8), 922. https://doi.org/10.3390/horticulturae9080922











