1. Introduction
Within the
Rosaceae family, there are several agronomically and economically important stone fruit plants, such as almonds, plums, peaches, and nectarines [
1]. The
Prunus genus alone includes about 400 to 430 species, including peaches (
Prunus persica L., 2n = 2x= 16), apricots (
Prunus armeniaca L., 2n = 2x = 16), sweet cherries (
Prunus avium L., 2n = 2x = 16), and sour cherries (
Prunus cerasus L., 2n = 4x = 32) [
1,
2,
3,
4,
5]. Cherries are indigenous to Europe and Asia, while sour cherries are native to the Near East center (Asia Minor, Iran, Iraq, Syria) [
6].
Prunus cerasus L. is an allotetraploid species that evolved from the natural hybridization of
Prunus avium L. (2x = 16) and
Prunus fruticosa L. (4x = 32) both originating from the same area [
7,
8].
Several theories have been proposed regarding the arrival of sour cherries in Europe, but the most widely accepted suggestion is that various ecotypes have developed over the years to adapt to different weather conditions [
9]. As a result of this broad adaptation, sour cherries can be grown in various areas, including Hungary. Additionally, Hrotkó et al. [
10] explored the genetic background of
Prunus fruticosa Pall. in various hybrid derivatives, revealing that the female parent of sour cherry must be
Prunus fruticosa. For natural hybridization to occur, both species need to share the same habitat and flowering time, providing an opportunity for normal
Prunus fruticosa gametes to encounter unreduced
Prunus avium gametes. This finding was further supported by Wöhner et al. [
11], who provided evidence through long-read chromosome-level draft genome assembly.
The vegetation period of sour cherries is between 200 and 240 days, with the blooming period starting in the middle of April and lasting until the beginning of May. Moreover, these fruits exhibit excellent adaptability to different climates and they can tolerate temperatures as low as −35 °C during the winter period [
12]. Although other countries initiated their sour cherry breeding projects in the early 1900s, Hungary only established its foundation in the 1950s when Pál Maliga aimed to select clones that ripened earlier than Pándy clones, were self-fertile, and possessed multi-purpose utilization [
13]. Until that time, Pándy dominated the market, despite being self-incompatible and having a low yield [
12]. However, there are no perfect varieties that can be used for every purpose, and breeders need to consider their customers’ demands. Furthermore, due to climate change, there is a need to improve and develop new, modern varieties that can adapt to the new environmental conditions and meet the interests of consumers and breeders [
14,
15,
16]. To accomplish this, traits are separated into various categories according to different priorities. For example, from the customers’ perspective, the primary group of traits includes ripening time, fruit size, skin color, taste, etc., while the secondary group encompasses kernel size, vitamin content, flesh color, etc. [
12].
Several studies have demonstrated that molecular markers are useful tools that allow breeders to select from their collections, reduce time and costs, and increase potential profit [
17,
18]. Molecular markers are widely used to assess genetic variation in germplasm collections [
19], evolutionary studies [
20], ecological and phylogenetic studies, as well as in various fields such as taxonomy and genetic engineering [
21,
22,
23].
One of the most popular molecular markers are microsatellites, or simple sequence repeats (SSRs), which can be detected using simple reproducible assays. They are co-dominant, abundant, multi-allelic, and uniformly distributed throughout the genome [
24]. SSRs can be classified as mono-, di-, tri-, tetra-, penta-, and hexanucleotide repeats, with lengths ranging from 1 to 6 nucleotides [
25]. Typically, they consist of tandem repeats of 5–20 times in the genome with a minimum repeat length of twelve base pairs [
26,
27,
28] and are widely applied in research [
29,
30,
31,
32,
33]. Microsatellites have been successfully used in
Prunus species since the early 2000s [
2,
34,
35,
36,
37], and were primarily developed in peaches, but have also been tested and used in other species [
2]. Moreover, SSRs have been developed and applied in sour cherries, not only in
Prunus cerasus L., but also in other members of the
Rosaceae family [
38,
39]. For instance, Downey and Iezzoni [
40] used two sour cherry microsatellites in black cherry along with sweet cherry and peach primers. Furthermore, microsatellites have evolved to be suitable not only for diversity evaluation [
31,
41]; they have also helped to preserve the germplasm [
42], assess the pollinizer success rate [
43], and supported further studies regarding, for instance, sex determination in the case of dioecious plants [
44] within or outside of the
Rosaceae family.
