Complete Chloroplast Genome Sequences of Five Ormosia Species: Molecular Structure, Comparative Analysis, and Phylogenetic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and DNA Extraction
2.2. Chloroplast Genome Sequencing and Assembly
2.3. Repeat Sequences and SSRs
2.4. Variations and Divergent Hotspot Regions of cp Genomes
2.5. Phylogenomic Reconstruction Based on cp Genomes
3. Results
3.1. Overall cp Genome Features of Five Ormosia Species
3.2. IR Expansion and Contraction
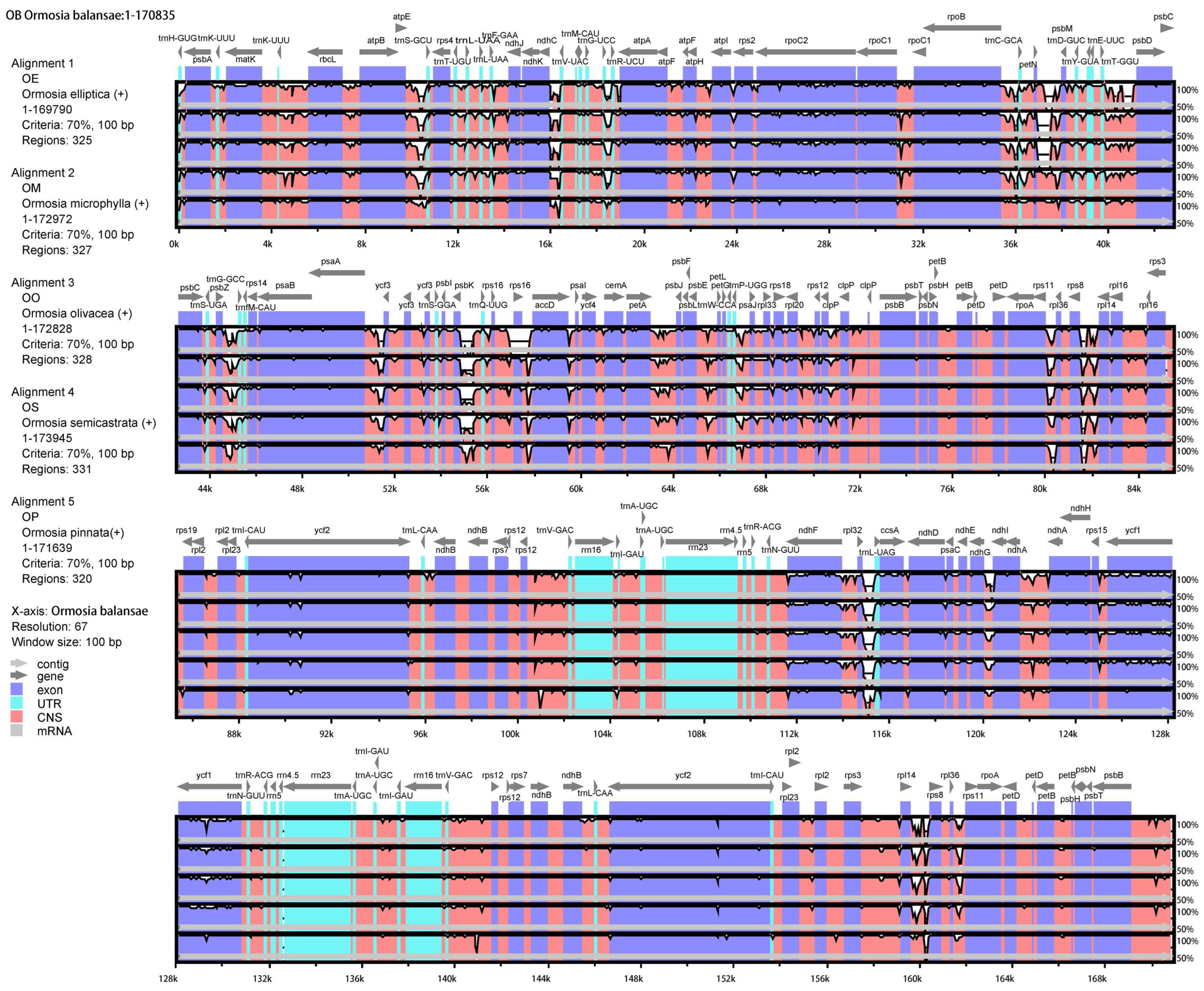
3.3. Variations and Divergence Hotspot Regions of cp Genomes
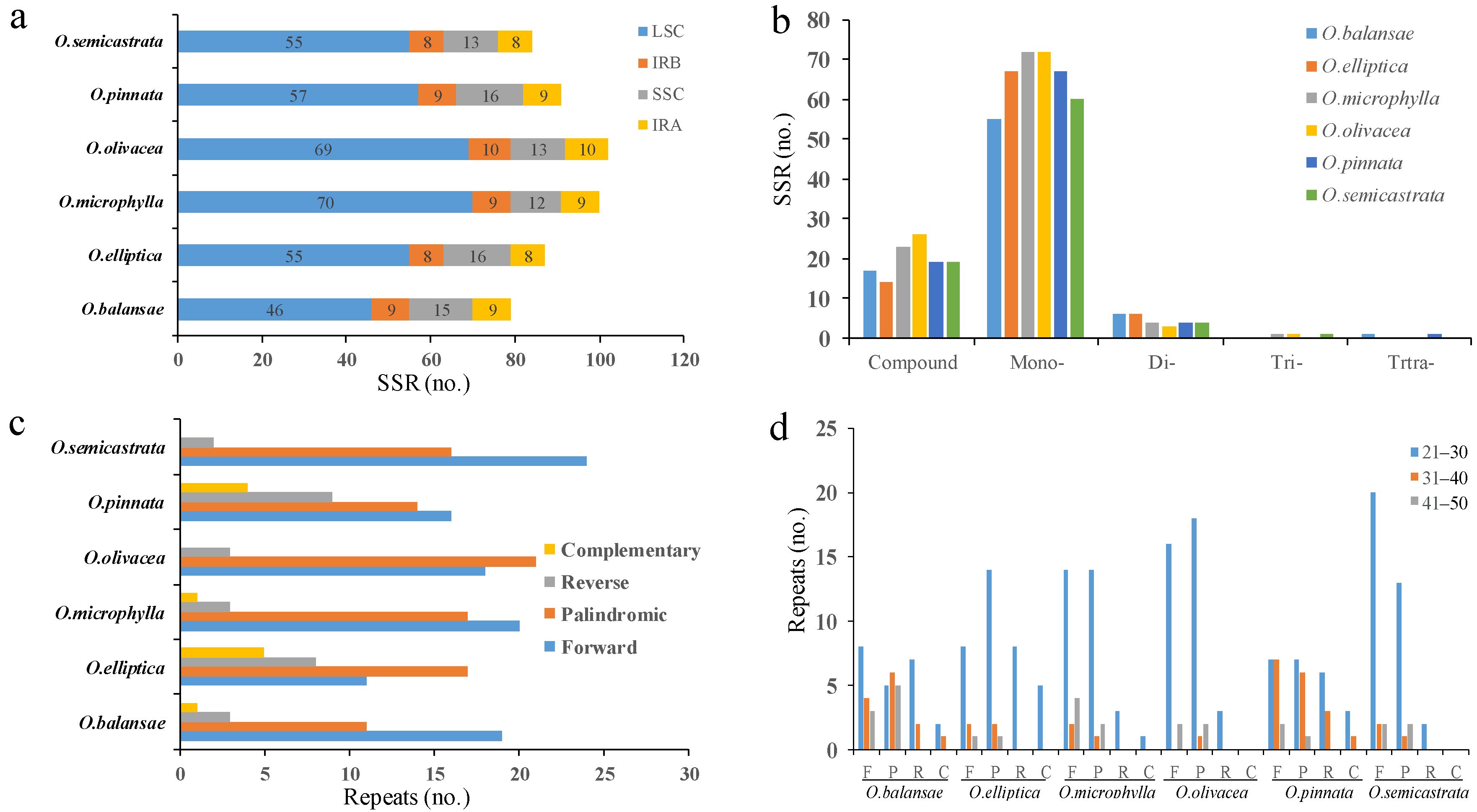
3.4. Repeat Analysis
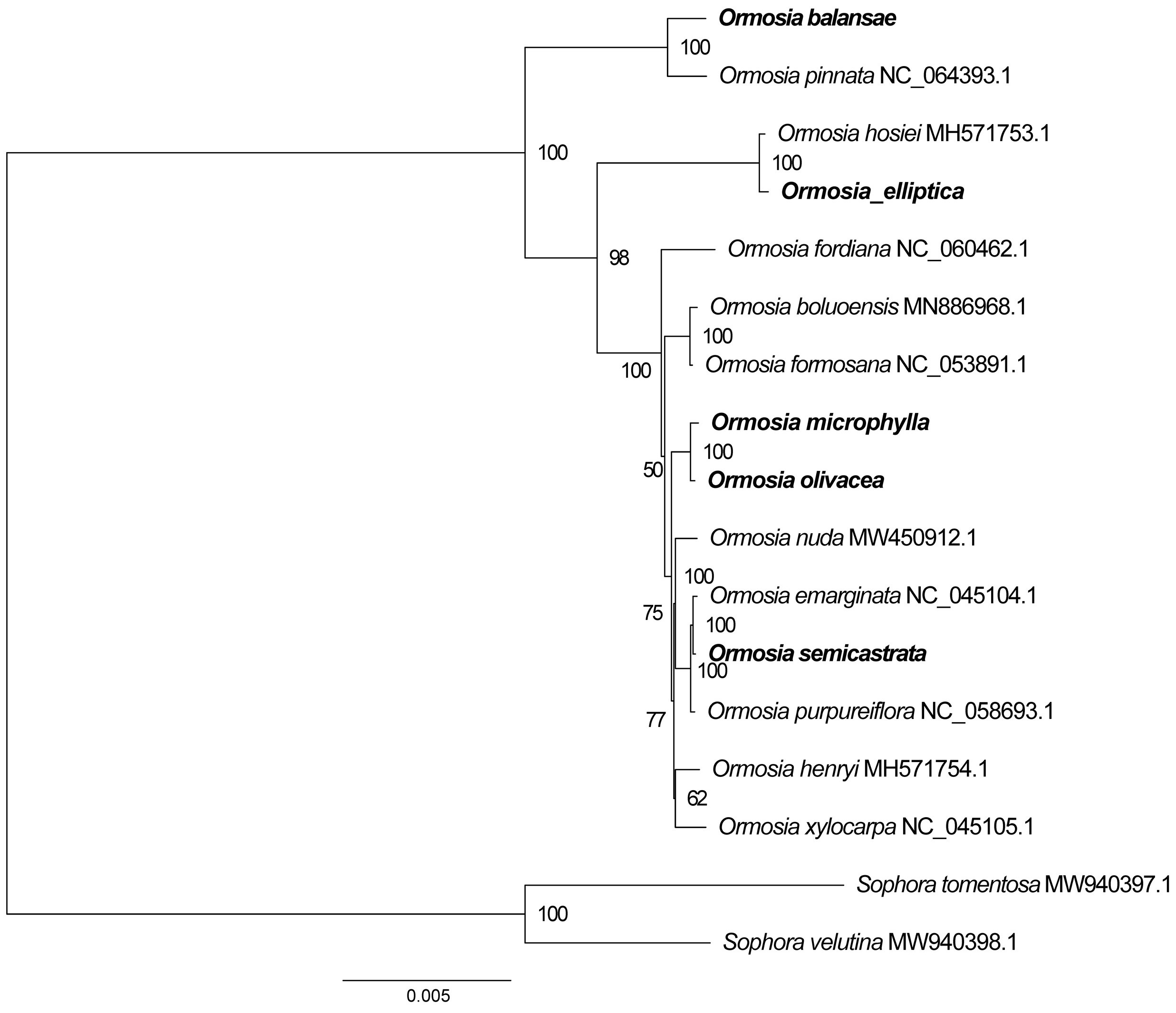
3.5. Phylogenomic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Azani, N.; Babineau, M.; Bailey, C.D.; Banks, H.; Barbosa, A.R.; Pinto, R.B.; Boatwright, J.S.; Borges, L.M.; Brown, G.K.; Bruneau, A. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny: The Legume Phylogeny Working Group (LPWG). Taxon 2017, 66, 44–77. [Google Scholar] [CrossRef]
- Kirkbride, J.H.; Gunn, C.R.; Weitzman, A.L. Fruits and Seeds of Genera in the Subfamily Faboideae (Fabaceae); US Department of Agriculture, Agricultural Research Service: Colombia, WA, USA, 2003; Volume 1.
