Genome-Wide Identification Reveals That BZR1 Family Transcription Factors Involved in Hormones and Abiotic Stresses Response of Lotus (Nelumbo)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Abiotic Stress Treatment
2.2. Identification of BZR1 Genes in N. nucifera (Nelumbo nucifera) Genome and N. lutea (Nelumbo lutea) Genome
2.3. Predicted Subcellular Localization of BZR1 Protein
2.4. Phylogenetic Analysis and Classification
2.5. Chromosomal Locations and Gene Structures of BZR1 Genes
2.6. Structural Analysis of Promoter Cis-Reactions
2.7. Covariance Analysis
2.8. RNA Isolation, cDNA Synthesis and Quantitative Real-Time PCR Analysis
3. Results
3.1. Identification of BZR1 TFs in Lotus
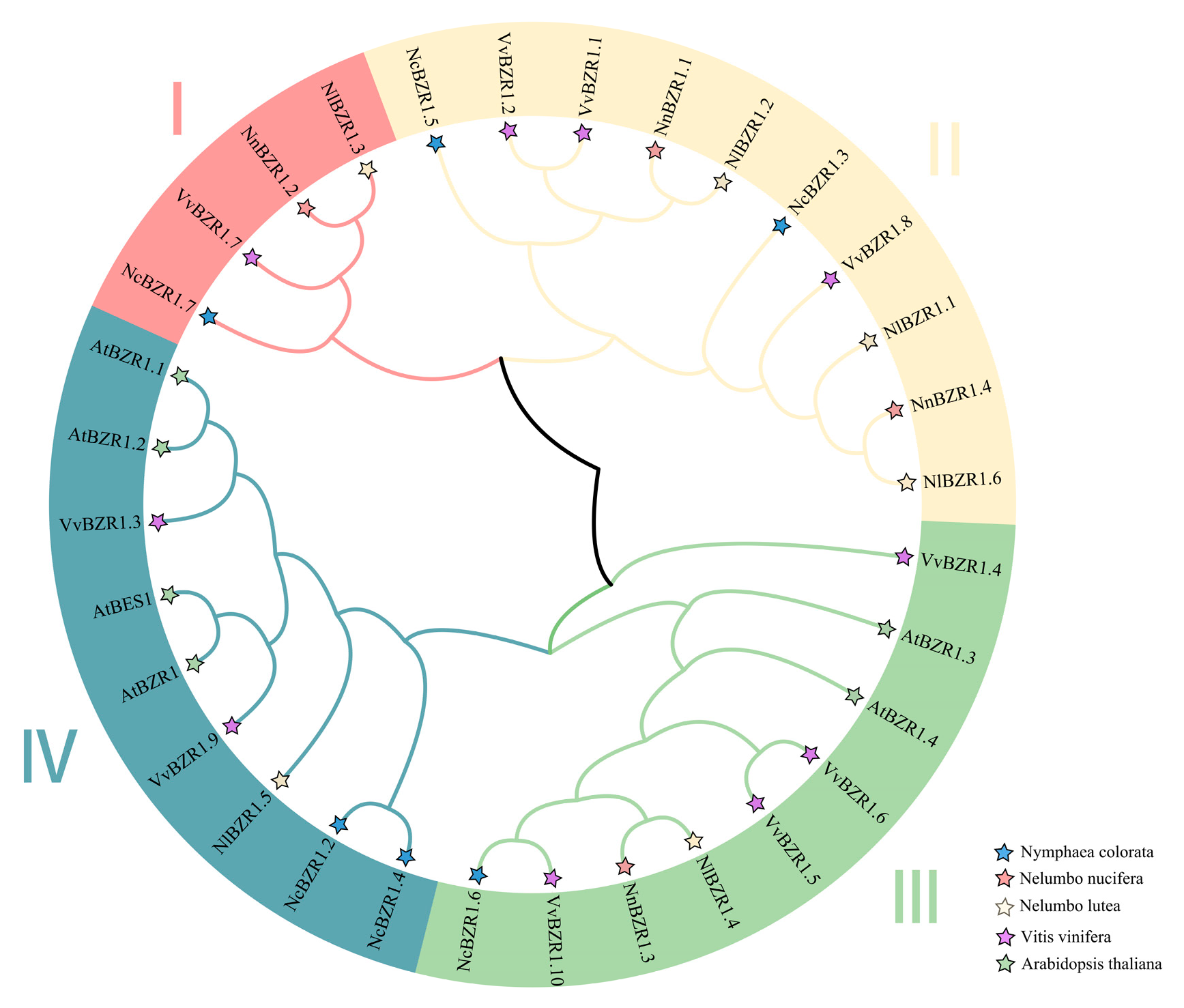
3.2. Phylogenetic Classification and Analysis of BZR1 TFs in Lotus
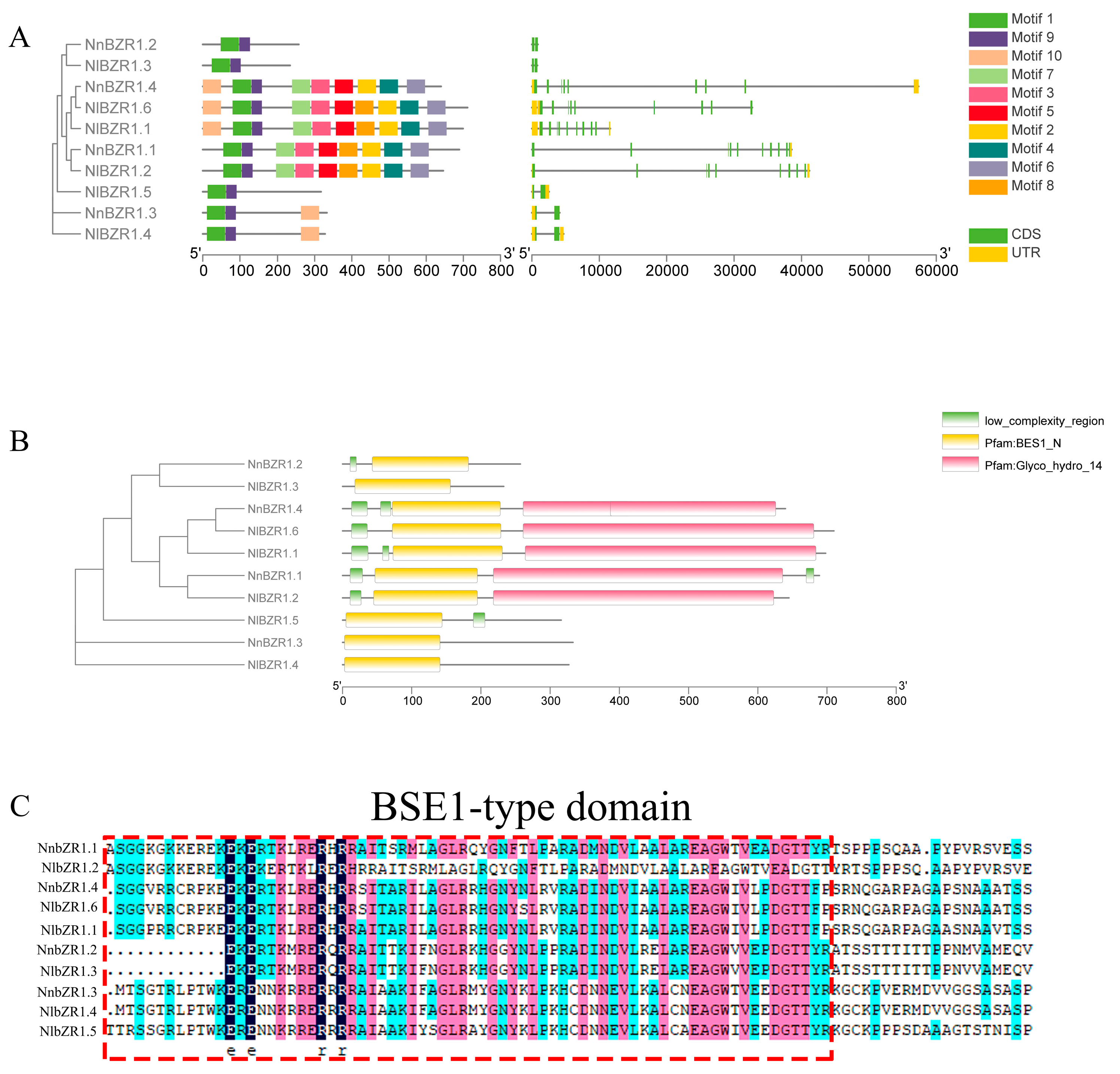
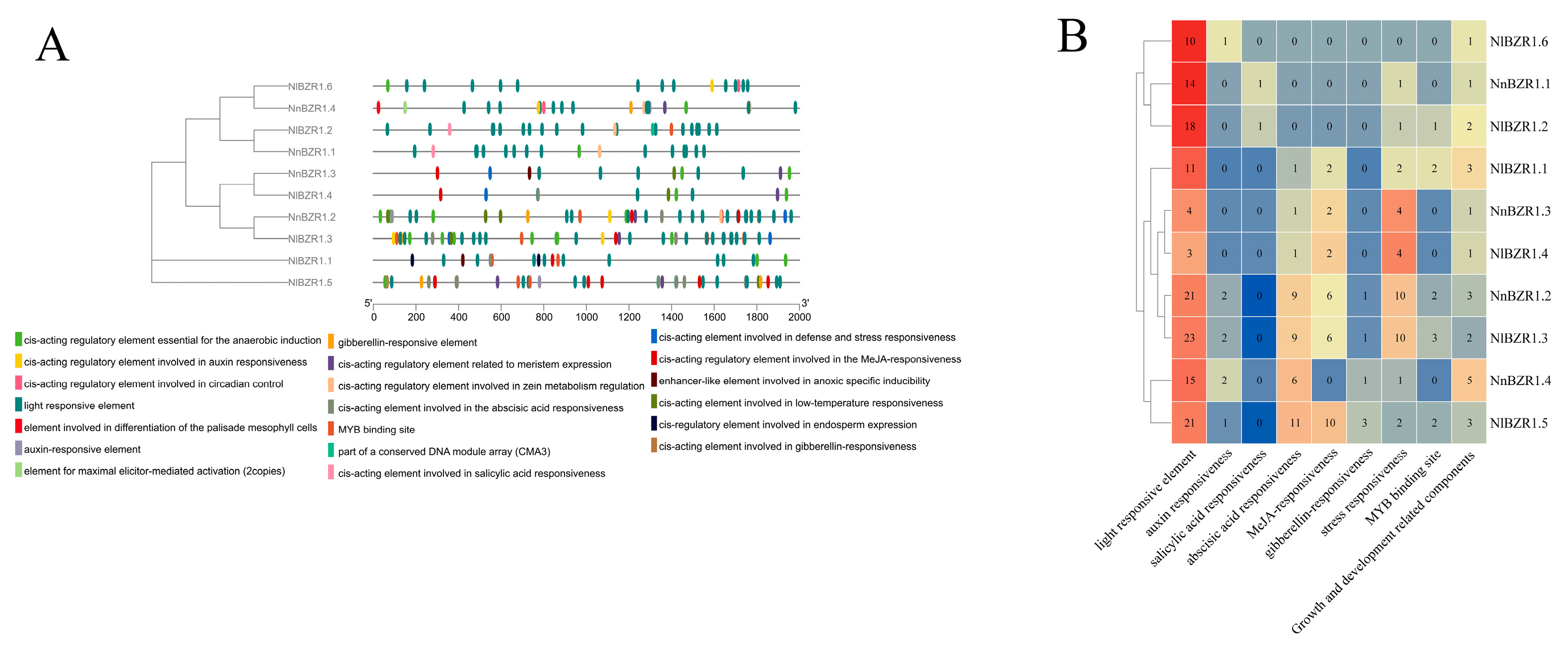
3.3. The Structure, Protein Motif, and Cis-Element Analysis of BZR1 TFs in Lotus
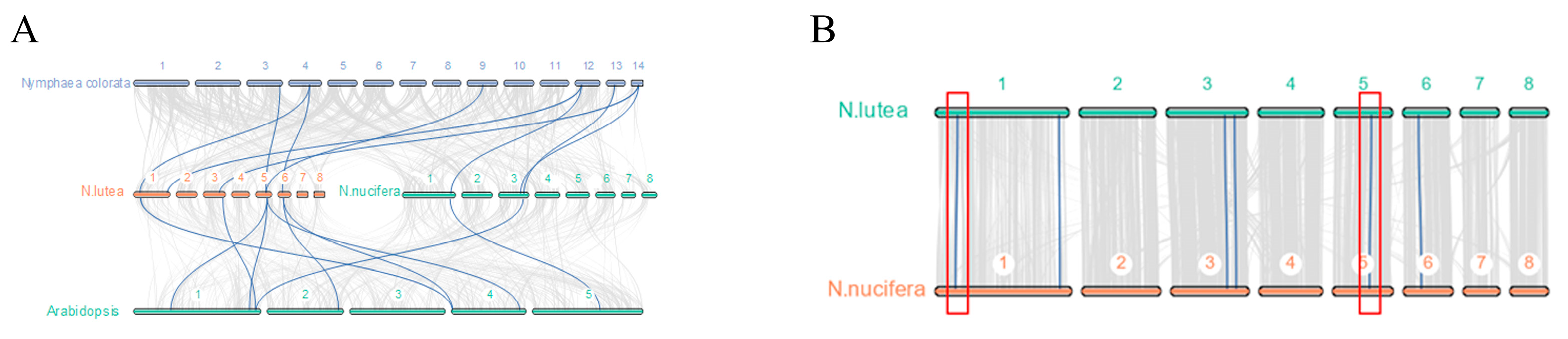
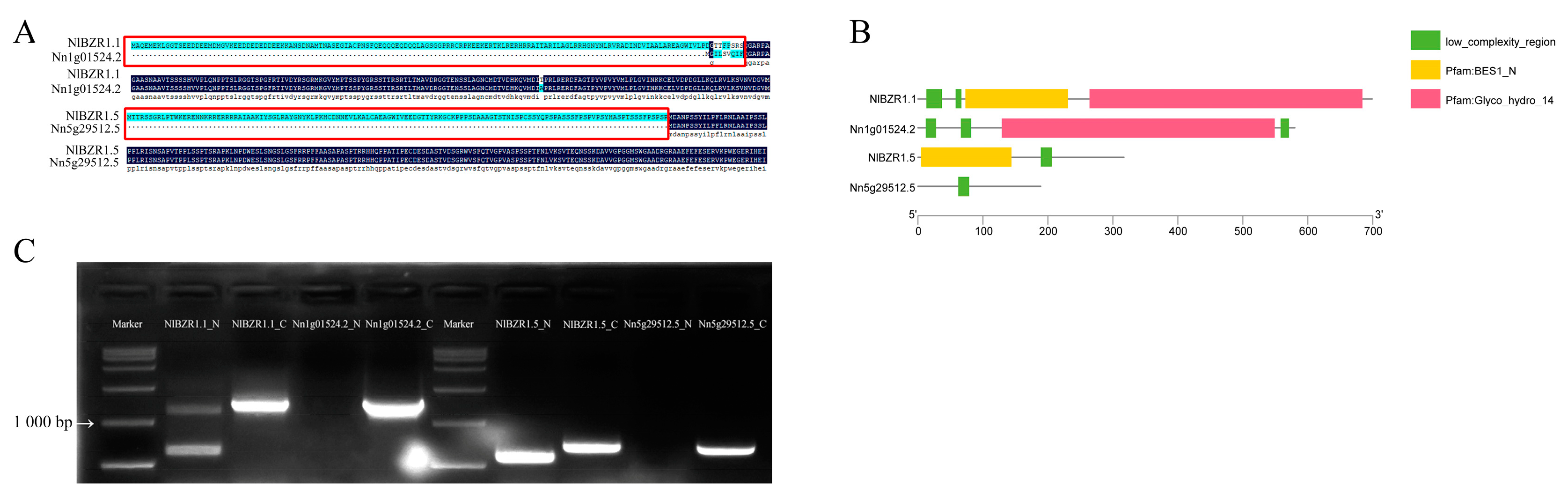
3.4. Collinearity Analysis of NnBZR1 Genes and NlBZR1 Genes
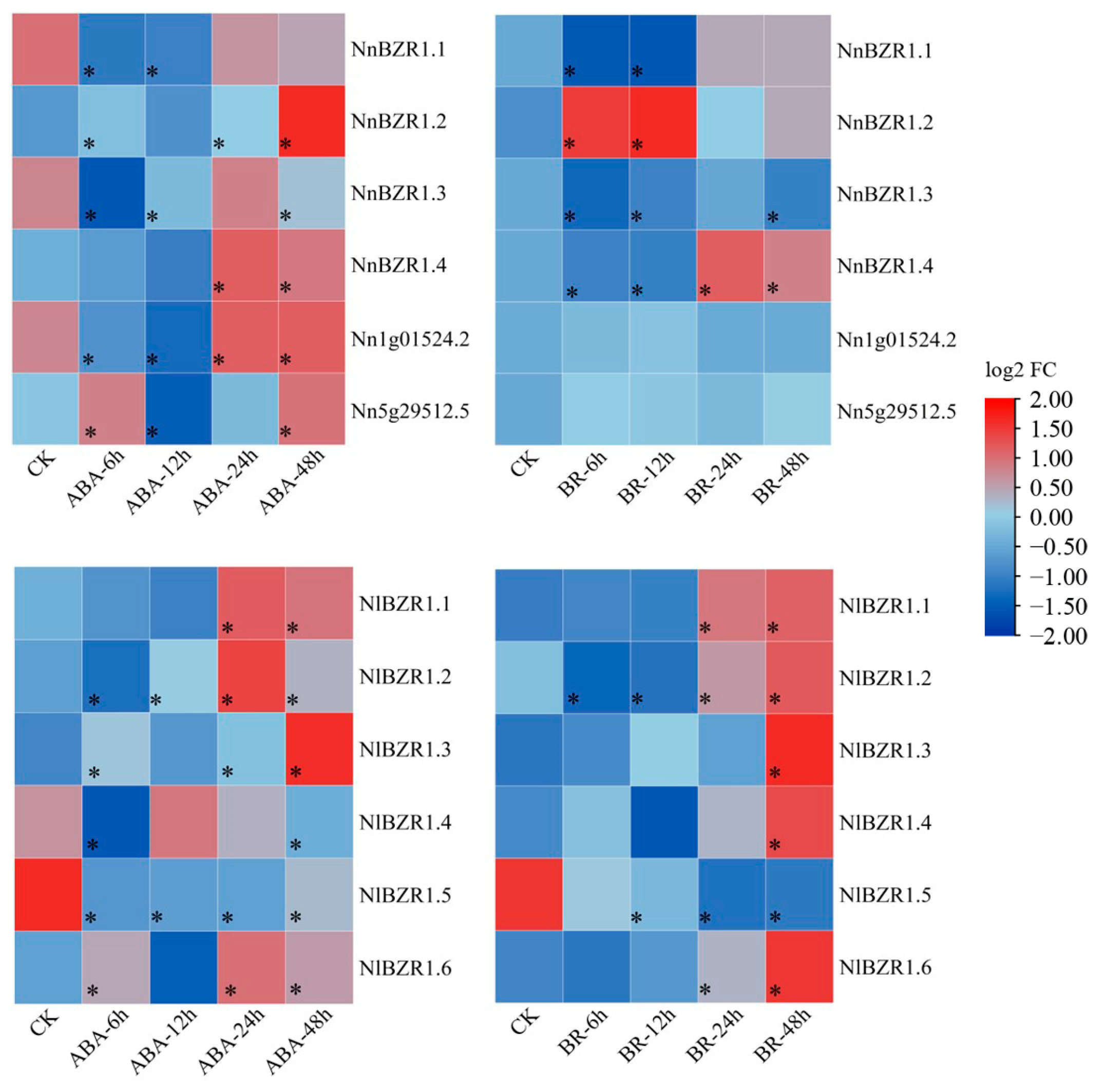
3.5. Expression Pattern of BZR1 Genes under BR and ABA Treatments
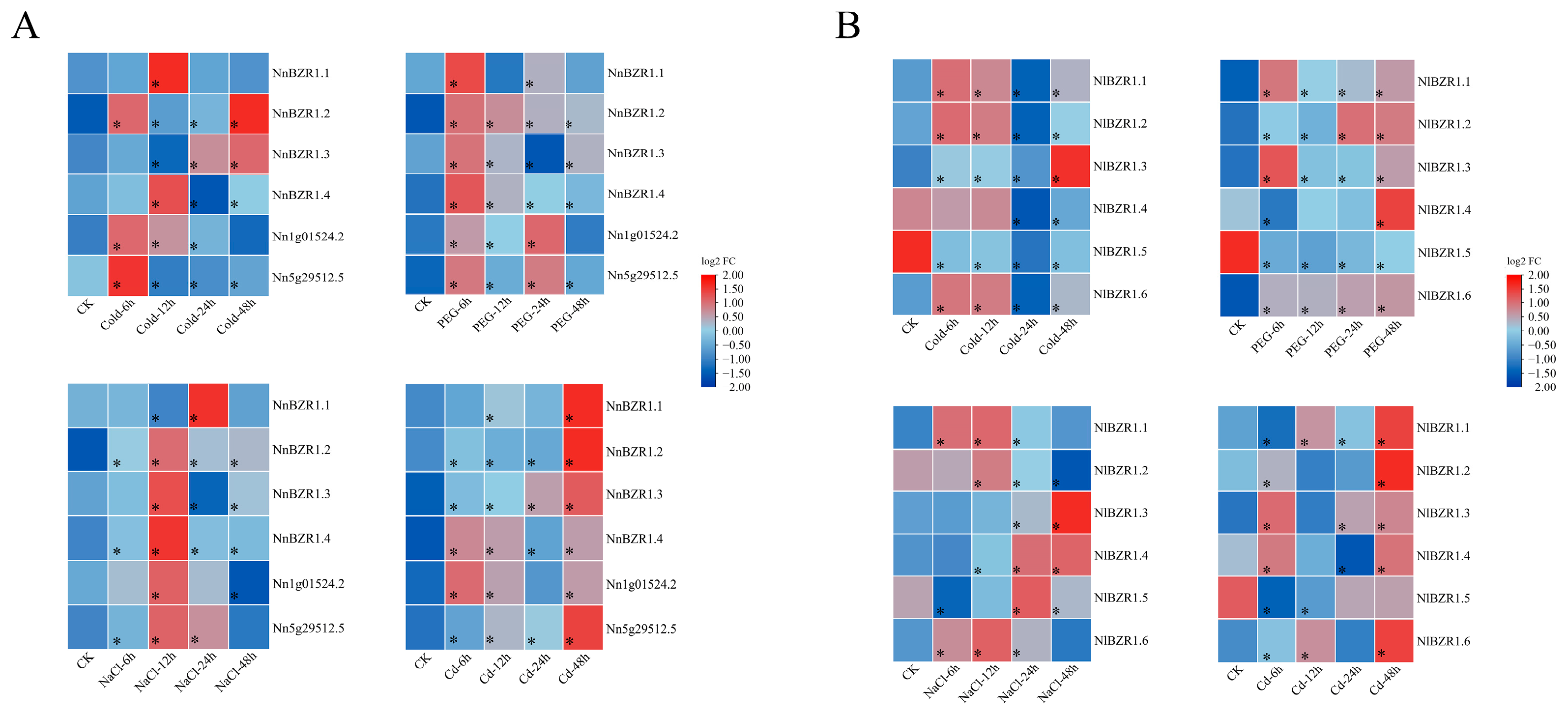
3.6. Expression Pattern of BZR1 Genes (under Different Stress Conditions)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grove, M.D.; Spencer, G.F.; Rohwedder, W.K.; Mandava, N.; Worley, J.F.; Warthen, J.D.; Steffens, G.L.; Flippen-Anderson, J.L.; Cook, J.C. Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 1979, 281, 216–217. [Google Scholar] [CrossRef]
- Mitchell, J.; Mandava, N.; Worley, J.; Plimmer, J.; Smith, M. Brassins—A new family of plant hormones from rape pollen. Nature 1970, 225, 1065–1066. [Google Scholar] [CrossRef]
