Abstract
As an important temperate gum source plant, Eucommia ulmoides is widely distributed in China, but the low yield of Eucommia ulmoides gum considerably affects its application as a natural rubber in practical production. The small rubber particle protein (SRPP) gene is an influential participant in the Eucommia ulmoides gum biosynthesis process, and its expression affects the gum content. In this study, the promoter activity of the Eucommia ulmoides SRPP (EuSRPP) gene was analyzed by molecular biology and bioinformatics. In order to understand the molecular regulation mechanism of the EuSRPP genes at the transcriptional level, we first obtained the promoter sequences of the EuSRPP1, 3, 4, 5, 6, and 7 genes via genome walking and PCR amplification experiments. Then, the T3 generation of the transgenic homozygous line was obtained via a genetic transformation of Arabidopsis thaliana mediated by Agrobacterium. The six EuSRPP promoters were expressed in transgenic plants and were stably expressed in the leaves, pollinated flowers, and mature pods. As the transgenic plant grows and develops, promoter activity in the root is barely expressed. In addition, after the transgenic Arabidopsis was treated with methyl jasmonate (1 mmol/L MeJA), gibberellin (1 mmol/L GA3), and drought (20% PEG6000), the activity expression of the six EuSRPP promoters increased first and then decreased. The difference, however, is that EuSRPP1, 3, and 4 reach their strongest GUS activity at 3 h of plant treatment, while EuSRPP5, 6, and 7 reach their strongest activity at 6 h of treatment. Based on all experimental results, for the first time, it has been shown that the expression loci of the six EuSRPP gene promoters were relatively consistent. Second, the expression activity of the promoters of the six EuSRPP genes was different under MeJA, GA3, and drought treatment, suggesting that the promoter activity of the EuSRPP genes was regulated by endogenous hormones and drought pathways.
1. Introduction
Eucommia ulmoides (E. ulmoides), also known as bakelite, is a plant of the Eucommiaceae family and precious medicinal tree species originating in China [1]. The bark, leaves, seeds, and stamens of E. ulmoides are abundant in lignans, flavonoids, phenols, iridoids, and other chemical constituents. Its main pharmacological effects include lowering blood pressure, acting as an anti-inflammatory and improving atherosclerosis, among others [2,3,4,5]. In addition, E. ulmoides contains a specific white filamentous material, Eucommia ulmoides gum, which is an important bio-based polymer material that is currently widely used in various areas of life, such as in the biomedical field, industrial production, and aerospace [6,7,8,9]. The chemical formula of E. ulmoides gum is trans-1,4-polyisoprene (TPI) [10]. The process of gum biosynthesis in E. ulmoides is relatively complex, with many genes involved. The REF/SRPP family plays an essential role in gum biosynthesis in E. ulmoides. The REF/SRPP family consists of 12 members named SRPP1 to SRPP7 and REF1 to REF5 according to the presence or absence of a specific C-terminal domain. The Eucommia ulmoides small rubber particle protein (EuSRPP) is particularly important for the synthesis and stability of E. ulmoides gum. The basic unit, isopentenyl pyrophosphate (IPP), of E. ulmoides gum is synthesized through the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway and/or the cytoplasmic valproic acid (MVA) pathway, and it reacts with 1,1-dimethyl-allyl pyrophosphate (DMAPP) through a series of condensation polymerization to generate geranyl diphosphate (GPP) and other initiators, and then the E. ulmoides gum is synthesized under the action of EuSRPP, EuREF, etc. [11,12,13].
Since its discovery, SRPP has been studied by many scholars in various species. In the early in vitro synthetic rubber experiment, the SRPP extracted from Parthenium hysterophorus was added to the experimental reaction system. It was later found that the rubber biosynthesis activity in the system was significantly increased, which also demonstrated that the SRPP gene could regulate rubber synthesis [14]. The overexpression of TkSRPP3 in Taraxacum kok-saghyz increased the rubber content in the roots [15], whereas there is relatively little research on SRPP in E. ulmoides. Among the seven EuSRPP genes, only the expression levels of EuSRPP1, 2, and 7 correspond to the accumulation rate of E. ulmoides gum [11]. It has been shown that the expression of EuSRPP1 can feedback-regulate the expression of upstream gum-synthesis-related genes of E. ulmoides, such as EuIPI, EuFPPS, and EuGGPPS [16]. In addition, overexpression of the SRPP3 gene in E. ulmoides increases the molecular weight of E. ulmoides gum, indicating a significant relationship between the expression of EuSRPP3 and the molecular weight of rubber [17]. Gene expression is a complex regulatory process governed by multiple factors. Promoters, as an important cis-regulatory element in this process, can control the initiation time of gene expression, tissue specificity, and expression levels [18]. The expression efficiency of endogenous or exogenous genes mainly depends on the activity of the promoters [19]. A recent study showed that HbWRKY14 binds to the HbSRPP promoters to regulate the expression of HbSRPP [20]. These results proved that the expression of the SRPP gene is strongly correlated with gum content. The TbbZIP transcription factor could bind to the SRPP promoter region and regulate the expression of SRPP under the regulation of abscisic acid-dependent regulation [21].
SRPP is not only one of the main components of the rubber granule membrane, but it is also closely associated with the biosynthesis of rubber [22]. Currently, there is no definite conclusion as to how the EuSRPP gene regulates the expression of E. ulmoides gum. The Brazilian Hevea tree produces cis-polyisoprene, which is the main source of commercial rubber. On the contrary, there are relatively few plant species producing trans-polyisoprene [23]. At present, trans-polyisoprene is mainly produced by synthesis, and few plant species are used for its commercial production [24]. E. ulmoides is the main plant used for trans-rubber production. It is of great significance to study the specific gum synthesis mechanism to alleviate the shortage of natural rubber resources and promote the sustainable and healthy development of the rubber industry.
In this study, the EuSRPP1, 3, 4, 5, 6, and 7 gene promoter sequences of E. ulmoides were obtained via genome walking and PCR; The T3 generation transgenic homozygous plants were obtained by genetic transformation of Arabidopsis thaliana mediated by Agrobacterium. Based on the analysis of the cis-acting elements of the EuSRPP gene promoters, the expression activities of different gene promoters under the conditions of methyl jasmonate, gibberellin, and drought induction were determined, and the results showed that the expression activities of the EuSRPP1, 3, 4, 5, 6, and 7 gene promoters were different, and the expression was regulated by the methyl jasmonate, gibberellin, and drought pathways to varying degrees. This study provides an understanding of the molecular regulatory mechanism of the EuSRPP genes at the transcriptional level, and further provides some data support for subsequent studies of the biosynthesis mechanism of E. ulmoides gum.
2. Experimental Materials and Methods
2.1. Experimental Materials
Wild-type Arabidopsis thaliana seeds (Columbia) were provided by the Tea College of Guizhou University; fresh Eucommia ulmoides leaves were collected from the farm of the Guizhou Academy of Agricultural Bioengineering.
