Elucidating Quadruplication Event of PHO1 Gene: A Key Regulator of Plant Phosphate Translocation in Brassica rapa
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Bacterial Artificial Chromosome (BAC)
2.2. Large-Construct BAC DNA Extraction
2.3. BAC DNA Precipitation
2.3.1. Preparation of Sequencing Library for MinION Sequencing
2.3.2. MinION Sequencing
2.3.3. MinION Data Analyses
2.4. Identification and Characterization of PHO1 Homolog Genes
2.4.1. Reverse Transcription Polymerase Chain Reaction (RT-PCR)
2.4.2. Cloning of PHO1 Transcripts
2.4.3. Restriction Analysis
2.4.4. Plasmid Sequencing
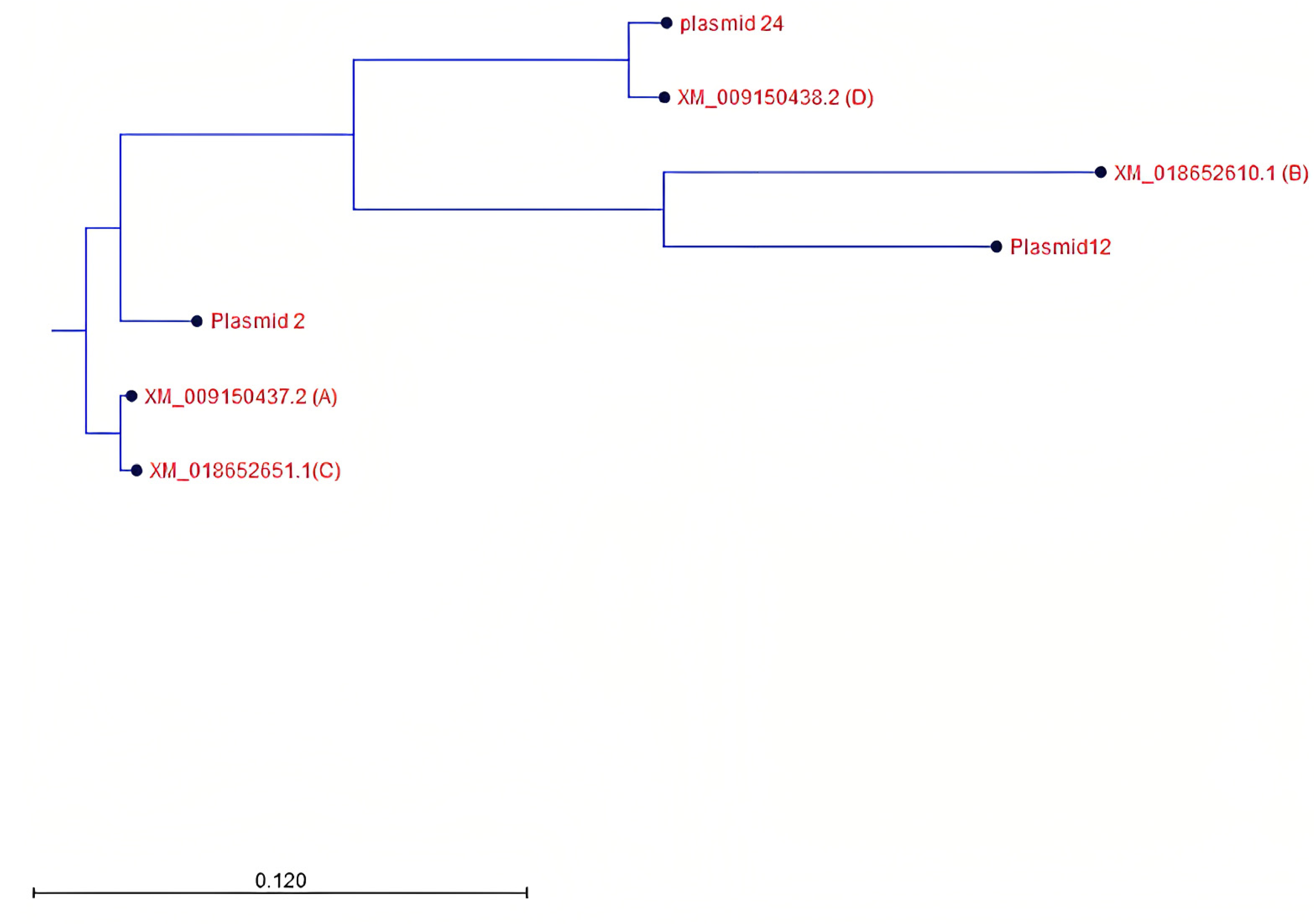
2.4.5. Phylogenetic Tree Construction
3. Results
3.1. PHO1 at Trans e-QTL Hotspots on Chromosome A06 B. rapa
3.2. Candidate Genes Underlying Trans-eQTL Hotspots
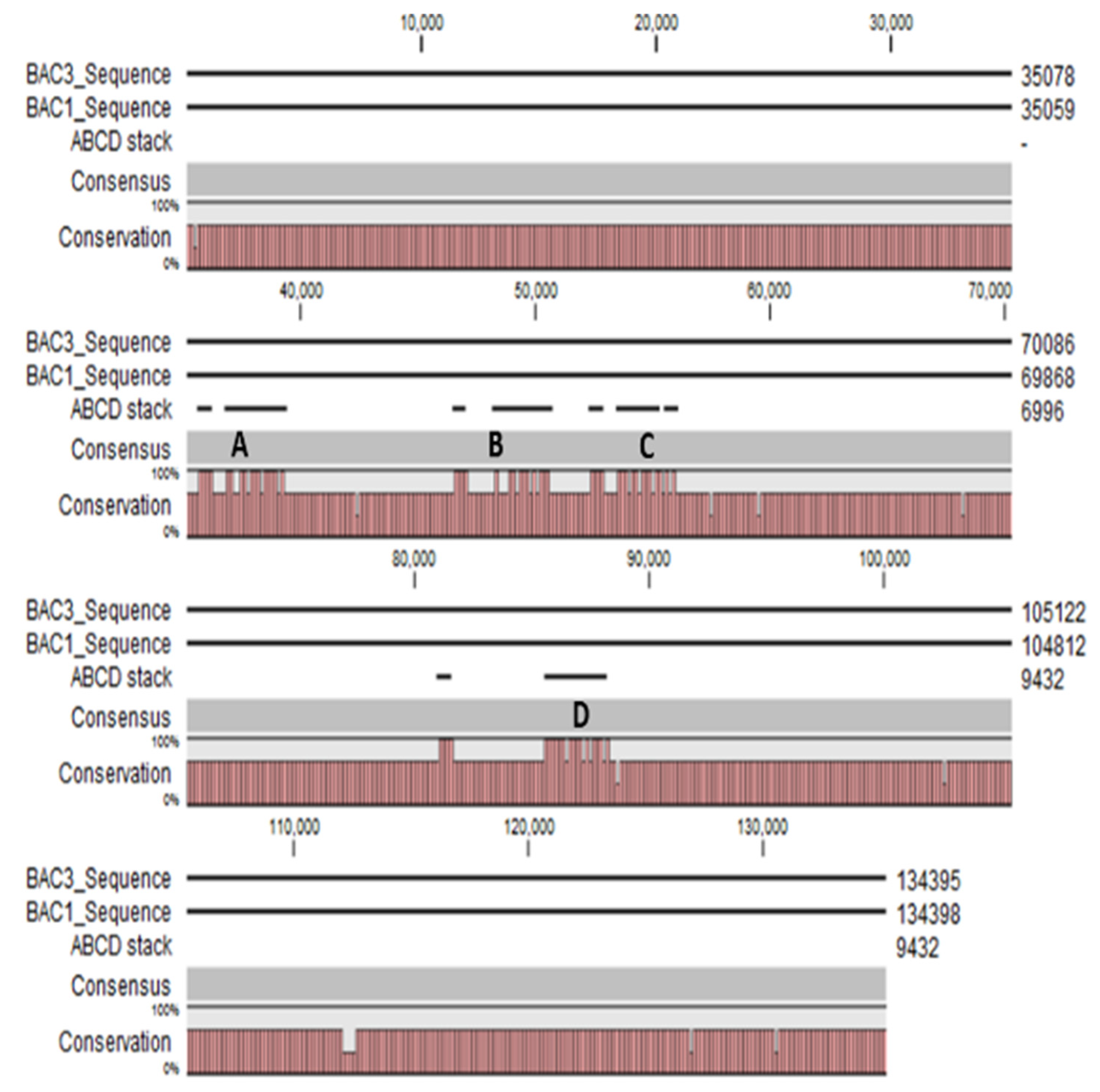
3.3. BAC DNA Sequencing Using MinION Sequencer from Oxford Nanopore Technologies
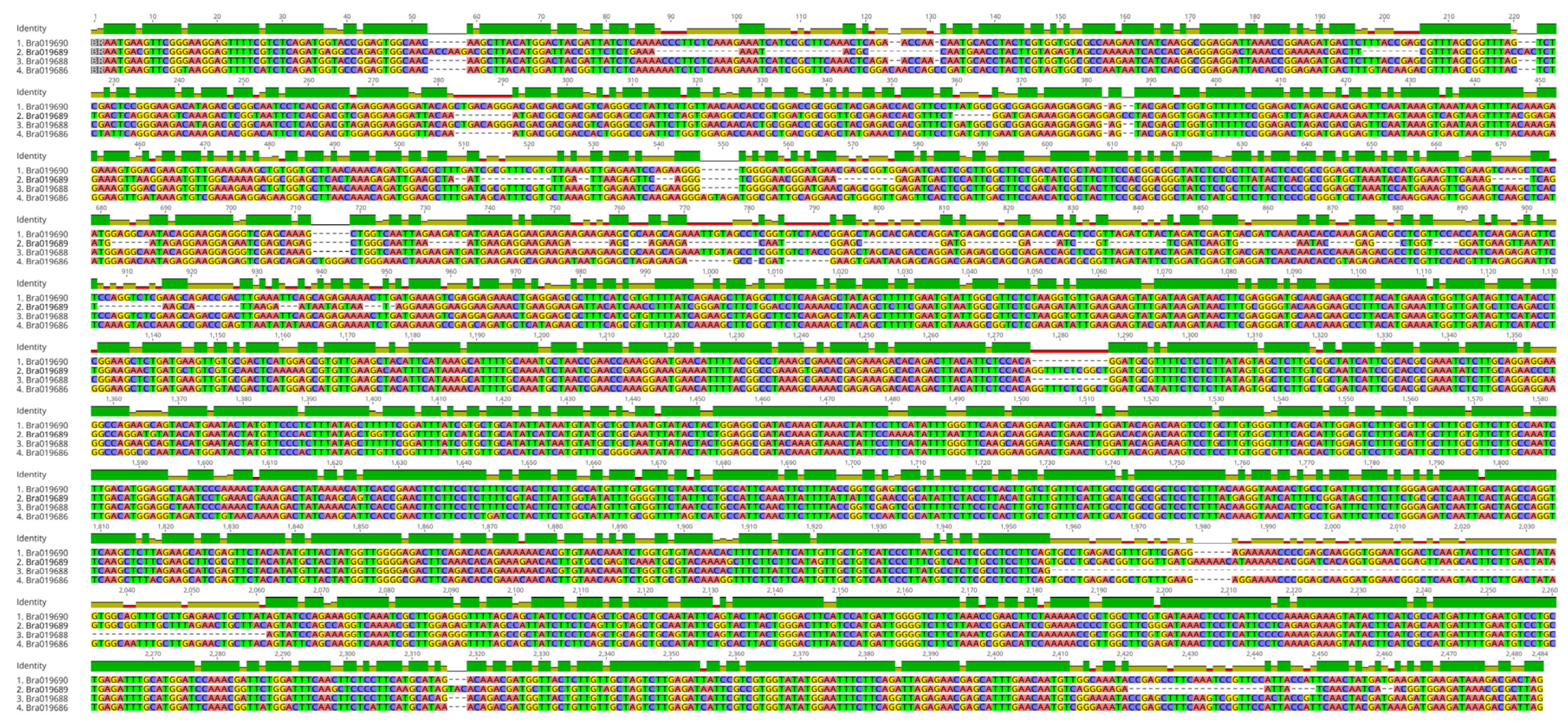
3.4. Identification and Characterization of PHO1 Homolog Transcripts
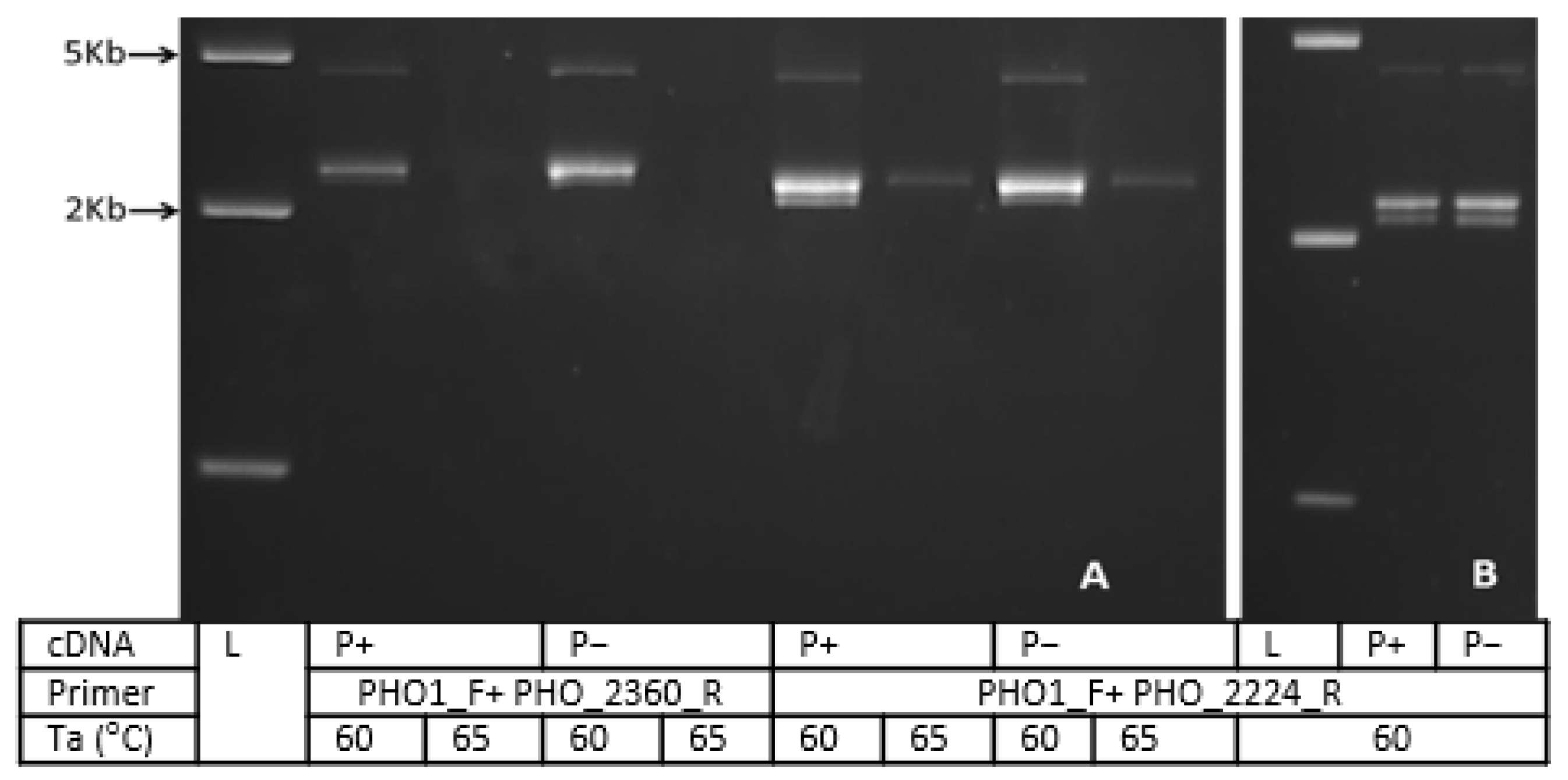
3.5. Colony PCR and Restriction Analysis
4. Discussion
4.1. Potential for Alternatively Spliced PHO1 Transcripts
4.2. Potential Structural Variation in PHO1 Paralogs
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cordell, D.; Drangert, J.O.; White, S. The story of phosphorus: Global food security and food for thought. Glob. Environ. Chang. 2009, 19, 292–305. [Google Scholar] [CrossRef]
- Vance, C.P.; Uhde-Stone, C.; Allan, D.L. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef] [PubMed]
- Schachtman, D.P.; Reid, R.J.; Ayling, S.M. Phosphorus uptake by plants: From soil to cell. Plant Physiol. 1998, 116, 447–453. [Google Scholar] [CrossRef] [PubMed]