- Kinghorn, A.D.; Hussain, R.A.; Robbins, E.F.; Balandrin, M.F.; Stirton, C.H.; Evans, S.V. Alkaloid distribution in seeds of Ormosia, Pericopsis and Haplormosia. Phytochemistry 1988, 27, 439–444. [Google Scholar] [CrossRef]
- Foster, M.S. Potential effects of arboreal and terrestrial avian dispersers on seed dormancy, seed germination and seedling establishment in Ormosia (Papilionoideae) species in Peru. J. Trop. Ecol. 2008, 24, 619–627. [Google Scholar] [CrossRef]
- Zhang, R.; Zhou, Z.; Du, K. Genetic diversity of natural populations of endangered Ormosia hosiei, endemic to China. Biochem. Syst. Ecol. 2012, 40, 13–18. [Google Scholar] [CrossRef]
- Lewis, G.P.; Schrire, B.; Mackinder, B.; Lock, M. Legumes of the World; Royal Botanic Gardens Kew: Richmond Kew, UK, 2005. [Google Scholar]
- Prain, D. The Asiatic species of Ormosia. J. Asiat. Soc. Bengal 1904, 73, 45–46. [Google Scholar]
- Prain, D. A list of the Asiatic species of Ormosia. J. Asiat. Soc. Bengal 1900, 69, 175–186. [Google Scholar]
- Merrill, E.D.; Chen, L. The Chinese and Indo-Chinese species of Ormosia. Sargentia 1943, 3, 77–120. [Google Scholar] [CrossRef]
- Torke, B.M.; Cardoso, D.; Chang, H.; Li, S.-J.; Niu, M.; Pennington, R.T.; Stirton, C.H.; Xu, W.-B.; Zartman, C.E.; Chung, K.-F. A dated molecular phylogeny and biogeographical analysis reveals the evolutionary history of the trans-pacifically disjunct tropical tree genus Ormosia (Fabaceae). Mol. Phylogenet. Evol. 2022, 166, 107329. [Google Scholar] [CrossRef]
- McCormack, J.E.; Hird, S.M.; Zellmer, A.J.; Carstens, B.C.; Brumfield, R.T. Applications of next-generation sequencing to phylogeography and phylogenetics. Mol. Phylogenet. Evol. 2013, 66, 526–538. [Google Scholar] [CrossRef]
- Saina, J.K.; Li, Z.-Z.; Gichira, A.W.; Liao, Y.-Y. The complete chloroplast genome sequence of tree of heaven (Ailanthus altissima (Mill.) (Sapindales: Simaroubaceae), an important pantropical tree. Int. J. Mol. Sci. 2018, 19, 929. [Google Scholar] [CrossRef]
- Liu, H.; Su, Z.; Yu, S.; Liu, J.; Yin, X.; Zhang, G.; Liu, W.; Li, B. Genome comparison reveals mutation hotspots in the chloroplast genome and phylogenetic relationships of Ormosia species. BioMed Res. Int. 2019, 2019. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wang, Z.-F.; Cao, H.-L. The complete chloroplast genome sequence of Ormosia boluoensis. Mitochondrial DNA Part B 2020, 5, 999–1000. [Google Scholar] [CrossRef]
- Wang, Z.-F.; Chang, L.-W.; Lian, J.-Y.; Cao, H.-L. The complete chloroplast genome sequence of Ormosia formosana. Mitochondrial DNA Part B 2020, 5, 2636–2637. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-Q.; Jiang, M.-Y.; Zhu, D.-D.; Zhang, J.-L.; Zheng, J.-Z.; Deng, C.-Y. Characterization of the complete chloroplast genome of an endemic species of pea family in China, Ormosia hosiei (Fabaceae). Conserv. Genet. Resour. 2019, 11, 443–446. [Google Scholar] [CrossRef]
- Wang, Z.-F.; Yu, E.-P.; Zeng, Q.-S.; Deng, H.-G.; Cao, H.-L.; Li, Z.-A.; Wei, X.; Lee, S.Y. The complete chloroplast genome of Ormosia purpureiflora (Fabaceae). Mitochondrial DNA Part B 2021, 6, 3327–3328. [Google Scholar] [CrossRef]