2.2. Methods
2.2.1. EuSRPP Promoter Sequence Amplification of E. ulmoides
There were seven EuSRPP genes in the genome of E. ulmoides [11], but the promoter sequence of EuSRPP2 gene was not successfully amplified by PCR and walking techniques. The additional six EuSRPP gene promoter subsequences were successfully obtained. Firstly, the promoter sequences of EuSRPP1, EuSRPP3, EuSRPP4, EuSRPP5, EuSRPP6, and EuSRPP7 were obtained from the E. ulmoides genome database, and the 2000 bp sequence upstream of the start codon was selected as the candidate sequence of EuSRPP gene promoters. The EuSRPP1 gene promoter sequence found in E. ulmoides genome database had a large number of uncertain base pairs, so it could not be obtained through ordinary PCR amplification. Ultimately, the sequence was obtained using genome walking. The amplified sequences of EuSRPP3, 4, 5, 6, and 7 promoters were obtained by PCR.
2.2.2. Bioinformatics Analysis of EuSRPP Promoter Sequences
The online websites PlantCARE and SoftBerry were used to predict the possible cis-acting elements and transcription initiation sites in the promoter sequences of EuSRPP1, EuSRPP3, EuSRPP4, EuSRPP5, EuSRPP6, and EuSRPP7. The cis-acting elements and transcription initiation sites of the EuSRPP promoters were visualized using the analysis software TBtools (v1.115).
2.2.3. Construction of EuSRPP Promoter Plant Expression Vectors
The six EuSRPP promoter sequences obtained by gel electrophoresis were first attached to the cloning vector pCAMBIA1300-GUS; this vector was then digested with EcoRI/SalI, and the plant expression vector pCAMBIA1300-EuSRPP-GUS was constructed by T4 ligase; this vector was transformed into E.coli and sequenced for validation; finally, the target vector was transformed into Agrobacterium for subsequent experiments (Supplementary Material File S1).
2.2.4. Agrobacterium-Mediated Genetic Transformation of EuSRPP Promoters into Arabidopsis thaliana
The six EuSRPP promoters’ positive Agrobacterium single colonies were selected and inoculated into YEP liquid medium (containing Rif 50 mg/L and Kan 100 mg/L) to expand the culture range. Place them in a constant temperature shaking incubator and avoid light for 4–5 h (28 °C, 200 rpm/min) until the OD600 value reaches 0.8–1.2. The bacterial precipitate was fully suspended using MS suspension with Silwet-77 added at a concentration of 0.02%; subsequently, wild-type Arabidopsis inflorescences were inoculated in a bacterial solution for 1 min, after which the plant was covered with clingfilm to maintain humidity above 90%. The plant was first cultured under 25 °C in dark for 24 h. After dark cultivation, remove the cling film and continue to cultivate under 23 °C, humidity of 65%, and light/dark conditions with a photoperiod of 16 h/8 h. The gestation period is seven days, with a total of three impregnation episodes; after normal cultivation of the transformed plants, generation T0 seeds were obtained. The generation T0 seeds were screened using hygromycin to obtain T1 generation transgenic plants. PCR identification was performed on positive transgenic plants using specific primers Hyg-F/R. After collecting seeds from generation T1, transgenic plants from generation T2 and T3 were screened and identified using the same method, until homozygous transgenic lines were obtained for subsequent experimental materials.
2.2.5. GUS Histochemical Staining of EuSRPP Promoters in Transgenic Arabidopsis Plants
The T3 transgenic Arabidopsis plants were placed in a centrifuge tube, and 200 μL GUS staining solution was added to completely soak the leaves. Place the centrifuge tube in a 37 °C incubator for overnight cultivation; pour out the GUS dye solution and decolorize the leaves in ethanol solutions of different concentrations (50%, 75%, 95%) until the remaining green color on the leaves completely fades; observe the plants under a stereomicroscope and photographed.
2.2.6. PCR Identification of EuSRPP Promoters in Transgenic Arabidopsis Plants
Extract DNA from transgenic Arabidopsis plants, use PCR for detection, and design specific primers using hygromycin resistance genes. PCR reaction products were detected via electrophoresis using 1% agarose gel, and wild-type Arabidopsis plants DNA and water were used as controls.
2.3. Analysis of the Active of EuSRPP Promoters
The obtained homozygous T3 generation EuSRPP1, 3, 4, 5, 6, and 7 promoters of the transgenic Arabidopsis plants were subjected to GUS chemical tissue staining at different growth stages to analyze the expression characteristics of six EuSRPP gene promoters. (1) Cotyledon stage: sampling was conducted after the T3 generation positive homozygous seeds grew cotyledons on the screening medium for hygromycin resistance. Then, GUS histochemical staining was performed. After decolorization with varying concentrations of ethanol, the plates were observed and photographed under stereomicroscopy. (2) True leaf stage: when the plants have grown to 4–6 true leaves, samples are taken for GUS histochemical staining; (3) Sexual reproduction stage: GUS histochemical staining is carried out when the plants begin to grow inflorescences, flowers and seed pods.
2.4. Determination of GUS Activity in EuSRPP Promoters in Transgenic Arabidopsis Plants under Different Treatment Conditions
First, EuSRPP1, 3, 4, 5, 6 and 7 homozygous transgenic Arabidopsis plants grown for approximately 30 days with consistent GUS staining were selected for treatment experiments. Spray the plant with 1 mmol/L MeJA solution, 1 mmol/L GA3 solution, and 20% PEG6000 solution in a dosage that makes the leaves completely moist and has droplets on the leaves. Cultivate in an artificial climate chamber with light/darkness at 23 °C, 16 h/8 h, and then take samples at 0 h, 3 h, 6 h, 12 h, and 24 h, respectively. The samples are frozen in liquid nitrogen and stored in an −80 °C ultra-low temperature refrigerator. The GUS enzymes were then extracted using a GUS enzyme-linked immunosorbent assay kit, their activity measured, and then the expression activity of the six EuSRPP was analyzed.
2.5. Data Analysis and Processing
All data in this experiment were processed and charted using Excel 2019, GraphPad Prism 8.0.2, and SPASS 8.1 software.
3. Results and Analysis
3.1. Obtaining EuSRPP Promoter Sequences
As early as 2018, Wuyun et al. performed evolutionary comparisons and a comprehensive transcriptome analysis of the genes involved in rubber biosynthesis in E. ulmoides. The results showed that 12 members of the REF/SRPP gene family were identified in the genome of E. ulmoides. Among them, there were seven members of SRPP, and they were named EuSRPP1, 2, 3, 4, 5, 6, and 7 [11]. Then, we placed EuSRPP1–7 in the E. ulmoides genome database for gene ID search, and we obtained the following corresponding gene ID numbers: GWHTAAAL026592 (EuSRPP1), GWHTAAAL026594 (EuSRPP2), GWHTAAAL026593 (EuSRPP3), GWHTAAAL003923 (EuSRPP4), GWHTAAAL003925 (EuSRPP5), GWHTAAAL021482 (EuSRPP6), and GWHTAAAL025875 (EuSRPP7). We then selected a base sequence of 2000 bp upstream of the onset codon as a candidate for the EuSRPP promoter. Based on the resulting candidate sequences, specific primers were designed, and the promoter sequence of EuSRPP3-7 was obtained via PCR amplification. The sequence lengths were 1498 bp, 1080 bp, 1475 bp, 1994 bp, and 2011 bp (Supplementary Material File S2). However, there are many uncertain base pairs in the EuSRPP1 promoter sequences. The sequence could not be obtained with PCR amplification. Therefore, the promoter sequence was obtained through the genome walking experiment. First, we obtained a promoter sequence of 1226 bp for EuSRPP1 through the first gene step, followed by a promoter sequence of 834 bp through the second gene step. We integrated the two steps to remove duplicate sequences, and finally, we obtained a promoter sequence of 1895 bp in length (Supplementary Material File S3). Only the promoter sequence of EuSRPP2 gene in E. ulmoides was not amplified, which may be due to the complexity of its own structure or significant differences between its genome database sequence and its actual sequence.