Therefore, the aim of this study was to assess the genetic diversity within sour cherry genotypes cultivated in Hungary. The subject of this present research is also to assess the correlation between genotypic data and phenotypic traits to provide background information and a tool that is suitable for selecting plants during breeding, thereby helping the work of breeders. This examination also assesses the transferability of microsatellite markers developed in other species to sour cherry.
3. Results
In this study, nineteen sour cherry varieties (
Table 1) were fingerprinted using twelve microsatellite markers, six newly applied pairs of SSR (Simple Sequence Repeat) primers, alongside with six previously used microsatellites together (
Table 4, [
55]) to assess their genetic diversity and relatedness.
A total of 56 alleles were found, with the number of alleles per locus ranging from 2 to 10, and a mean of 4.67 alleles per locus. The highest number of alleles was observed in BPPCT007 (
Table 4).
None of the twelve examined SSR loci exhibited a monomorphic pattern. We evaluated our SSR results based on a study by Amiryousefi et al. [
51] (
Table 5). The loci Ma39a, UCDCH17 and UDP 96 005 showed the highest heterozygosity index (H = 0.50), while CPPCT022 had the lowest (H = 0.35). These three primer pairs have the highest probability of predicting a heterozygous individual for the given locus. The average heterozygosity index across the twelve markers was 0.46. Among the twelve markers tested, CPPCT022 (PIC = 0.43), BPPCT041 (PIC = 0.42), and BPPCT030 (PIC = 0.41) exhibited the highest PIC values in