- White, P.J.; Hammond, J.P. The sources of phosphorus in the waters of Great Britain. J. Environ. Qual. 2009, 38, 13–26. [Google Scholar] [CrossRef]
- Savci, S. Investigation of effect of chemical fertilizers on environment. APCBEE Procedia 2012, 1, 287–292. [Google Scholar] [CrossRef]
- Schindler, D.W.; Carpenter, S.R.; Chapra, S.C.; Hecky, R.E.; Orihel, D.M. Reducing phosphorus to curb lake eutrophication is a success. Environ. Sci. Technol. 2016, 50, 8923–8929. [Google Scholar] [CrossRef]
- Cooper, J.; Lombardi, R.; Boardman, D.; Carliell-Marquet, C. The future distribution and production of global phosphate rock reserves. Resour. Conserv. Recycl. 2011, 57, 78–86. [Google Scholar] [CrossRef]
- Koppelaar, R.H.E.M.; Weikard, H.P. Assessing phosphate rock depletion and phosphorus recycling options. Glob. Environ. Chang. 2013, 23, 1454–1466. [Google Scholar] [CrossRef]
- Wang, X.R.; Shen, J.B.; Liao, H. Acquisition or utilization, which is more critical for enhancing phosphorus efficiency in modern crops? Plant Sci. 2010, 179, 302–306. [Google Scholar] [CrossRef]
- Chiou, T.J.; Lin, S.I. Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 2011, 62, 185–206. [Google Scholar] [CrossRef] [PubMed]
- Ramaiah, M.; Jain, A.; Baldwin, J.C.; Karthikeyan, A.S.; Ragothama, K.G. Characterization of the phosphate starvation-induced glycerol-3-phosphate permease gene family in Arabidopsis. Plant Physiol. 2011, 157, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Rubio, V.; Linhares, F.; Solano, R.; Martin, A.C.; Iglesias, J.; Leyva, A.; Paz-Ares, J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133. [Google Scholar] [CrossRef] [PubMed]