3.2. Analysis of Cis-Acting Elements of EuSRPP Promoters
We submitted the EuSRPP1, 3, 4, 5, 6, and 7 promoter sequences to the online sites PlantCARE and SoftBerry, respectively. The specific results are shown in Figure 1 and Table 1. The promoters EuSRPP1, EuSRPP4, and EuSRPP5 had two transcription initiation sites, and both EuSRPP1 and EuSRPP4 had a site close to the initiation codon. The promoters EuSRPP6 and EuSRPP7 only had one transcription initiation site, while the promoter EuSRPP3 had three transcription initiation sites. The common feature of these three promoters were that they also contain a site close to the initiation codon. In addition, two enhancer sites were detected in EuSRPP1 and EuSRPP3. The cis-acting elements in the promoter region of EuSRPP include not only basic elements such as the TATA box, which enables precise transcription initiation, and the CAAT box, which controls transcription frequency, but also many relevant elements relating to light, plant hormones, plant growth and development, and environmental responses.
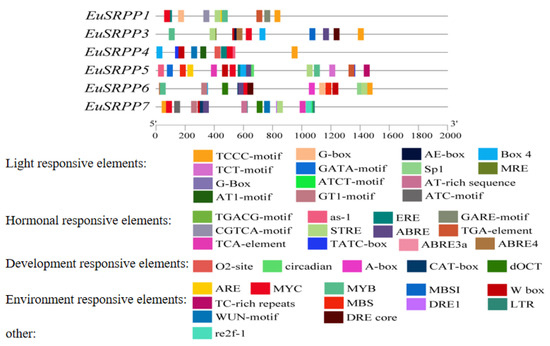
Figure 1.
Analysis of cis-acting elements of EuSRPP1, 3, 4, 5, 6, and 7 promoters.

Table 1.
EuSRPP1, 3, 4, 5, 6, and 7 promoter transcription binding sites.
3.3. Identification of EuSRPP Promoter Transgenic Arabidopsis Strains
After infecting the inflorescences, the wild-type Arabidopsis was genetically modified to produce T0 seeds. The seeds were screened on a hormone-resistant screening medium containing 30 mg/L. The seeds were able to grow normally and develop seedlings on resistant screening media, suggesting that the plant expression vector containing the target promoter fragment may have been transferred into Arabidopsis plants. The positive plants were subsequently identified by GUS chemical staining and PCR. Six EuSRPP promoters were then obtained as T3 generation homozygous transgenic positive Arabidopsis plants after multiple generations of screening and PCR testing. The specific results are as follows (Supplementary Material File S4): EuSRPP1 promoter Arabidopsis plants were detected by PCR with 14 positive bands, indicating that the promoter screened 14 transgenic-positive Arabidopsis lines; the 17 transgenic-positive Arabidopsis strains were obtained from the EuSRPP3 promoter; EuSRPP4 obtained 12 transgenic-positive Arabidopsis strains; EuSRPP5 obtained 11 transgenic-positive Arabidopsis strains; EuSRPP6 obtained 23 transgenic-positive Arabidopsis strains; EuSRPP7 obtained 11 transgenic-positive Arabidopsis strains.
3.4. Analysis of the Active Expression of EuSRPP Promoters at Different Development Stages in Transgenic Arabidopsis
First, strains with consistent growth status of EuSRPP1, 3, 4, 5, 6, and 7 promoters were selected for the experiments. Then, GUS histochemical staining was performed on tissue materials from transgenic Arabidopsis at different growth stages to observe the expression patterns of six EuSRPP promoters at different developmental stages. The specific experimental results are as follows:
EuSRPP1 (Figure 2): The seedlings were stained blue, and the GUS activity of the EuSRPP1 promoter was expressed in the cotyledons, hypocotyls, and roots of the transgenic Arabidopsis thaliana (Figure 2a). As the plant slowly grew to 4–6 true leaves, its staining began to change. The leaves were distinctly blue, while the blue of the roots is very light. This suggests that GUS activity is primarily expressed in Arabidopsis leaves during the true leaf stage, where the level of expression is higher than in cotyledons. In addition, there was also a small amount of GUS activity in the roots (Figure 2b). The stems, inflorescences, and stamens of the sexual reproduction period appeared blue after staining (Figure 2c). The newly formed pods did not appear blue, but with the continuous development, the gradually mature pods became blue after staining (Figure 2d). Based on the above results, it is concluded that the activity of EuSRPP1 gene promoters is mainly expressed in leaves and mature sexual reproductive organs.
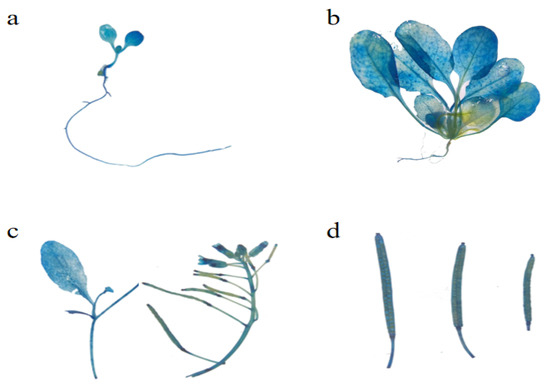
Figure 2.
GUS active expression of EuSRPP1 promoter at different development stages in transgenic Arabidopsis. (a) Whole plant of seedling in cotyledon stage; (b) whole plant in true leaf stage; (c) stem and inflorescence in sexual reproduction stage; (d) seed pod in sexual reproduction stage.
EuSRPP3 (Figure 3): Transgenic Arabidopsis seedlings appeared blue in their cotyledons, hypocotyls, and roots. Among them, the staining of cotyledons was deeper than that of roots, indicating that the GUS activity of cotyledons is higher than that of roots (Figure 3a). During the true leaf stage, the GUS staining of true leaves was stronger than that of cotyledons, and there was almost no blue color in the roots (Figure 3b). At the sexual reproduction stage, the pollinated flowers and lateral leaves on the inflorescence turn blue after dyeing, but the GUS dyeing intensity at the upper end of the inflorescence stem was lower than that at the lower end (Figure 3c). Mature seed pods and seeds had darker staining than immature ones, indicating higher GUS activity expression than immature seed pods (Figure 3d). In summary, the active expression sites of the EuSRPP3 promoter are located mainly in the leaf and mature sexual reproduction organs. The expression in the inflorescence stem is unstable. At the same time, there is almost no active expression in the roots from the true leaf stage.