- Wang, Y.-H. The complete chloroplast genome of Ormosia nuda (fabaceae), an endemic species in China. Mitochondrial DNA Part B 2021, 6, 2095–2096. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.i.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Shahzadi, I.; Mehmood, F.; Ali, Z.; Malik, M.S.; Waseem, S.; Mirza, B.; Ahmed, I.; Waheed, M.T. Comparative analyses of chloroplast genomes among three Firmiana species: Identification of mutational hotspots and phylogenetic relationship with other species of Malvaceae. Plant Gene 2019, 19, 100199. [Google Scholar]
- Xie, D.-F.; Yu, Y.; Deng, Y.-Q.; Li, J.; Liu, H.-Y.; Zhou, S.-D.; He, X.-J. Comparative analysis of the chloroplast genomes of the Chinese endemic genus Urophysa and their contribution to chloroplast phylogeny and adaptive evolution. Int. J. Mol. Sci. 2018, 19, 1847. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, T.; Duan, D.; Yang, J.; Feng, L.; Zhao, G. Comparative analysis of the complete chloroplast genomes of five Quercus species. Front. Plant Sci. 2016, 7, 959. [Google Scholar] [CrossRef]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; Depamphilis, C.W.; Leebens-Mack, J.; Müller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef]
- Win, P.P.; Li, X.; Chen, L.-Q.; Tan, Y.-H.; Yu, W.-B. Complete plastid genome of two Dalbergia species (Fabaceae), and their significance in conservation and phylogeny. Mitochondrial DNA Part B 2020, 5, 1967–1969. [Google Scholar] [CrossRef]
- Shahzadi, I.; Mehmood, F.; Ali, Z.; Ahmed, I.; Mirza, B. Chloroplast genome sequences of Artemisia maritima and Artemisia absinthium: Comparative analyses, mutational hotspots in genus Artemisia and phylogeny in family Asteraceae. Genomics 2020, 112, 1454–1463. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhang, L.; Qi, J.; Zhang, L. Complete chloroplast genome sequence of Hibiscus cannabinus and comparative analysis of the Malvaceae family. Front. Genet. 2020, 11, 227. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, G.L.; Dorman, H.E.; Buchanan, A.; Challagundla, L.; Wallace, L.E. A review of the prevalence, utility, and caveats of using chloroplast simple sequence repeats for studies of plant biology. Appl. Plant Sci. 2014, 2, 1400059. [Google Scholar] [CrossRef] [PubMed]
- Chumley, T.W.; Palmer, J.D.; Mower, J.P.; Fourcade, H.M.; Calie, P.J.; Boore, J.L.; Jansen, R.K. The complete chloroplast genome sequence of Pelargonium× hortorum: Organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 2006, 23, 2175–2190. [Google Scholar] [CrossRef]
- Wang, R.-J.; Cheng, C.-L.; Chang, C.-C.; Wu, C.-L.; Su, T.-M.; Chaw, S.-M. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol. 2008, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Nguyen, V.B.; Dong, J.; Wang, Y.; Park, J.Y.; Lee, S.-C.; Yang, T.-J. Evolution of the Araliaceae family inferred from complete chloroplast genomes and 45S nrDNAs of 10 Panax-related species. Sci. Rep. 2017, 7, 4917. [Google Scholar] [CrossRef]
- Gielly, L.; Taberlet, P. The use of chloroplast DNA to resolve plant phylogenies: Noncoding versus rbcL sequences. Mol. Biol. Evol. 1994, 11, 769–777. [Google Scholar]
- Shen, X.; Guo, S.; Yin, Y.; Zhang, J.; Yin, X.; Liang, C.; Wang, Z.; Huang, B.; Liu, Y.; Xiao, S. Complete chloroplast genome sequence and phylogenetic analysis of Aster tataricus. Molecules 2018, 23, 2426. [Google Scholar] [CrossRef]
- Akashi, H.; Osada, N.; Ohta, T. Weak selection and protein evolution. Genetics 2012, 192, 15–31. [Google Scholar] [CrossRef]