- She, J.; Han, Z.; Kim, T.-W.; Wang, J.; Cheng, W.; Chang, J.; Shi, S.; Wang, J.; Yang, M.; Wang, Z.-Y. Structural insight into brassinosteroid perception by BRI1. Nature 2011, 474, 472–476. [Google Scholar] [CrossRef]
- Clouse, S.D.; Langford, M.; McMorris, T.C. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996, 111, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.-J.; Russinova, E. Brassinosteroid signalling. Curr. Biol. 2020, 30, R294–R298. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ye, H.; Guo, H.; Yin, Y. Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc. Natl. Acad. Sci. USA 2010, 107, 3918–3923. [Google Scholar] [CrossRef] [PubMed]
- Ahammed, G.J.; Li, X.; Liu, A.; Chen, S. Brassinosteroids in plant tolerance to abiotic stress. J. Plant Growth Regul. 2020, 39, 1451–1464. [Google Scholar] [CrossRef]
- Li, Z.; He, Y. Roles of brassinosteroids in plant reproduction. Int. J. Mol. Sci. 2020, 21, 872. [Google Scholar] [CrossRef]
- Li, Q.-F.; Lu, J.; Yu, J.-W.; Zhang, C.-Q.; He, J.-X.; Liu, Q.-Q. The brassinosteroid-regulated transcription factors BZR1/BES1 function as a coordinator in multisignal-regulated plant growth. BBA Gene Regul. Mech. 2018, 1861, 561–571. [Google Scholar] [CrossRef]
- Nolan, T.; Chen, J.; Yin, Y. Cross-talk of Brassinosteroid signaling in controlling growth and stress responses. Bioch. J. 2017, 474, 2641–2661. [Google Scholar] [CrossRef]
- Friedrichsen, D.M.; Nemhauser, J.; Muramitsu, T.; Maloof, J.N.; Alonso, J.; Ecker, J.R.; Furuya, M.; Chory, J. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 2002, 162, 1445–1456. [Google Scholar] [CrossRef]