- Hamburger, D.; Rezzonico, E.; Petétot, J.M.-C.; Somerville, C.; Poirier, Y. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 2002, 14, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Mlodzinska, E.; Zboinska, M. Phosphate uptake and allocation—A closer look at Arabidopsis thaliana L. and Oryza sativa L. Front. Plant Sci. 2016, 7, 1198. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.Y.; Huang, T.K.; Chiou, T.J. Nitrogen limitation adaptation, a target of MicroRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 2013, 10, 4061–4074. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhou, J.; Zhou, H.J.; Wang, M.M.; Liu, M.M.; Ke, Y.Z.; Li, P.F.; Li, J.N.; DU, H. Global survey and expression of the phosphate transporter gene families in Brassica napus and their roles in phosphorus response. Int. J. Mol. Sci. 2020, 21, 1752. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ribot, C.; Rezzonico, E.; Poirier, Y. Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol. 2004, 135, 400–411. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, H.; Zhu, W.; Yan, L.; Chen, Q.; He, C. The PHOSPHATE1 genes participate in salt and Pi signaling pathways and play adaptive roles during soybean evolution. BMC Plant Biol. 2019, 19, 353. [Google Scholar] [CrossRef] [PubMed]
- Secco, D.; Wang, C.; Arpat, B.A.; Wang, Z.; Poirier, Y.; Tyerman, S.D.; Wu, P.; Shou, H.; Whelan, J. The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 2012, 193, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.F.; Li, L.Q.; Xu, Q.; Kong, Y.H.; Wang, H.; Wu, W.H. The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell 2009, 21, 3554–3566. [Google Scholar] [CrossRef]