Figure 3.
GUS active expression of EuSRPP3 promoter at different development stages in transgenic Arabidopsis. (a) Whole plant of seedling in cotyledon stage; (b) whole plant in true leaf stage; (c) inflorescence in sexual reproduction stage; (d) seed pod in sexual reproduction stage.
EuSRPP4 (Figure 4): The stained cotyledon stage seedlings typically had a lighter blue color, suggesting that the expression of EuSRPP4 promoter activity may be weaker in the plants (Figure 4a). After the plant was stained in the true leaf stage, the staining in the leaves was light, indicating low GUS expression activity in the leaves and essentially no expression in the petioles. Very few parts of the root are stained blue (Figure 4b). For plants in the sexual reproduction period, the flowering flowers on the inflorescence turn blue after dyeing, and the upper part of the flowering stem was darker than the lower part (Figure 4c). As the seed pods grew and developed, GUS staining became deeper, indicating a gradual increase in activity (Figure 4d). Based on the above results, we can determine that the active expression sites of the EuSRPP4 promoter are mainly located in the leaves, the mature seed pods, and the pollinated flowers, but overall, the expression activity is low.
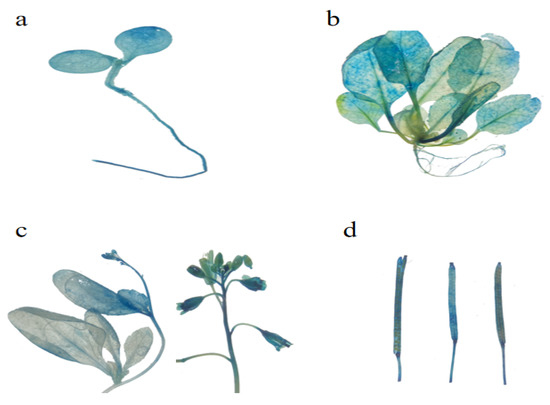
Figure 4.
GUS active expression of EuSRPP4 promoter at different development stages in transgenic Arabidopsis. (a) Whole plant of seedling in cotyledon stage; (b) whole plant in true leaf stage; (c) inflorescence in sexual reproduction stage; (d) seed pod in sexual reproduction stage.
EuSRPP5 (Figure 5): The GUS staining of transgenic plant seedlings during the cotyledon stage was significant (Figure 5a). As the plant grows, GUS staining was also deep during the true leaf stage, but the roots are not stained blue (Figure 5b). When the sexual reproduction organ developed, the inflorescence and its lateral leaves, mature seed pods, and seeds appear blue (Figure 5c,d). Based on the above experimental results, it can be inferred that the EuSRPP5 promoter is strongly active in all other parts of the plant except at the root where it is not expressed.
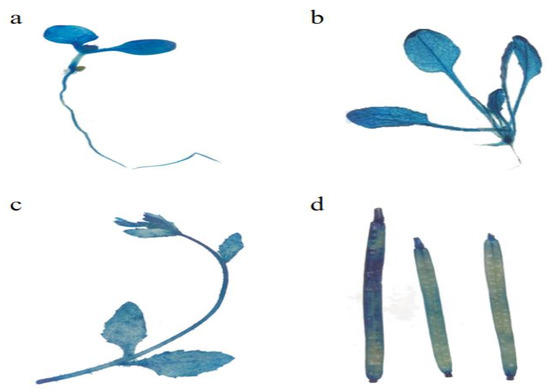
Figure 5.
GUS active expression of EuSRPP5 promoter at different development stages in transgenic Arabidopsis. (a) Whole plant of seedling in cotyledon stage; (b) whole plant in true leaf stage; (c) inflorescence in sexual reproduction stage; (d) seed pod in sexual reproduction stage.
EuSRPP6 (Figure 6): Cotyledons, hypocotyls and roots are all blue (Figure 6a). In the true leaf stage, the blue color of the plants was mainly concentrated in the middle of the leaves and petioles. Similarly, the GUS staining of the roots was lighter (Figure 6b). The GUS staining of flowers, pods and mature seeds at the reproductive stage is obvious, indicating that the promoter expression activity was strong (Figure 6c,d). According to the differences in GUS staining in different parts of the plant, the EuSRPP6 promoter has very low activity at the root, and the main expression sites are the mature sexual organs, the plant leaves, and the petioles, with the petioles having higher activity than the leaves.
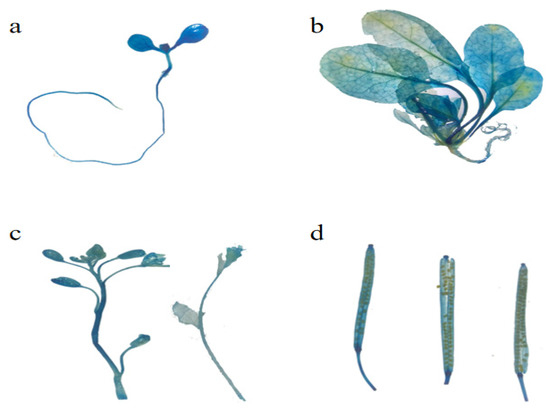
Figure 6.
GUS active expression of EuSRPP6 promoter at different development stages in transgenic Arabidopsis. (a) Whole plant of seedling in cotyledon stage; (b) whole plant in true leaf stage; (c) inflorescence in sexual reproduction stage; (d) seed pod in sexual reproduction stage.
EuSRPP7 (Figure 7): GUS staining could be detected in leaves, hypocotyls, and roots of seedlings at the cotyledon stage (Figure 7a). With the growth of plants, GUS staining was mainly concentrated in the veins and petioles, while the root activity was relatively shallow (Figure 7b). At the late stage of plant growth, GUS staining was obvious in inflorescence, flowers, stem lateral leaves, and mature pods (Figure 7c,d). Summarizing the above results, we can roughly infer that the EuSRPP7 promoter is primarily expressed in the veins, petioles, and mature gonads, with a small amount of active expression in the roots.
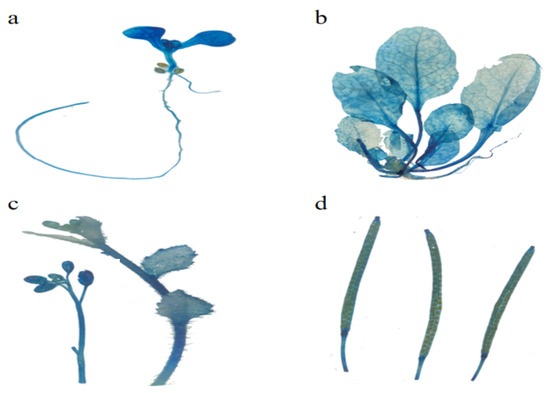
Figure 7.