Prunus cerasus L. The effective multiplex ratio (EMR), ranging from 0.55 to 6.23, reflects the effectiveness of the primer–marker system, with higher values indicating better effectiveness. The Marker Index (MI) is interpreted similarly to EMR, where higher values indicate better performance. According to the discriminating power, BPPCT041 (DP = 0.93) and UDP 98 410 (DP = 0.89) have the best ability to distinguish individuals in a population, thus reducing the probability of confusion between individuals, meanwhile CPPCT022 has the poorest ability at 0.41 (
Table 5).
The Jaccard index, among the examined
Prunus cerasus L. genotypes, indicates that Pándy clones are identical to each other. Also, Újfehértói fürtös, Debreceni bőtermő, and Kántorjánosi 3 exhibit high similarity (
Table 6). The genotypes based on SSR data that bear the closest resemblance to Érdi bőtermő are Favorit, Hibrid 3/48, and Maliga emléke.
On the other hand, Pipacs genotypes show the least similarity to the analyzed samples according to the Jaccard index (
Table 6).
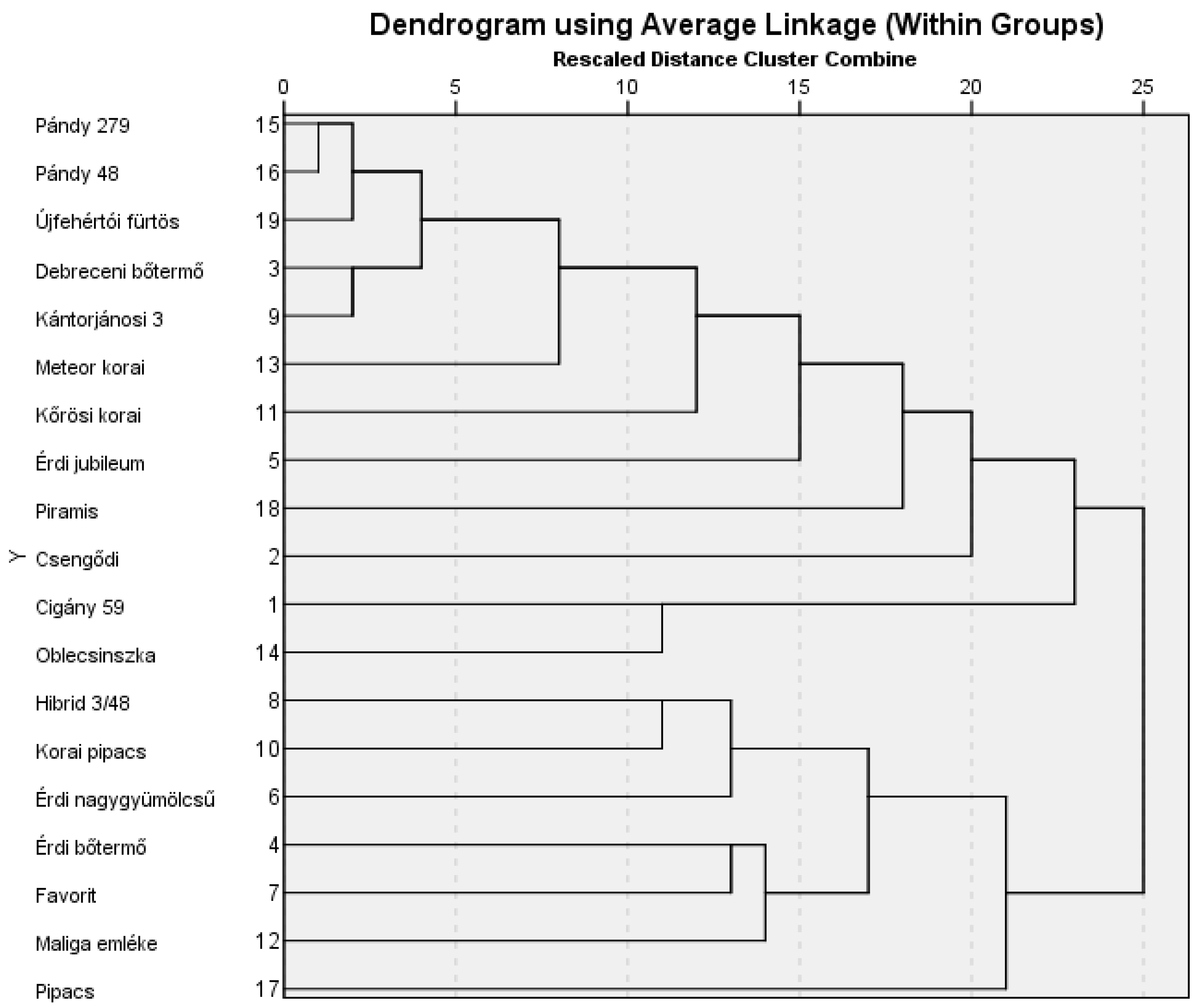
To determine genetic relatedness, a dendrogram was constructed based on the SSR data of the nineteen sour cherry cultivars using 12 microsatellite markers, as shown in
Figure 1. The dendrogram reveals two main groups. The first and largest group contains Pándy clones, Érdi jubileum, a Cigány clone, and Debreceni bőtermő, among others. The second main group consists of Korai pipacs, Favorit, and the other two Érdi genotypes, among others. Pándy 48 and Pándy 279 cannot be distinguished from each other based on the dendrogram. Additionally, Pipacs, Cigány 59, and Oblecsinszka genotypes are genetically the furthest from the analyzed samples within their respective cluster, as indicated in
Figure 1.