- Wege, S.; Khan, G.A.; Jung, J.-Y.; Vogiatzaki, E.; Pradervand, S.; Aller, I.; Meyer, A.J.; Poirier, Y. The EXS Domain of PHO1 participates in the response of shoots to phosphate deficiency via a root-to-shoot signal. Plant Physiol. 2016, 170, 385–400. [Google Scholar] [CrossRef] [PubMed]
- Arpat, A.B.; Magliano, P.; Wege, S.; Rouached, H.; Stefanovic, A.; Poirier, Y. Functional expression of PHO1 to the Golgi and trans-Golgi network and its role in export of inorganic phosphate. Plant J. 2012, 71, 479–491. [Google Scholar] [CrossRef]
- Stefanovic, A.; Ribot, C.; Rouached, H.; Wang, Y.; Chong, J.; Belbahri, L.; Delessert, S.; Poirier, Y. Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to the shoot, and are regulated by phosphate deficiency via distinct pathways. Plant J. 2007, 50, 982–994. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Zhao, M.; Wang, Y.; Gai, J.; He, C. Phylogeny, structural evolution and functional diversification of the plant PHOSPHATE1 gene family: A focus on Glycine max. BMC Evol. Biol. 2013, 13, 103. [Google Scholar] [CrossRef] [PubMed]
- Hammond, J.P.; Mayes, S.; Bowen, H.C.; Graham, N.S.; Hayden, R.M.; Love, C.G.; Spracklen, W.P.; Wang, J.; Welham, S.J.; White, P.J.; et al. Regulatory hotspots are associated with plant gene expression under varying soil phosphorus supply in Brassica rapa. Plant Physiol. 2011, 156, 1230–1241. [Google Scholar] [CrossRef] [PubMed]
- Kliebenstein, D. Quantitative genomics: Analyzing intraspecific variation using global gene expression polymorphisms or eQTLs. Annu. Rev. Plant Biol. 2009, 60, 93–114. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; MillerI, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, T.; He, X.; Cai, X.; Lin, R.; Liang, J.; Wu, J.; King, G.; Wang, X. BRAD V3: An upgraded Brassicaceae database. Nucleic Acids Res. 2022, 50, D1432–D1441. [Google Scholar] [CrossRef] [PubMed]