GUS active expression of EuSRPP7 promoter at different development stages in transgenic Arabidopsis. (a) Whole plant of seedling in cotyledon stage; (b) whole plant in true leaf stage; (c) inflorescence in sexual reproduction stage; (d) seed pod in sexual reproduction stage.
3.5. Activity Analysis of EuSRPP Promoters under Methyl Jasmonic Acid (MeJA), Gibberellin (GA3), and Drought Treatment
According to the previous prediction of cis-acting elements in the EuSRPP promoters, we understood that the EuSRPP1, 3, 4, 5, 6, and 7 gene promoters may be greatly regulated by plant hormones, so we selected the hormone regulatory elements jasmonic acid methyl ester (MeJA), gibberellin (GA3), and drought response elements shared by the six promoters for induction activity analysis. Six EuSRPP promoters in transgenic Arabidopsis plants were sprayed with a solution containing 1 mmol/L MeJA, 1 mmol/L GA3, and 20% PEG6000. After treatment at 0, 3, 6, 12, and 24 h, the activity expression of the promoters was analyzed by measuring GUS enzyme activity. The concrete experimental results are shown in Figure 8.
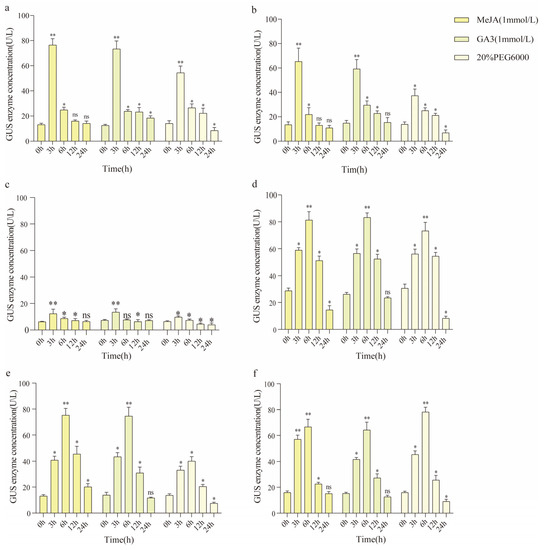
Figure 8.
GUS enzymatic activity of EuSRPP1, 3, 4, 5, 6, and 7 promoters treated with (1 mmol/L) MeJA, (1 mmol/L) GA3, and (20%) PEG6000. (a) EuSRPP1 GUS enzymatic activity; (b) EuSRPP3 GUS enzymatic activity; (c) EuSRPP4 GUS enzymatic activity; (d) EuSRPP5 GUS enzymatic activity; (e) EuSRPP6 GUS enzymatic activity; (f) EuSRPP7 GUS enzymatic activity. “**” Represents a significant difference compared to before treatment (p < 0.01); “*” represents a significant difference compared to before treatment (p < 0.05); “ns” represents no significant difference.
For the treatment of endogenous hormone signaling molecules, the GUS activity expression trends of the six EuSRPP promoters were divided into two cases. The first type was the transgenic plant that increases with time after treatment with MeJA and GA3. At 3 h, the GUS activity of EuSRPP1, EuSRPP3, and EuSRPP4 increases significantly, peaking at this time and then decreasing gradually; however, EuSRPP5, EuSRPP6, and EuSRPP7 showed the strongest activity after 6 h of treatment, and they decreased after 6 h. Using 0 h as a control, the GUS enzyme activity of EuSRPP1, EuSRPP3, and EuSRPP4 transgenic Arabidopsis increased by 4.86, 3.86, and 0.95 times after MeJA treatment for 3 h, while GA3 treatment increased by 4.92 times, 3.01 times, and 0.82 times, respectively. The GUS enzyme activity of the EuSRPP5, EuSRPP6, and EuSRPP7 transgenic Arabidopsis plants increased by 1.68, 4.78, and 3.20 times, respectively, after 6 h of MeJA treatment, and by 2.30, 4.40, and 3.22 times, respectively, after GA3 treatment. Therefore, it is determined that MeJA and GA3 were inducers of the EuSRPP gene promoters, and the GUS activity of the six EuSRPP promoters was induced by MeJA (1 mmol/L) and GA3 (1 mmol/L) and showed a trend of first increasing and then decreasing.
For drought treatment, the GUS enzyme activity of six EuSRPP promoters in transgenic Arabidopsis plants treated with 20% PEG6000 also show a trend of first increasing and then decreasing over time. Similarly, after treatment, EuSRPP1, EuSRPP3, and EuSRPP4 exhibited the highest GUS enzyme activity at 3 h, while EuSRPP5, EuSRPP6, and EuSRPP7 also exhibit the highest enzyme activity at 6 h. The GUS enzyme activity of Arabidopsis plants with EuSRPP1, EuSRPP3, and EuSRPP4 promoters significantly increased by 2.89, 1.86, and 0.40 times when treated for 3 h compared to 0 h, respectively; EuSRPP5, 6, and 7 showed a significant increase of 1.57, 1.94, and 3.82 times at 6 h. The results suggest that drought stress can induce the expression of GUS activity in the EuSRPP gene promoter.
The GUS enzyme activity of transgenic Arabidopsis plants with EuSRPP4 promoter before and after treatment was lower than that of the other five promoters, indicating that the expression regulation of EuSRPP4 gene in the plant may not be very significant; EuSRPP5 had the highest overall GUS enzyme activity. When untreated, GUS enzyme activity was above 20 U/L. As MeJA, GA3, and drought treatment time increased, GUS activity continued to increase and began to decrease after 6 h. This indicates that EuSRPP5 had a strong expression effect in plants, and its activity is greatly influenced by external stimuli. EuSRPP1 and EuSRPP3 showed the most rapid changes in activity under the regulation of the hormones MeJA and GA3. The overall expression activity of the EuSRPP1, EuSRPP5, and EuSRPP7 promoters was strongly influenced by this regulation in the drought environment induced by 20% PEG6000.
Based on the above results, it was demonstrated that endogenous hormones MeJA and GA3, and drought regulation, can all drive GUS activity, further indicating that the promoter activity of the EuSRPP gene was regulated by hormones and drought pathways.
4. Discussion
Eucommia ulmoides is widely distributed in China, but the low yield of E. ulmoides gum compared to that of rubber trees limits its use in practical production. Eucommia ulmoides gum has a duality of rubber and plastic. Based on the advantages of thermoplasticity, thermoelasticity, and rubber elasticity of E. ulmoides gum [25], it can be used in aerospace, national defense, medical treatment, transportation, construction, and other fields, with wide coverage and broad application prospects. In the plant kingdom, there are few plants that can synthesize TPI, such as E. ulmoides, balata, pistachio, sophora flavescens, and others. At present, the plants that produce trans-rubber in the scientific community are mainly concentrated in E. ulmoides, and there are few reports on other plants [26]. Previous research results showed that the REF/SRPP family is crucial for the biosynthesis and stability of rubber particles [27]. This study obtained the sequence of the EuSRPP gene promoters through specific experiments, predicted the presence of cis-acting elements using bioinformatics, measured the expression activity of the EuSRPP gene promoters, and further analyzed the expression of the EuSRPP gene to explore the regulatory mechanism. We obtain the promoter sequences for genes EuSRPP1, 3, 4, 5, 6, and 7 via genome walking and PCR amplification techniques, but EuSRPP2 was not successfully amplified. The reason for this may be that the secondary structure of its sequence is relatively complex, making it difficult to amplify the entire sequence and requiring segmented cloning and recombination. Another issue is that the sequence contains highly GC-rich fragments that can easily form anticrossings or hairpin structures, making template denaturation more difficult. Finally, during the experimental run, the sequence could not be amplified due to sub-optimal amplification of the primers and conditions, or the presence of excess ethanol in the sample DNA.