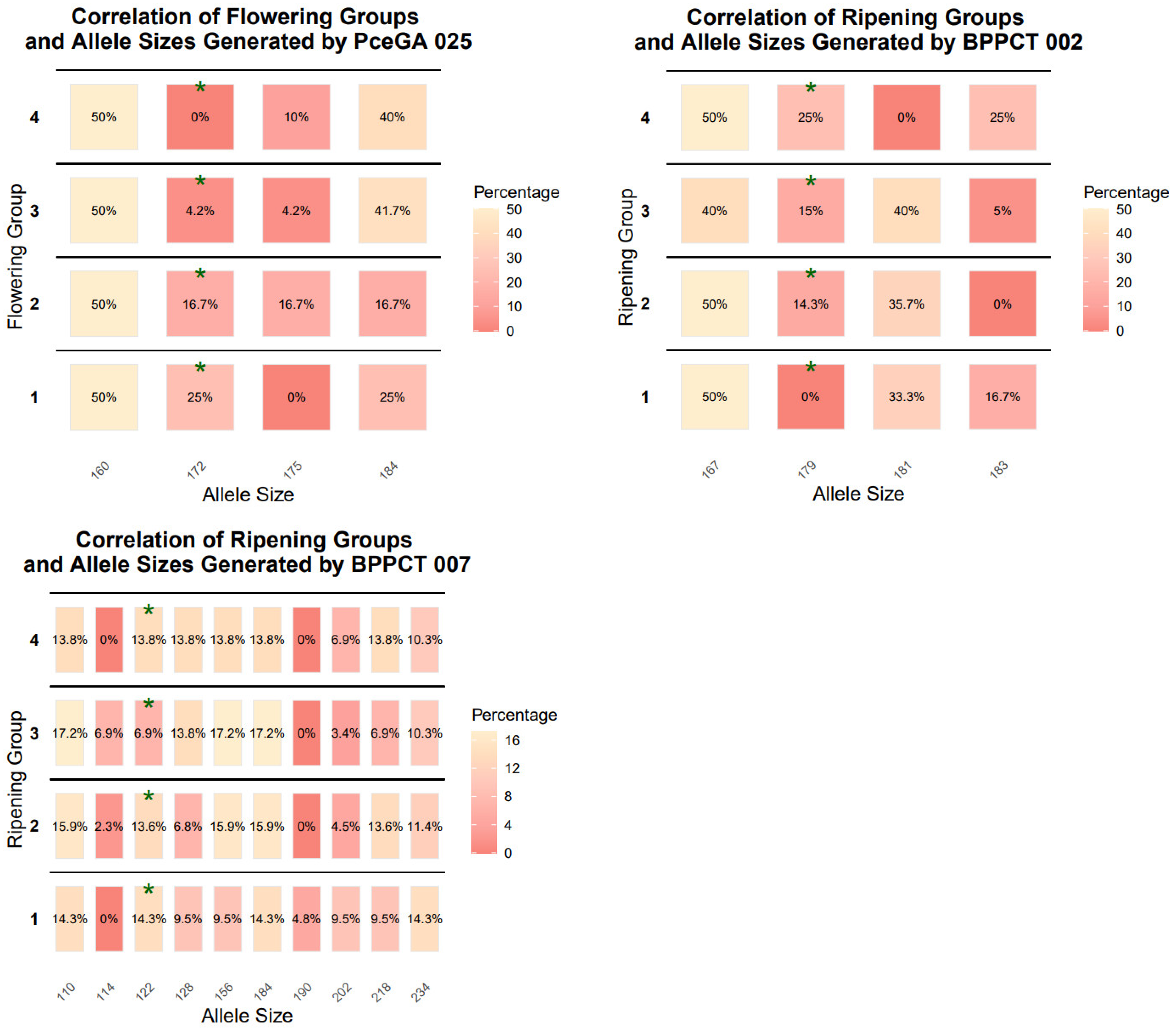
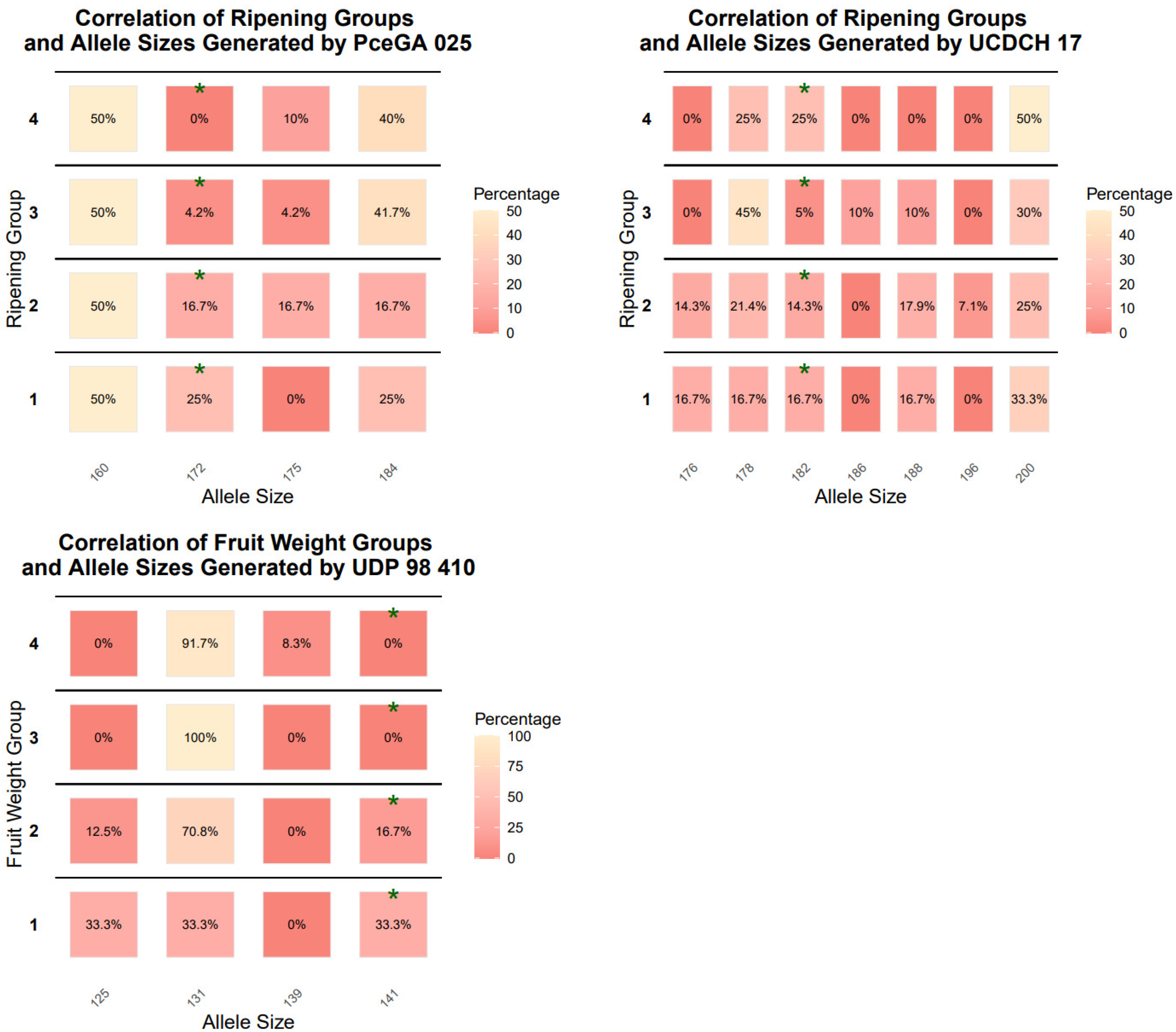
Additionally, based on their characteristics (flowering time, ripening time, and fruit weight) we classified the genotypes into 4–4 groups for each trait (
Table 2), then regression association analysis was performed to determine the linkage for each phenotypic trait and the microsatellite markers used. The aim of these analyses is to be able to examine populations to help breeders in the early selection phase. Thus, we have demonstrated the dispersion of allele sizes on heatmaps (
Figure 2). The statistically significant values were marked with green asterisks. Following this analysis, we carried out a Chi-square test. PceGA025
172 was found to be associated with flowering time. The marker alleles BPPCT002
179, BPPCT007
122, PceGA025
172, UCDCH17
182 showed an association with the ripening time. Also, UDP98 410
141 showed correlation with fruit weight. The highest correlation was shown by PceGA025
172 and UCDCH17
182 (R
2 = 0.795 and R
2 = 0.852, respectively)
. One of the markers, BPPCT007
122, had −0.753 as a standardized beta coefficient and showed statistically significant (t = −5.965,
p < 0.001) negative correlation with ripening time. BPPCT002
179 also had a statistically significant correlation with ripening time, where the standardized beta coefficient was 0.763 (t = 6.525,
p < 0.001) (
Table 7).