- Druka, A.; Potokina, E.; Luo, Z.; Jiang, N.; Chen, X.; Kearsey, M.; Waugh, R. Expression quantitative trait loci analysis in plants. Plant Biotechnol. J. 2010, 8, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Iniguez-Lui, F.L.; Lukens, L.; Farnham, M.W.; Amasino, R.M.; Osborn, T.C. Development of public immortal mapping populations, molecular markers, and linkage maps for rapid cycling Brassica rapa and Brassica oleracea. Theor. Appl. Genet. 2009, 120, 31–43. [Google Scholar] [CrossRef]
- Mardis, E.R. DNA sequencing technologies: 2006–2016. Nat. Protoc. 2017, 12, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Adewale, B.A. Will long-read sequencing technologies replace short-read sequencing technologies in the next 10 years? Afr. J. Lab. Med. 2020, 9, 1340. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Blencowe, B.J. Alternative splicing: New insights from global analyses. Cell 2006, 126, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Mount, S.M.; Salzberg, S.L. A computational survey of candidate exonic splicing enhancer motifs in the model plant Arabidopsis thaliana. BMC Bioinform. 2007, 8, 159. [Google Scholar] [CrossRef]
- Shang, X.; Cao, Y.; Ma, L. Alternative splicing in plant genes: A means of regulating the environmental fitness of plants. Int. J. Mol. Sci. 2017, 18, 432. [Google Scholar] [CrossRef]
- Zhang, G.; Guo, G.; Hu, X.; Zhang, Y.; Li, Q.; Li, R.; Zhuang, R.; Lu, Z.; He, Z.; Fang, X.; et al. Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res. 2010, 20, 646–654. [Google Scholar] [CrossRef]