The promoters EuSRPP1, 3, 4, 5, 6, and 7 were mainly found in the leaves of transgenic Arabidopsis at various growth stages, with the difference being that EuSRPP6 and EuSRPP7 had higher promoter activity in the petiole than in the leaves. EuSRPP4 had a lower overall promoter activity, while EuSRPP5 promoter had a higher activity throughout the entire growth period. The activity of EuSRPP4 and EuSRPP6 promoters in the flowering stem was lower than that of other promoters. However, the common feature of these six EuSRPP promoters is that their expression activity was relatively stable in sexual reproductive organs, especially in pollinated flowers and mature seed pods. The expression sites of the six EuSRPP promoters in Arabidopsis agree with the main synthetic sites of E. ulmoides gum, and they are stable in the leaves, while expression levels are low in the roots. It can be inferred that there may be transcription factors that activate the promoter in the leaves of the EuSRPP gene, while the expression of the promoter is suppressed in the roots. To verify this inference, follow-up experiments are needed. The above results indicate that the activity of EuSRPP promoters varies in different parts of the plants. Related studies had shown that the TkSRPP3, TkSRPP4, and TkSRPP5 genes of Hevea brasiliensis are expressed in latex, leaves, flowers, petioles, and roots, but their expression levels are relatively high in latex [28,29]. Our results bear similarities to theirs.
Various studies had shown that the biosynthesis of E. ulmoides gum is not a single straight chain process but the result of a combination of multiple pathways and related factors. Methyl jasmonic acid (MeJA) was found to be one of the most important plant hormones that regulate the main mechanism of plants under normal and stress conditions [30]. The transcriptional sites upstream of the six EuSRPP gene promoter sequences obtained in this experiment all contain significant MeJA response elements. When transgenic Arabidopsis was treated with 1 mmol/L MeJA, the activity of EuSRPP1, 3, 4, 5, 6, and 7 increased and then decreased. EuSRPP1, 3, and 4 exhibited the highest expression activity at 3 h of treatment, while EuSRPP5, 6, and 7 exhibited the highest promoter expression level at 6 h of treatment. One research result showed it was found that after MeJA treatment, the expression level of SRPP gene in Brazilian rubber trees was significantly higher than before treatment, with the highest SRPP gene expression level reached after 8 h of treatment [31]. Some researchers had analyzed the transcriptome data and showed that MeJA could induce the up-regulated expression of TkSRPP3 and 4 genes in Hevea brasiliensis; QRT PCR analysis showed that the expression level of the TkSRPP4 gene increased by 4.3- and 8.9-fold at 6 and 24 h, respectively [32]. These studies indicate that the expression of the SRPP gene in different species is induced by methyl jasmonic acid, which then affect the synthesis of rubber.
Gibberellin (GA3) is a plant growth regulator that can promote crop growth and development and facilitate the accumulation of metabolites [33]. Previous studies had found that the growth rate of aucubin content in the leaves of E. ulmoides reaches its maximum on the first day after hormone spraying, and that 50 mg/L of GA3 after treatment is significantly higher than that in the control; the content growth rates of geniposide and chlorogenic acid reached their maximum on the 19th day after spraying 50 mg/L GA3 [34]. Modern plant physiology research has proved that GA3 can promote the synthesis of auxin (IAA) and inhibit the activity of auxin oxidase. This feature may increase the amount of EUG by increasing the endogenous hormone content in E. ulmoides. After spraying GA3 on E. ulmoides leaves, the gum content in the leaves increased, and the molecular weight and molecular weight distribution of the gum increased [35]. In this study, EuSRPP transgenic Arabidopsis was treated with a 1 mmol/L GA3 spray, and the GUS activity of the EuSRPP1, 3, 4, 5, 6, and 7 gene promoters was found to increase over a certain period of time compared to the activity in the untreated sample. The difference was that EuSRPP1, 3, and 4 reached their strongest activity at 3 h of treatment, and EuSRPP5, 6, and 7 had the highest GUS activity at 6 h. Among these six EuSRPP promoters, EuSRPP1 was the most sensitive to GA3, and the GUS activity increased 4.92 times compared to 0 h after treatment for 3 h. This may relate to the fact that EuSRPP1 contains more GA3 response elements than other promoters, and more auxin elements. Based on the results, it can be concluded that GA3 has a significant regulatory effect on the expression of the EuSRPP gene, which varies due to structural differences in the gene itself.
We simulated drought stress in transgenic Arabidopsis with 20% PEG6000 and found that EuSRPP1, 3, and 4 had the highest GUS enzyme activity at 3 h. EuSRPP1 had a 2.89-fold increase in GUS enzyme activity compared to 0 h, which is larger than the increases in EuSRPP3 and 4. EuSRPP5, 6, and 7 had the highest GUS enzyme activity at 6 h, which is 3.82 times higher than that of EuSRPP7 at 0 h and more than that of EuSRPP5 and 6. In addition, the GUS activity of the six EuSRPP promoters showed a trend of first increasing and then decreasing under drought stress. The difference was that the activity of EuSRPP1, 5, and 7 was greatly regulated by drought. The above results indicated that under drought stress, the EuSRPP gene had strong stress tolerance, and there were differences in the activity expression of the EuSRPP gene promoters, which may be caused by the structure of the gene itself, and it was speculated that the up-regulation of the EuSRPP gene in stress ensures the synthesis of gum. This conclusion was in line with some previous findings. For example, researchers had transferred SRPP into tobacco and induced drought, and the results also indicated that the SRPP gene has a role in enhancing plant drought tolerance [36]. Overexpressed CaSRP1 also showed stronger drought resistance in Arabidopsis [37].