4. Discussion
Simple Sequence Repeats (SSRs) were primarily developed and tested in peaches in the
Rosaceae family. However, multiple studies have shown that although they were generated in
Prunus persica L., some of them can also be used in other species within the
Rosaceae family [
2,
32,
56].
It is a difficult challenge to analyze sour cherries because they are tetraploid and most of the statistical analysis techniques and software used molecular genetics are designed for diploid species or dominant features. Moreover, diversity analyses in sour cherries are usually based on morphological characteristics [
57]; only a few studies have applied genetic markers to examine them [
7,
58]. In the present study, twelve molecular markers were used to analyze and characterize the genetic diversity of sour cherries cultivated in Hungary and we attempted to overcome the associated limitations. Of these markers, ten originated from peach, one from sweet cherry, and one from sour cherry (
Table 3). Among these twelve microsatellites, only Ma039a has never been previously applied in
Prunus cerasus L.; however, we have proved that this marker is applicable in this genus. Moreover, this marker could be link to flowering time [
59].
There are studies in which the same markers were used in sour cherry, but detailed data on the generated alleles are not available [
60,
61]. However, our data are within or close to the size ranges reported in other analyses. Dirlewanger et al. [
2] developed several
Prunus microsatellites in peach and tested them in species within the
Rosaceae family and outside of the family. They reported the exact allele sizes for
Prunus persica L. and
Prunus avium L. However, for other tested genotypes, there were cases in which the markers worked, partially worked, or did not amplify at all. In the case of sour cherry, markers BPPCT002, BPPCT015, BPPCT030, and BPPCT041 generated amplicons in the tested genotypes, while only BPPCT007 did not yield fragments for all individuals. Additionally, the BPPCT007 microsatellite appears to be multilocal since more than four fragments were detected in approximately half of the samples. It has been reported that this locus is also multilocal in sweet cherry, which is a parent of sour cherry, explaining the shared characteristics.
Although Dirlewanger et al. [
2] did not report allele sizes, subsequent studies have used these markers and reported them. Wünsch et al. [
62] applied BPPCT002 and BPPCT007, but only size ranges were published. In the case of BPPCT002, the expected amplicon length was between 166 bp and 180 bp. Comparing our results (
Table 4) to theirs, the generated allele sizes are within or slightly above their range. In the case of BPPCT007, the lowest range is quite similar to theirs, but in our case the highest size is approximately 50 bp longer. Antonius et al. [
63] in agreement with Khadiv-Khub et al. [
7,
64,
65] also applied these to markers in sour cherry. While their results showed similar fragment lengths for BPPCT002, the highest value for BPPCT007 was 187 bp and 189 bp, respectively, compared to our result of 234 bp. Khadivi-Khub [
65] has indicated that the 234 bp fragment of BPPCT007 is associated with doubled fruit, but this trait was not the focus of our research. Nevertheless, we observed this length in almost half of our examined genotypes.
Cantini et al. [
38], Pedersen et al. [
39], Lacis et al. [
66], and Najafzadeh et al. [
67] reported the use of PceGA025. Our allele results are within the size range reported by these authors. Moreover, Pedersen et al. [
39] have analyzed four genotypes that are consistent with our analyzed varieties. In the case of the Favorit genotype, our results match with theirs, but for Érdi jubileum, Oblacsinszka, and Újfehértói fürtös, we observed one fewer allele. Even though we used the same percentage for the polyacrylamide gel, we used different running conditions and a different visualization method. The other researchers used dried gel that was exposed to Kodak BioMax film and in our case, a computer program detected the fragments. It might not be able to sense the allele sizes that are too close to each other; thus, the different technique could be the reason for the lower number of alleles.