- Baker, W.J.; Hedderson, T.A.; Dransfield, J. Molecular phylogenetics of subfamily Calamoideae (Palmae) based on nrDNA ITS and cpDNA rps16 intron sequence data. Mol. Phylogenet. Evol. 2000, 14, 195–217. [Google Scholar] [CrossRef] [PubMed]
- Park, I.; Yang, S.; Kim, W.J.; Song, J.-H.; Lee, H.-S.; Lee, H.O.; Lee, J.-H.; Ahn, S.-N.; Moon, B.C. Sequencing and comparative analysis of the chloroplast genome of Angelica polymorpha and the development of a novel indel marker for species identification. Molecules 2019, 24, 1038. [Google Scholar] [CrossRef] [PubMed]
- Gitzendanner, M.A.; Soltis, P.S.; Wong, G.K.S.; Ruhfel, B.R.; Soltis, D.E. Plastid phylogenomic analysis of green plants: A billion years of evolutionary history. Am. J. Bot. 2018, 105, 291–301. [Google Scholar] [CrossRef]
- Dong, W.; Xu, C.; Li, C.; Sun, J.; Zuo, Y.; Shi, S.; Cheng, T.; Guo, J.; Zhou, S. ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 2015, 5, 8348. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Liu, J.; Yu, J.; Wang, L.; Zhou, S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE 2012, 7, e35071. [Google Scholar] [CrossRef]



| Ormosia balansae | Ormosia microphylla | Ormosia elliptica | Ormosia olivacea | Ormosia semicastrata | ||
|---|---|---|---|---|---|---|
| Total | Length (bp) | 170,836 | 172,973 | 169,797 | 172,829 | 173,946 |
| GC% | 35.92 | 35.83 | 36.16 | 35.84 | 35.71 | |
| LSC | Length (bp) | 71,096 | 73,163 | 70,849 | 73,055 | 73,937 |
| GC% | 33.79 | 33.62 | 33.98 | 33.65 | 33.45 | |
| Length (%) | 41.62 | 42.30 | 41.73 | 42.27 | 42.51 | |
| IR | Length (bp) | 81,038 | 81,178 | 80,676 | 81,154 | 81,224 |
| GC% | 39.21 | 39.18 | 39.42 | 39.18 | 39.16 | |
| Length (%) | 47.44 | 46.93 | 47.51 | 46.96 | 46.70 | |
| SSC | Length (bp) | 18,702 | 18,632 | 18,272 | 18,620 | 18,785 |
| GC% | 29.74 | 29.86 | 30.24 | 29.87 | 29.73 | |
| Length (%) | 10.95 | 10.77 | 10.76 | 10.77 | 10.80 | |
| Protein-coding | Length (bp) | 81,768 | 81,777 | 81,445 | 80,838 | 76,590 |
| GC% | 37.42 | 37.46 | 37.53 | 37.37 | 37.29 | |
| Length (%) | 47.86 | 47.28 | 47.97 | 46.77 | 44.03 | |
| tRNA | Length (bp) | 2793 | 2737 | 2792 | 2795 | 2804 |
| GC% | 53.35 | 53.49 | 53.51 | 53.42 | 53.35 | |
| Length (%) | 1.63 | 1.58 | 1.64 | 1.62 | 1.61 | |
| rRNA | Length (bp) | 9062 | 9062 | 9062 | 9056 | 9054 |
| GC% | 55.40 | 55.44 | 55.44 | 55.43 | 55.47 | |
| Length (%) | 5.30 | 5.24 | 5.34 | 5.24 | 5.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, J.; Zou, R.; Wei, X.; Li, D. Complete Chloroplast Genome Sequences of Five Ormosia Species: Molecular Structure, Comparative Analysis, and Phylogenetic Analysis. Horticulturae 2023, 9, 796. https://doi.org/10.3390/horticulturae9070796
Tang J, Zou R, Wei X, Li D. Complete Chloroplast Genome Sequences of Five Ormosia Species: Molecular Structure, Comparative Analysis, and Phylogenetic Analysis. Horticulturae. 2023; 9(7):796. https://doi.org/10.3390/horticulturae9070796
Chicago/Turabian StyleTang, Jianmin, Rong Zou, Xiao Wei, and Dianpeng Li. 2023. "Complete Chloroplast Genome Sequences of Five Ormosia Species: Molecular Structure, Comparative Analysis, and Phylogenetic Analysis" Horticulturae 9, no. 7: 796. https://doi.org/10.3390/horticulturae9070796
APA StyleTang, J., Zou, R., Wei, X., & Li, D. (2023). Complete Chloroplast Genome Sequences of Five Ormosia Species: Molecular Structure, Comparative Analysis, and Phylogenetic Analysis. Horticulturae, 9(7), 796. https://doi.org/10.3390/horticulturae9070796