5. Conclusions
In this study, the promoter sequence of EuSRPP gene was amplified, cis-acting elements and transcription start sites were predicted, and T3 transgenic Arabidopsis homozygous lines were obtained by Agrobacterium-mediated genetic transformation of Arabidopsis. The expression pattern of EuSRPP promoters in transgenic Arabidopsis and its expression under methyl jasmonate, gibberellin, and PEG6000 drought treatment were analyzed. The conclusion drawn is as follows: EuSRPP1, 3, 4, 5, 6, and 7 promoter activity is mainly expressed in leaves. At the same time, there is expression activity in inflorescences, open flowers, pollen, and mature seed pods. The six EuSRPP promoters have almost no activity in mature roots. Under the treatment of methyl jasmonic acid (1 mmol/L MeJA), gibberellin (1 mmol/L GA3), and drought (20% PEG6000), the activity expression of six EuSRPP promoters first increased and then decreased. The difference, however, is that the expression activity of EuSRPP1, 3, and 4 peaks at 3 h of plant treatment, while EuSRPP5, 6, and 7 showed their strongest expression activity at 6 h of treatment. Based on all the experimental results, it is evident that the expression sites of the six EuSRPP promoters are in relatively good agreement. Second, differences in the expression activity of six EuSRPP gene promoters are observed under MeJA, GA3, and drought treatment, suggesting that the activity of EuSRPP gene promoters in Eucommia ulmoides is regulated by endogenous hormones and drought pathways. China is a big consumer of natural rubber, so it is of great significance to study the specific expression regulation mechanism of the EuSRPP gene at the promoters level, and then to lay a foundation for exploring the specific synthesis pathway of E. ulmoides gum, which is of great significance to alleviate the serious shortage of natural rubber resources in China and the sustainable and healthy development of China’s rubber industry.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae9080856/s1, File S1: Construction diagram of EuSRPP promoter plant expression vector; File S2: Sequence amplification experiment of EuSRPP3, 4, 5, 6, and 7; File S3: EuSRPP1 sequence amplification experiment and gel map; File S4: PCR Identification of EuSRPP Promoter Transgenic plant.
Author Contributions
L.L. and X.Y. conducted experimental designs and reviewed the manuscript. H.Z. carried out the experiments, analyzed the data, and wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Guizhou Province Science and Technology Planning Project, grant number Qiankehe Support (2021) General 111; Natural Science Foundation of China, grant number 3226180451; Education Department of Guizhou Province Scientific Research Program of Higher Education Institutions (Young Scientific Program), grant number Qianjiaoji (2022) No. 118; and Research Funds for Introduced Talents of Guizhou University, grant number Guizhou University renjihezi (2021) No. 34.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhu, M.Q.; Sun, R.C. Eucommia ulmoides Oliver: A Potential Feedstock for Bioactive Products. J. Agric. Food Chem. 2018, 66, 5433. [Google Scholar] [CrossRef]
- Xing, Y.-F.; He, D.; Wang, Y.; Zeng, W.; Zhang, C.; Lu, Y.; Su, N.; Kong, Y.-H.; Xing, X.-H. Chemical constituents, biological functions and pharmacological effects for comprehensive utilization of Eucommia ulmoides Oliver—ScienceDirect. Food Sci. Hum. Wellness 2019, 8, 12. [Google Scholar] [CrossRef]
- Wei, X.; Peng, P.; Peng, F.; Dong, J. Natural Polymer Eucommia Ulmoides Rubber: A Novel Material. J. Agric. Food Chem. 2021, 69, 3797–3821. [Google Scholar] [CrossRef]
- Li, R.; Huang, T.; Nie, L.; Jia, A.; Zhang, L.; Yuan, Y.; Hong, Y.; Wang, J.; Hu, X. Chemical Constituents from Staminate Flowers of Eucommia ulmoides Oliver and Their Anti-Inflammation Activity in Vitro. Chem. Biodivers. 2021, 18, e2100331. [Google Scholar] [CrossRef]
- Xiang, H.; Zheng, H.X.; Du, L.S.; Zhang, Z.J. The Accumulated Variation of the Active Ingredient in Eucommia ulmoides Leaves. J. Anhui Agric. Sci. 2016, 44, 182–183. [Google Scholar]
- Cao, R.; Deng, L.; Feng, Z.; Zhao, X.; Li, X.; Zhang, L. Preparation of natural bio-based Eucommia ulmoides gum/styrene-butadiene rubber composites and the evaluation of their damping and sound absorption properties. Polymer 2021, 213, 123292. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, H.; Ma, C.; Han, H.; Sun, R.; Xie, M. Binary Modification of Eucommia ulmoides Gum Toward Elastomer with Tunable Mechanical Properties and Good Compatibility. J. Polym. Sci. Part A Polym. Chem. 2019, 57, 1247–1255. [Google Scholar] [CrossRef]
- Tsukada, G.; Kato, R.; Tokuda, M. Intraoral Temperature Triggered Shape-Memory Effect and Sealing Capability of A Transpolyisoprene-Based Polymer. Polymers 2015, 7, 2259–2275. [Google Scholar] [CrossRef]
- Qi, X.; Zhao, X.; Li, Y.; Zhang, J.; Zhang, L.; Yue, D. A high toughness elastomer based on natural Eucommia ulmoides gum. J. Appl. Polym. Sci. 2021, 138, 50007. [Google Scholar] [CrossRef]
- Gao, R.; Zhang, H.; Li, B.; Guo, H.; Li, H.; Xiong, L.; Chen, X. Extraction ofEucommia ulmoidesgum and microbial lipid fromEucommia ulmoides Oliverleaves by dilute acid hydrolysis. Biotechnol. Lett. 2023, 45, 619–628. [Google Scholar] [CrossRef]