UDP 96 001 marker was studied in flowering cherries by Ohta et al. [
60] and in
Prunus rootstock by Turkoglu et al. [
68]. They obtained almost identical results to ours. However, Najafzadeh et al. [
67] have also applied this primer pair, and their reported size range was slightly broader, with triple the number of alleles compared to ours. They also analyzed their samples with a UDP 96 005 microsatellite, and in that case, their size range was much larger (75–180 bp) than ours (102–134 bp). Nonetheless, Wünsch et al., Turkoglu et al. and Khadivi-Khub et al. reported similar allele sizes and ranges to ours [
7,
62,
68]. Khadivi-Khub [
65] suggested that the 122 bp length of UDP 96 005 is correlated with fruit weight, fruit length, and fruit diameter. We did not obtain this specific allele size in our study; the closest size we obtained was 118 bp. However, it was present in all genotypes regardless of their fruit weight and length (
Table 2). Thus far, we have only had the opportunity to examine individuals and Khadivi-Khub [
65] has used only five wild cherry (Mazzard) and four sour cherry genotypes; it is important and necessary to investigate segregating generations to prove and establish a correlation.
The data of Kompetenzzentrum [
69] include four markers (CPPCT022, UCDCH17, UDP 96 001, UDP 98 410) applied in sour cherry, and three of them (UCDCH17, UDP 96 001, UDP 98 410) produced exact sizes in their study, with four genotypes (Favorit, Kánotjánosi 3, Maliga emléke, Újfehértói fürtös) matching with ours. Regarding UCDCH17 and UDP 96 001, although our data are close to their size range, all common genotypes had one additional allele. In the case of UDP 98 410, the two datasets are the same. However, differences in the technique (Multiplex PCR), polymerase (GE Taq Polymerase), and running conditions (sequencer for analyzing fragments) used in the research may account for the slight discrepancies in exact allele sizes. For example, UDP 98 410 results show that in the case of Kompetenzzentrum [
69], they obtained 127 bp,135 bp and 127 bp for Favorit and Maliga emléke, respectively, while in our dataset, the same genotypes had 131 bp,139 bp, and 131 bp, respectively.
The twelve primer pairs used to screen the nineteen sour cherry genotypes generated a total of 52 distinct alleles. The average allele number per locus was 4.67, ranging from 2 to 10 (
Table 4).
In conclusion, in our research, the lowest heterozygosity index was observed for CPPCT022 (H = 0.35), while the highest values were observed for Ma39a(H = 0.50), UCDCH17(H = 0.50) and UDP 96 005 (H = 0.50). On the other hand, in the case of PIC the lowest applies to Ma39a (PIC = 0.37), UCDCH17 (PIC = 0.37), and UDP 96 005 (PIC = 0.37). The highest values were 0.43 for CPPCT022, followed by 0.42 for BPPCT041. Regarding Effective Multiplex Ratio (EMR), the values ranged from 0.55 to 6.23, with BPPCT 007 having the highest value and BPPCT 041 having the lowest. Similar results were observed for the Marker Index (MI). Additionally, according to our results, BPPCT041 (DP = 0.93) and UDP 98 410 (DP = 0.89) have a higher chance of distinguishing a genotype, while CPPCT022 (DP = 0.41) has the least discriminative power (
Table 5).
Regarding the Jaccard index (
Table 6), the lowest average similarity was observed for Pipacs, followed by Csengődi and Cigány 59, respectively. All the genotypes selected are from different regions. Correspondingly, the dendrogram partially supported the results, as genetically, the furthest genotypes were Pipacs, Piramis, and Csengődi (
Figure 1).
The regression association analysis showed that PceGA025
172, BPPCT002
179, BPPCT007
122, UCDCH17
182, and UDP98410
141 had statistically significant values regarding flowering time, ripening time, and fruit weight (
Table 2). In the case of peach genotypes, BPPCT007 was associated with fruit weight [
70,
71]; in our case, statistically it was more in line with ripening time. Dirlewanger et al. [
59] found in the case of apricot UDP 98 410, BPPCT007, Ma39a, and CPPCT022, and in the case of sweet cherry BPPCT002, UDP 98 410, and CPPCT022 that they could relate to flowering time.
In summary, based on our results, the most efficient primers used to analyze the nineteen sour cherry genotypes were BPPCT 007, and BPPCT 002, while the least efficient was BPPCT 041.
Additionally, this type of research can help breeders to overcome limitations and may assist with early selection. Moreover, monilia and blumeriella leaf spot are common diseases of cherries [
72], so the breeding of varieties resistant or tolerant to these diseases is crucial, and the analysis of additional morphological characteristics could contribute a lot to the work of breeders.