- Liu, N.; Shang, W.; Li, C.; Jia, L.; Wang, X.; Xing, G.; Zheng, W. Evolution of the SPX gene family in plants and its role in the response mechanism to phosphorus stress. Open Biol. 2018, 8, 170231. [Google Scholar] [CrossRef]
- Duan, K.; Yi, K.; Dang, L.; Huang, H.; Wu, W.; Wu, P. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 2008, 54, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Yang, C.; Ding, J.; Shi, L.; Xu, F. Genome wide identification and characterization in SPX do-main-containing members and their responses to Pi deficiency in Brassica napus. Front. Plant Sci. 2017, 8, 35. [Google Scholar] [CrossRef]
- Puga, M.I.; Mateos, I.; Charukesi, R.; Wang, Z.; Franco-Zorrilla, J.M.; De Lorenzo, L.; Irigoyen, M.L.; Masiero, S.; Bustos, R.; RodrÍguez, J.; et al. SPX1 is a phosphate-dependent inhibitor of PHOSPHATE STARVATION RESPONSE 1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14947–14952. [Google Scholar] [CrossRef] [PubMed]
- Rouached, H. Multilevel coordination of phosphate and sulfate homeostasis in plants. Plant Signal. Behav. 2011, 6, 952–955. [Google Scholar] [CrossRef]
- Wang, Z.; Ruan, W.; Shi, J.; Zhang, L.; Xiang, D.; Yang, C.; Li, C.; Wu, Z.; Liu, Y.; Yu, Y.; et al. Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. USA 2014, 111, 14953–14958. [Google Scholar] [CrossRef] [PubMed]
- Yoshitake, Y.; Yoshimoto, K. Intracellular phosphate recycling systems for survival during phosphate starvation in plants. Front. Plant. Sci. 2022, 13, 1088211. [Google Scholar] [CrossRef]
- Hammond, J.P.; Bennett, M.J.; Bowen, H.C.; Broadley, M.R.; Eastwood, D.C.; May, S.T.; Rahn, C.; Swarup, R.; Woolaway, K.E.; White, P.J. Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol. 2003, 132, 578–596. [Google Scholar] [CrossRef] [PubMed]