- Li, J.; Chory, J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 1997, 90, 929–938. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Nam, K.H. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 2002, 295, 1299–1301. [Google Scholar] [CrossRef]
- Li, J.; Wen, J.; Lease, K.A.; Doke, J.T.; Tax, F.E.; Walker, J.C. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef] [PubMed]
- He, J.-X.; Gendron, J.M.; Sun, Y.; Gampala, S.S.; Gendron, N.; Sun, C.Q.; Wang, Z.-Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 2005, 307, 1634–1638. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar] [CrossRef]
- Ruan, J.; Chen, H.; Zhu, T.; Yu, Y.; Lei, Y.; Yuan, L.; Liu, J.; Wang, Z.-Y.; Kuang, J.-F.; Lu, W.-J. Brassinosteroids repress the seed maturation program during the seed-to-seedling transition. Plant Physiol. 2021, 186, 534–548. [Google Scholar] [CrossRef]
- Jia, C.; Zhao, S.; Bao, T.; Zhao, P.; Peng, K.; Guo, Q.; Gao, X.; Qin, J. Tomato BZR/BES transcription factor SlBZR1 positively regulates BR signaling and salt stress tolerance in tomato and Arabidopsis. Plant Sci. 2021, 302, 110719. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Nakano, T.; Gendron, J.; He, J.; Chen, M.; Vafeados, D.; Yang, Y.; Fujioka, S.; Yoshida, S.; Asami, T. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2002, 2, 505–513. [Google Scholar] [CrossRef]
- Zhang, L.e.; Han, Q.; Xiong, J.; Zheng, T.; Han, J.; Zhou, H.; Lin, H.; Yin, Y.; Zhang, D. Sumoylation of BRI1-EMS-SUPPRESSOR 1 (BES1) by the SUMO E3 ligase SIZ1 negatively regulates brassinosteroids signaling in Arabidopsis thaliana. Plant Cell Physiol. 2019, 60, 2282–2292. [Google Scholar] [CrossRef]