- Wuyun, T.-N.; Wang, L.; Liu, H.; Wang, X.; Zhang, L.; Bennetzen, J.L.; Li, T.; Yang, L.; Liu, P.; Du, L.; et al. The Hardy Rubber Tree Genome Provides Insights into the Evolution of Polyisoprene Biosynthesis. Mol. Plant 2018, 11, 429. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, Y.; Takeda, T.; Suzuki, N.; Hayashi, T.; Harada, Y.; Bamba, T.; Kobayashi, A. Histochemical study of trans-polyisoprene accumulation by spectral confocal laser scanning microscopy and a specific dye showing fluorescence solvatochromism in the rubber-producing plant, Eucommia ulmoides Oliver. Planta 2013, 238, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Bamba, T.; Murayoshi, M.; Gyoksen, K.; Nakazawa, Y.; Okumoto, H.; Katto, H.; Fukusaki, E.; Kobayashi, A. Contribution of Mevalonate and Methylerythritol Phosphate Pathways to Polyisoprenoid Biosynthesis in the Rubber-Producing Plant Eucommia ulmoides Oliver. Z. Naturforschung C 2010, 65, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Collins-Silva, J.; Nural, A.T.; Skaggs, A.; Scott, D.; Hathwaik, U.; Woolsey, R.; Schegg, K.; McMahan, C.; Whalen, M.; Cornish, K.; et al. Altered levels of the Taraxacum kok-saghyz (Russian dandelion) small rubber particle protein, TkSRPP3, result in qualitative and quantitative changes in rubber metabolism. Phytochemistry 2012, 79, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Ponciano, G.; McMahan, C.M.; Xie, W.; Lazo, G.R.; Coffelt, T.A.; Collins-Silva, J.; Nural-Taban, A.; Gollery, M.; Shintani, D.K.; Whalen, M.C. Transcriptome and gene expression analysis in cold-acclimated guayule (Parthenium argentatum) rubber-producing tissue. Phytochemistry 2012, 79, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Ran, X.; Liu, Y.; Zhao, D. The relationship between EuSRPP1 gene expression and rubber biosynthesis in Eucommia ulmoides Oliver (Du-zhong). Ind. Crops Prod. 2022, 175, 114246. [Google Scholar] [CrossRef]
- Wang, Y. Study on the Effect of Eucommia ulmoides Small Rubber Granular Protein EuSRPP3 Gene on the Molecular Size of Eucommia ulmoides Gum; Guizhou University: Guizhou, China, 2021. [Google Scholar]
- Liu, L.; Gallagher, J.; Arevalo, E.D.; Chen, R.; Skopelitis, T.; Wu, Q.; Bartlett, M.; Jackson, D. Enhancing grain-yield-related traits by CRISPR–Cas9 promoter editing of maize CLE genes. Nat. Plants 2021, 7, 477. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, G.; Liu, S.; Guo, X.; Zheng, C. Isolation and functional analysis of a strong specific promoter in photosynthetic tissues. Sci. China Ser. C-Life Sci. 2003, 46, 651–660. [Google Scholar] [CrossRef]
- Li, H.-L.; Qu, L.; Guo, D.; Wang, Y.; Zhu, J.-H.; Peng, S.-Q. Histone deacetylase interacts with a WRKY transcription factor to regulate the expression of the small rubber particle protein gene from Hevea brasiliensis. Ind. Crops Prod. 2019, 145, 111989. [Google Scholar] [CrossRef]
- Julia, F.; Andrea, H.; Twyman, R.M.; Prufer, D.; Gronover, C.S. Abscisic Acid-Dependent Regulation of Small Rubber Particle Protein Gene Expression in Taraxacum brevicorniculatum is Mediated by TbbZIP1. Plant Cell Physiol. 2013, 54, 448–464. [Google Scholar] [CrossRef]
- Bröker, J.N.; Laibach, N.; Müller, B.; Prüfer, D.; Gronover, C.S. Taraxacum brevicorniculatum rubber elongation factor TbREF associates with lipid droplets and affects lipid turn-over in yeast. Biotechnol. Rep. 2018, 20, e00290. [Google Scholar] [CrossRef] [PubMed]
- Cornish, W.K. Microstructure of Purified Rubber Particles. Int. J. Plant Sci. 2000, 161, 435. [Google Scholar] [CrossRef]
- Teng, F.; Wu, J.; Su, B.; Wang, Y. High-speed tribological properties of Eucommia ulmoides gum/natural rubber blends: Experimental and molecular dynamics simulation study. Tribol. Int. 2022, 171, 107542. [Google Scholar] [CrossRef]
- Peters, B.; Casey, J.; Aidley, J.; Zohrab, S.; Borg, M.; Twell, D.; Brownfield, L. A Conserved cis-Regulatory Module Determines Germline Fate through Activation of the Transcription Factor DUO1 Promoter. Plant Physiol. 2017, 173, 280–293. [Google Scholar] [CrossRef]
- Lin, T.; Xu, X.; Ruan, J.; Liu, S.; Wu, S.; Shao, X.; Wang, X.; Gan, L.; Qin, B.; Yang, Y.; et al. Genome analysis of Taraxacum kok-saghyz Rodin provides new insights into rubber biosynthesis. Natl. Sci. Rev. 2018, 5, 78–87. [Google Scholar] [CrossRef]
- Wang, D.; Xie, Q.; Sun, Y.; Tong, Z.; Chang, L.; Yu, L.; Zhang, X.; Yuan, B.; He, P.; Jin, X.; et al. Proteomic Landscape Has Revealed Small Rubber Particles Are Crucial Rubber Biosynthetic Machines for Ethylene-Stimulation in Natural Rubber Production. Int. J. Mol. Sci. 2019, 20, 5082. [Google Scholar] [CrossRef]
- Huang, L.; Huang, J.; Jiang, Y.C.; Zhang, H.; Ni, J. Cloning and bioinformatics analysis of the SRPP5 gene from Hevea brasiliensis. Mol. Plant Breed. 2020, 18, 4624–4628. [Google Scholar]
- Zhang, R.Z.; Jiang, Y.C.; Huang, J.; Yan, J. Cloning and expression analysis of SRPP2 gene from Hevea brasiliensis. Biotechnol. Bull. 2020, 36, 9–14. [Google Scholar]
- Esmaielzadeh, S.; Fallah, H.; Niknejad, Y.; Mahmoudi, M.; Tari, D.B. Methyl jasmonate increases aluminum tolerance in rice by augmenting the antioxidant defense system, maintaining ion homeostasis, and increasing nonprotein thiol compounds. Environ. Sci. Pollut. Res. 2022, 29, 46708–46720. [Google Scholar] [CrossRef]
- Wang, Y.; Kang, G.; Li, Y.; Qin, H.; Zeng, R. Expression Analysis of Key Genes REF, SRPP, HRT1 and HRT2 Related to Rubber Biosynthesis of Hevea brasiliensis. Genom. Appl. Biol. 2018, 37, 3933–3943. [Google Scholar]
- Huang, J.; Jiang, Y.; Zhang, Y.; Yan, J. Genome-wide identification and expressional analysis of SRPP/REF gene family in Taraxacum kok-saghyz. Zhiwu Shengli Xuebao/Plant Physiol. J. 2020, 56, 1541–1552. [Google Scholar] [CrossRef]
- Wen, B.; Song, W.; Sun, M.; Chen, M.; Mu, Q.; Zhang, X.; Wu, Q.; Chen, X.; Gao, D.; Wu, H. Identification and characterization of cherry (Cerasus pseudocerasus G. Don) genes responding to parthenocarpy induced by GA3 through transcriptome analysis. BMC Genet. 2019, 20, 58. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Su, Y.; Peng, J.; Li, D.; He, W.; Zhang, Q. The Effect of Exogenous Hormones on Content and Molecular Weight of Eucommia ulmoides Gum. J. Northwest For. Univ. 2008, 23, 184–188. [Google Scholar] [CrossRef]
- Peng, J.N.; Su, Y.Q.; Sun, Q.S.; Yue, J.; Yoshihisa, N.; Sun, R.C. Content and molecular-weight distribution of EU rubber from Eucommia ulmoides leaves. For. Prod. J. 2007, 57, 65–67. [Google Scholar] [CrossRef]
- Laibach, N.; Schmidl, S.; Müller, B.; Bergmann, M.; Prüfer, D.; Gronover, C.S. Small rubber particle proteins from Taraxacum brevicorniculatum promote stress tolerance and influence the size and distribution of lipid droplets and artificial poly(cis -1,4-isoprene) bodies. Plant J. Cell Mol. Biol. 2018, 93, 1045. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.Y.; Seo, Y.S.; Lee, H.; Kim, W.T. Constitutive expression of CaSRP1, a hot pepper small rubber particle protein homolog, resulted in fast growth and improved drought tolerance in transgenic Arabidopsis plants. Planta 2010, 232, 71–83. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).