- Misson, J.; Raghothama, K.G.; Jain, A.; Jouhet, J.; Block, M.A.; Bligny, R.; Ortet, P.; Creff, A.; Somerville, S.; Rolland, N.; et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl Acad. Sci. USA 2005, 102, 11934–11939. [Google Scholar] [CrossRef]
- Morcuende, R.; Bari, R.; Gibon, Y.; Zheng, W.; Pant, B.D.; Blasing, O.; Usadel, B.; Czechowski, T.; Udvardi, M.K.; Stitt, M.; et al. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 2007, 30, 85–112. [Google Scholar] [CrossRef]
- Pan, Y.; Song, Y.; Zhao, L.; Chen, P.; Bu, C.; Liu, P.; Zhang, D. The genetic basis of phosphorus utilization efficiency in plants provide new insight into woody perennial plants improvement. Int. J. Mol. Sci. 2022, 23, 2253. [Google Scholar] [CrossRef] [PubMed]




| Barcode ID | Primer Sequence (5′-3′) | BAC ID |
|---|---|---|
| NB01 | AAGAAAGTTGTCGGTGTCTTTGTG | KBrB-029J08 |
| NB02 | TCGATTCCGTTTGTAGTCGTCTGT | KBrB-3E10 |
| NB03 | GAGTCTTGTGTCCCAGTTACCAGG | KBrB-63F11 |
| NB04 | TTCGGATTCTATCGTGTTTCCCTA | KBrH-38K12 |
| NB05 | CTTGTCCAGGGTTTGTGTAACCTT CCTA | KBrH-102C10 |
| Sequence (5′ to 3′) |
| NB01 | AAGAAAGTTGTCGGTGTCTTTGTG |
| NB02 | TCGATTCCGTTTGTAGTCGTCTGT |
| NB03 | GAGTCTTGTGTCCCAGTTACCAGG |
| NB04 | TTCGGATTCTATCGTGTTTCCCTA |
| NB05 | CTTGTCCAGGGTTTGTGTAACCTT CCTA |
| Expected size of cDNA (bp) |
| PHO1_A | 2224 |
| PHO1_B | 2044 |
| PHO1_C | 2152 |
| PHO1_D | 2227 |
| ID | BAC | Predicted BAC Size (bp) | BAC Length (bp) |
|---|---|---|---|
| 1 | KBrB-63F11 | 134,576 | 184,651 |
| 2 | KBrH-102C10 | 152,113 | 153,335 |
| 3 | KBrB-029J08 | 148,972 | 185,935 |
| 4 | KBrH-38K12 | 155,059 | 165,146 |
| 5 | KBrB-3E10 | 136,929 | 45,485 |
| Transcript No. (NCBI) | Plasmid | Nucleotide Length | Peptide Length | Closest BLASTN Hits | Closest BLASTP Hits | Predicted PHO1 |