- Chen, L.-G.; Gao, Z.; Zhao, Z.; Liu, X.; Li, Y.; Zhang, Y.; Liu, X.; Sun, Y.; Tang, W. BZR1 family transcription factors function redundantly and indispensably in BR signaling but exhibit BRI1-independent function in regulating anther development in Arabidopsis. Mol. Plant 2019, 12, 1408–1415. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Fan, X.-Y.; Cao, D.-M.; Tang, W.; He, K.; Zhu, J.-Y.; He, J.-X.; Bai, M.-Y.; Zhu, S.; Oh, E. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 2010, 19, 765–777. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, L.; Zola, J.; Aluru, M.; Ye, H.; Foudree, A.; Guo, H.; Anderson, S.; Aluru, S.; Liu, P. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011, 65, 634–646. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, Z.-Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef]
- Li, H.; Ye, K.; Shi, Y.; Cheng, J.; Zhang, X.; Yang, S. BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol. Plant 2017, 10, 545–559. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.-Y.; Gao, Y.; Guo, J.; Yu, T.-F.; Zheng, W.-J.; Liu, Y.-W.; Chen, J.; Xu, Z.-S.; Ma, Y.-Z. BES/BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1. Plant Physiol. 2019, 180, 605–620. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Jin, Q.; Qian, P.; Wang, Y.; Wang, X.; Jiang, H.; Yao, D.; Liu, X.; Liu, F.; Li, J. Genetic resources of lotus (Nelumbo) and their improvement. Ornam. Plant Res. 2022, 2, 1–16. [Google Scholar] [CrossRef]
- Yu, H.; Feng, W.; Sun, F.; Zhang, Y.; Qu, J.; Liu, B.; Lu, F.; Yang, L.; Fu, F.; Li, W. Cloning and characterization of BES1/BZR1 transcription factor genes in maize. Plant Growth Regul. 2018, 86, 235–249. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Holzberg, S.; Brosio, P.; Gross, C.; Pogue, G.P. Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 2002, 30, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Khaliq, A.; Lu, S.; Xie, M.; Ma, Z.; Mao, J.; Chen, B. Genome-wide identification and characterization of the BES1 gene family in apple (Malus domestica). Plant Biol. 2020, 22, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Ma, X.; Li, C.; Hu, J.; Yang, Q.; Wang, T.; Wang, L.; Wang, J.; Guo, D.; Ge, W. Comprehensive analyses of the BES1 gene family in Brassica napus and examination of their evolutionary pattern in representative species. BMC Genom. 2018, 19, 346. [Google Scholar] [CrossRef] [PubMed]
- Sarwar, R.; Geng, R.; Li, L.; Shan, Y.; Zhu, K.-M.; Wang, J.; Tan, X.-L. Genome-Wide Prediction, Functional Divergence and characterization of stress-responsive BZR transcription factors in B. napus. Front. Plant Sci. 2022, 12, 790655. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wu, Y.; Li, L.; Li, C. Comprehensive analysis of the BES1 gene family and its expression under abiotic stress and hormone treatment in Populus trichocarpa. Plant Physiol. Biochem. 2022, 173, 1–13. [Google Scholar] [CrossRef]
- Martinez, M. Plant protein-coding gene families: Emerging bioinformatics approaches. Trends Plant Sci. 2011, 16, 558–567. [Google Scholar] [CrossRef]
- Saha, G.; Park, J.-I.; Jung, H.-J.; Ahmed, N.U.; Kayum, M.A.; Kang, J.-G.; Nou, I.-S. Molecular characterization of BZR transcription factor family and abiotic stress induced expression profiling in Brassica rapa. Plant Physiol. Biochem. 2015, 92, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Li, S.; Liu, X.; Wen, B.; Wang, N.; Zhang, R.; Li, D.; Chen, X.; Fu, X.; Xiao, W. Genome-wide identification and characterization of the MdBZR1 gene family in apple and their roles in improvement of drought tolerance. Sci. Hortic. 2021, 288, 110359. [Google Scholar] [CrossRef]
- Su, D.; Xiang, W.; Wen, L.; Lu, W.; Shi, Y.; Liu, Y.; Li, Z. Genome-wide identification, characterization and expression analysis of BES1 gene family in tomato. BMC Plant Biol. 2021, 21, 161. [Google Scholar] [CrossRef]
- Wu, P.; Song, X.; Wang, Z.; Duan, W.; Hu, R.; Wang, W.; Li, Y.; Hou, X. Genome-wide analysis of the BES1 transcription factor family in Chinese cabbage (Brassica rapa ssp. pekinensis). Plant Growth Regul. 2016, 80, 291–301. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, X.; Zhu, W.; Lin, H.; Chen, X.; Li, Y.; Ye, W.; Yin, Z. Molecular Traits and Functional Exploration of BES1 Gene Family in Plants. Int. J. Mol. Sci. 2022, 23, 4242. [Google Scholar] [CrossRef]
- Roy, S.W.; Penny, D. A very high fraction of unique intron positions in the intron-rich diatom Thalassiosira pseudonana indicates widespread intron gain. Mol. Biol. Evol. 2007, 24, 1447–1457. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lyu, H.M.; Zhu, K.; Van de Peer, Y.; Cheng, Z.M. The emergence and evolution of intron-poor and intronless genes in intron-rich plant gene families. Plant J. 2021, 105, 1072–1082. [Google Scholar] [CrossRef] [PubMed]
- Lei, P.; Wei, X.; Gao, R.; Huo, F.; Nie, X.; Tong, W.; Song, W. Genome-wide identification of PYL gene family in wheat: Evolution, expression and 3D structure analysis. Genomics 2021, 113, 854–866. [Google Scholar] [CrossRef]
- Huo, S.; Li, Y.; Li, R.; Chen, R.; Xing, H.; Wang, J.; Zhao, Y.; Song, X. Genome-wide analysis of the MADS-box gene family in Rhododendron hainanense Merr. and expression analysis under heat and waterlogging stresses. Ind. Crop. Prod. 2021, 172, 114007. [Google Scholar] [CrossRef]