|---|---|---|---|---|---|---|
| XM_009150437.2 | 2 | 2236 | 744 | XM_009150437.2 | XP_009148685.1 | A |
| XM_009150437.2 | 7 | 2150 | 552 | XM_009150437.2 | XP_018508167.1 | A |
| XM_009150437.2 | 23 | 2130 | 629 | XM_009150437.2 | XP_009148685.1 | A |
| XM_009150437.2 | 26 | 2317 | 746 | XM_009150437.2 | XP_009148685.1 | A |
| XM_009150437.2 | 22 | 2073 | 490 | XM_009150437.2 | XP_018508167.1 | A |
| XM_018652610.2 | 12 | 2092 | 697 | XM_018652610 | XP_018508125 | B |
| XM_009150438.3 | 24 | 2042 | 516 | XM_009150438 | XP_009148686 | D |
| XM_009150438.3 | 16 | 513 | 170 | XM_009150438 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahbuddin, D.; Rusly, R.; Shuid, A.N.; Ahmad Khair, A.B. Elucidating Quadruplication Event of PHO1 Gene: A Key Regulator of Plant Phosphate Translocation in Brassica rapa. Horticulturae 2023, 9, 845. https://doi.org/10.3390/horticulturae9070845
Shahbuddin D, Rusly R, Shuid AN, Ahmad Khair AB. Elucidating Quadruplication Event of PHO1 Gene: A Key Regulator of Plant Phosphate Translocation in Brassica rapa. Horticulturae. 2023; 9(7):845. https://doi.org/10.3390/horticulturae9070845
Chicago/Turabian StyleShahbuddin, Dahlia, Rosazlina Rusly, Ahmad Naqib Shuid, and Ahmad Bukhary Ahmad Khair. 2023. "Elucidating Quadruplication Event of PHO1 Gene: A Key Regulator of Plant Phosphate Translocation in Brassica rapa" Horticulturae 9, no. 7: 845. https://doi.org/10.3390/horticulturae9070845
APA StyleShahbuddin, D., Rusly, R., Shuid, A. N., & Ahmad Khair, A. B. (2023). Elucidating Quadruplication Event of PHO1 Gene: A Key Regulator of Plant Phosphate Translocation in Brassica rapa. Horticulturae, 9(7), 845. https://doi.org/10.3390/horticulturae9070845