- Kim, T.-W.; Guan, S.; Sun, Y.; Deng, Z.; Tang, W.; Shang, J.-X.; Sun, Y.; Burlingame, A.L.; Wang, Z.-Y. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 2009, 11, 1254–1260. [Google Scholar] [CrossRef]
- Tang, W.; Yuan, M.; Wang, R.; Yang, Y.; Wang, C.; Oses-Prieto, J.A.; Kim, T.-W.; Zhou, H.-W.; Deng, Z.; Gampala, S.S. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 2011, 13, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Tan, W.; Yang, F.; Han, Q.; Deng, X.; Guo, H.; Liu, B.; Yin, Y.; Lin, H. A BIN2-GLK1 signaling module integrates brassinosteroid and light signaling to repress chloroplast development in the dark. Dev. Cell 2021, 56, 310–324. [Google Scholar] [CrossRef]
- Yue, Z.-L.; Liu, N.; Deng, Z.-P.; Zhang, Y.; Wu, Z.-M.; Zhao, J.-L.; Sun, Y.; Wang, Z.-Y.; Zhang, S.-W. The receptor kinase OsWAK11 monitors cell wall pectin changes to fine-tune brassinosteroid signaling and regulate cell elongation in rice. Cur. Biol. 2022, 32, 2454–2466. [Google Scholar] [CrossRef]

| Species | Gene Name | Gene ID | Chromosome Location | Protein Length (aa) | MW (Da) | pI | AI | GRAVY | Predicted Location(s) |
|---|---|---|---|---|---|---|---|---|---|
| Nelumbo nucifera | NnBZR1.1 | Nn1g09318.4 | chr1:204,191,887-204,230,451(+) | 689 | 77,942.6 | 6.36 | 70.94 | −0.548 | Cytoplasm, Nucleus |
| NnBZR1.2 | Nn3g20241.1 | chr3:74,526,375-74,527,263(−) | 257 | 27,833.27 | 9.4 | 50.54 | −0.661 | Nucleus | |
| NnBZR1.3 | Nn3g21067.3 | chr3:94,419,885-94,424,008(−) | 333 | 35,471.42 | 8.59 | 56.61 | −0.578 | Nucleus | |
| NnBZR1.4 | Nn6g33434.9 | chr6:30,291,446-30,348,832(+) | 640 | 71,770.73 | 5.64 | 74.22 | −0.507 | Cytoplasm, Nucleus | |
| Nelumbo lutea | NlBZR1.1 | Al05735 | chr1:38,701,447-38,713,085(−) | 710 | 80,357.63 | 5.65 | 73.62 | −0.467 | Cytoplasm, Nucleus |
| NlBZR1.2 | Al21137 | chr1:211,166,400-211,207,570(+) | 698 | 78,611.41 | 5.57 | 73.51 | −0.518 | Cytoplasm, Nucleus | |
| NlBZR1.3 | Al22218 | chr3:76,546,305-76,547,121(−) | 327 | 34,780.6 | 8.43 | 55.26 | −0.597 | Nucleus | |
| NlBZR1.4 | Al12740 | chr3:96,705,633-96,710,336(−) | 316 | 34,086.92 | 9.22 | 56.58 | −0.660 | Nucleus | |
| NlBZR1.5 | Al13506 | chr5:65,454,507-65,457,060(+) | 645 | 72,936.19 | 6.46 | 72.29 | −0.511 | Nucleus | |
| NlBZR1.6 | Al00702 | chr6:30,635,376-30,668,093(+) | 233 | 24,827.91 | 9.47 | 50.3 | −0.547 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, P.; Jiang, H.; Li, J.; Jin, Q.; Wang, Y.; Xu, Y. Genome-Wide Identification Reveals That BZR1 Family Transcription Factors Involved in Hormones and Abiotic Stresses Response of Lotus (Nelumbo). Horticulturae 2023, 9, 882. https://doi.org/10.3390/horticulturae9080882
Zhou P, Jiang H, Li J, Jin Q, Wang Y, Xu Y. Genome-Wide Identification Reveals That BZR1 Family Transcription Factors Involved in Hormones and Abiotic Stresses Response of Lotus (Nelumbo). Horticulturae. 2023; 9(8):882. https://doi.org/10.3390/horticulturae9080882
Chicago/Turabian StyleZhou, Ping, Huiyan Jiang, Jingwen Li, Qijiang Jin, Yanjie Wang, and Yingchun Xu. 2023. "Genome-Wide Identification Reveals That BZR1 Family Transcription Factors Involved in Hormones and Abiotic Stresses Response of Lotus (Nelumbo)" Horticulturae 9, no. 8: 882. https://doi.org/10.3390/horticulturae9080882
APA StyleZhou, P., Jiang, H., Li, J., Jin, Q., Wang, Y., & Xu, Y. (2023). Genome-Wide Identification Reveals That BZR1 Family Transcription Factors Involved in Hormones and Abiotic Stresses Response of Lotus (Nelumbo). Horticulturae, 9(8), 882. https://doi.org/10.3390/horticulturae9080882





