Two Bacterial Bioagents Boost Onion Response to Stromatinia cepivora and Promote Growth and Yield via Enhancing the Antioxidant Defense System and Auxin Production
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of the Causal Agent of Onion White Rot Disease
2.1.1. Pathogenicity Test
2.1.2. Molecular Identification of S. cepivora
2.1.3. Phylogenetic Analysis
2.2. Isolation of Plant Growth-Promoting Rhizobacterium and Endophytic Bacterium
Molecular Identification of S. maltophilia and S. liqufaciens
2.3. In Vitro Antifungal Activity of S. maltophilia and S. liqufaciens
2.3.1. Dual Culture Assay
2.3.2. Culture Filtrate Assay
2.4. Field Experiments
2.4.1. Disease Assessment
2.4.2. Vegetative and Yield Parameters
2.4.3. Total Soluble Phenolic and Flavonoid Compounds
2.4.4. Enzymatic Activity
2.5. Gas Chromatography–Mass Spectrophotometry (GC-MS) Analysis
2.5.1. Chemical Composition of Bacterial Culture Filtrates
2.5.2. Quantification of Indole-3-Acetic Acid (IAA) and Tryptophan Using GC-MS
2.6. Statistical Analysis
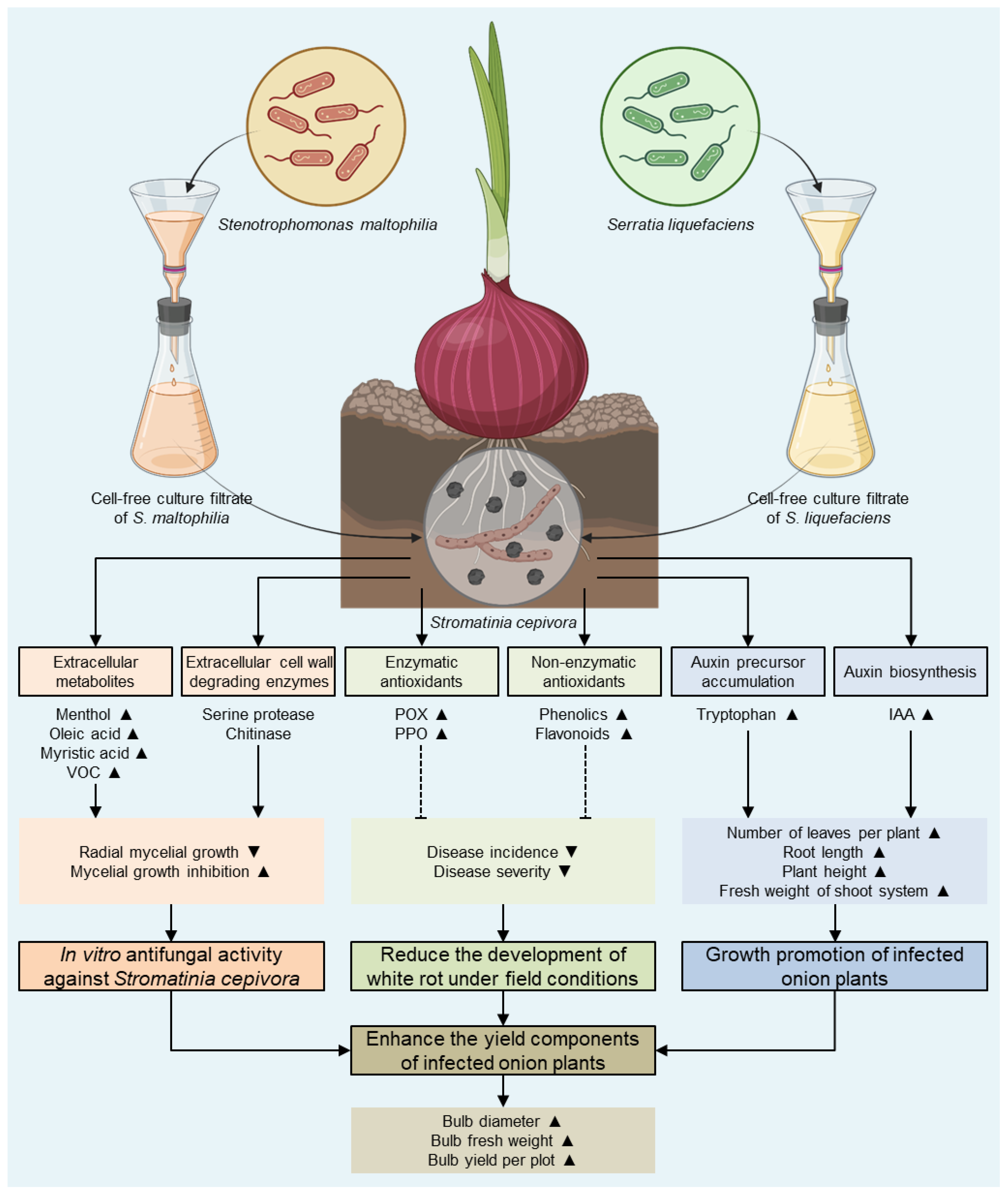
3. Results
3.1. Isolation and Identification of S. cepivora
3.2. Isolation and Identification of Bacterial Bioagent
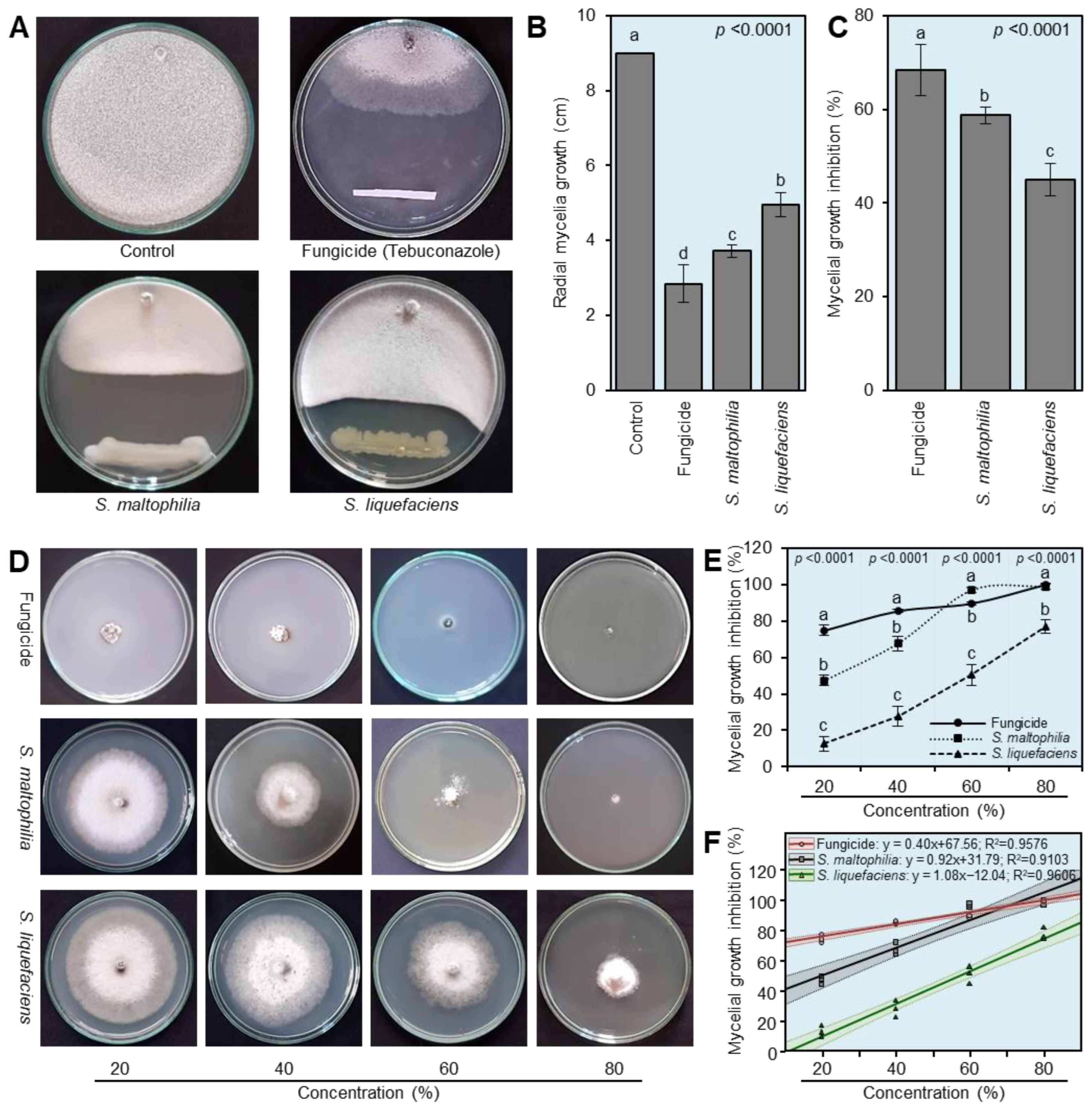
3.3. S. maltophilia and S. Liquefaciens Inhibited the Mycelial Growth of S. cepivora
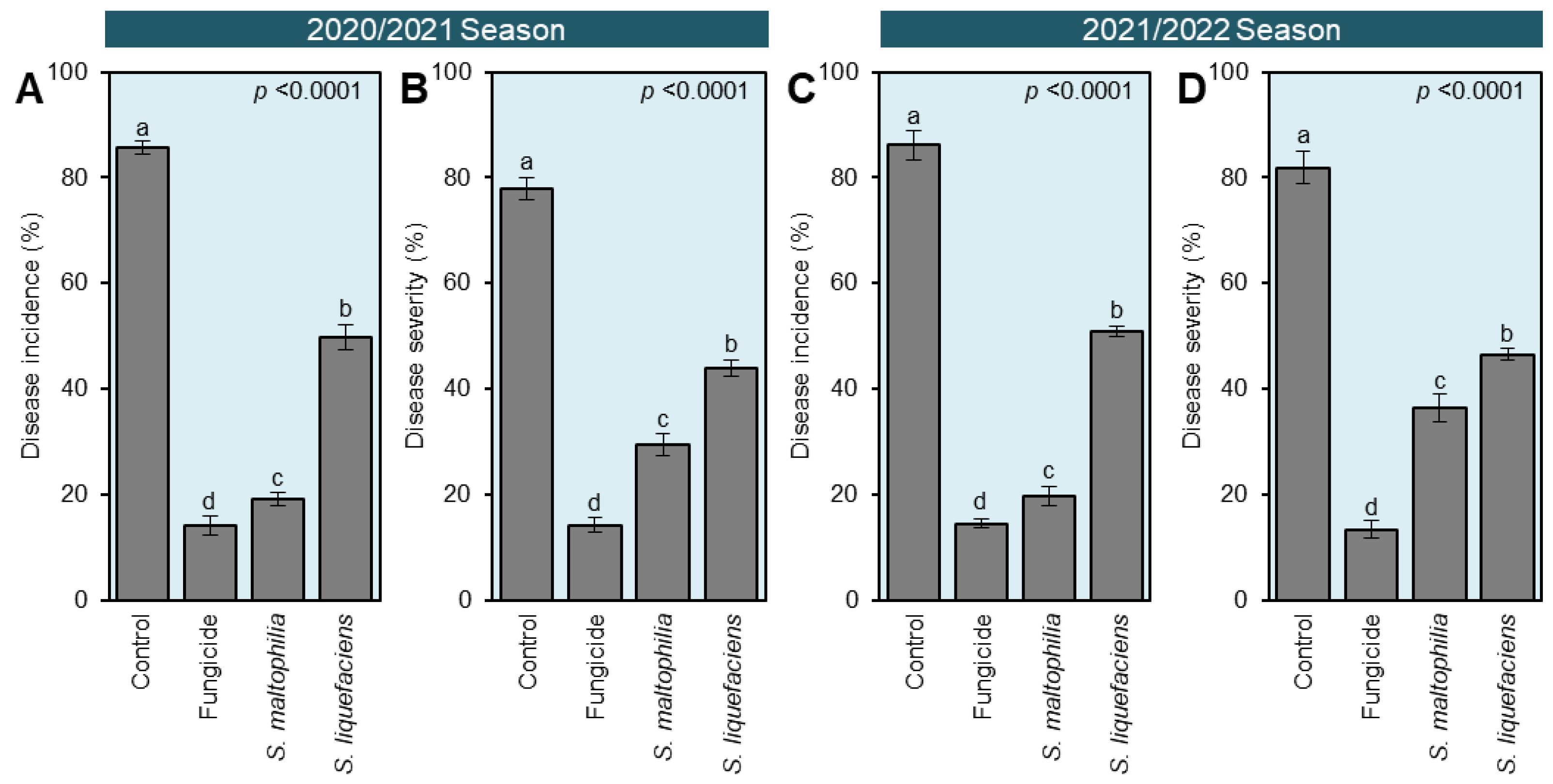
3.4. Culture filtrates of S. maltophilia and S. liquefaciens Reduced the Development of White Rot Disease
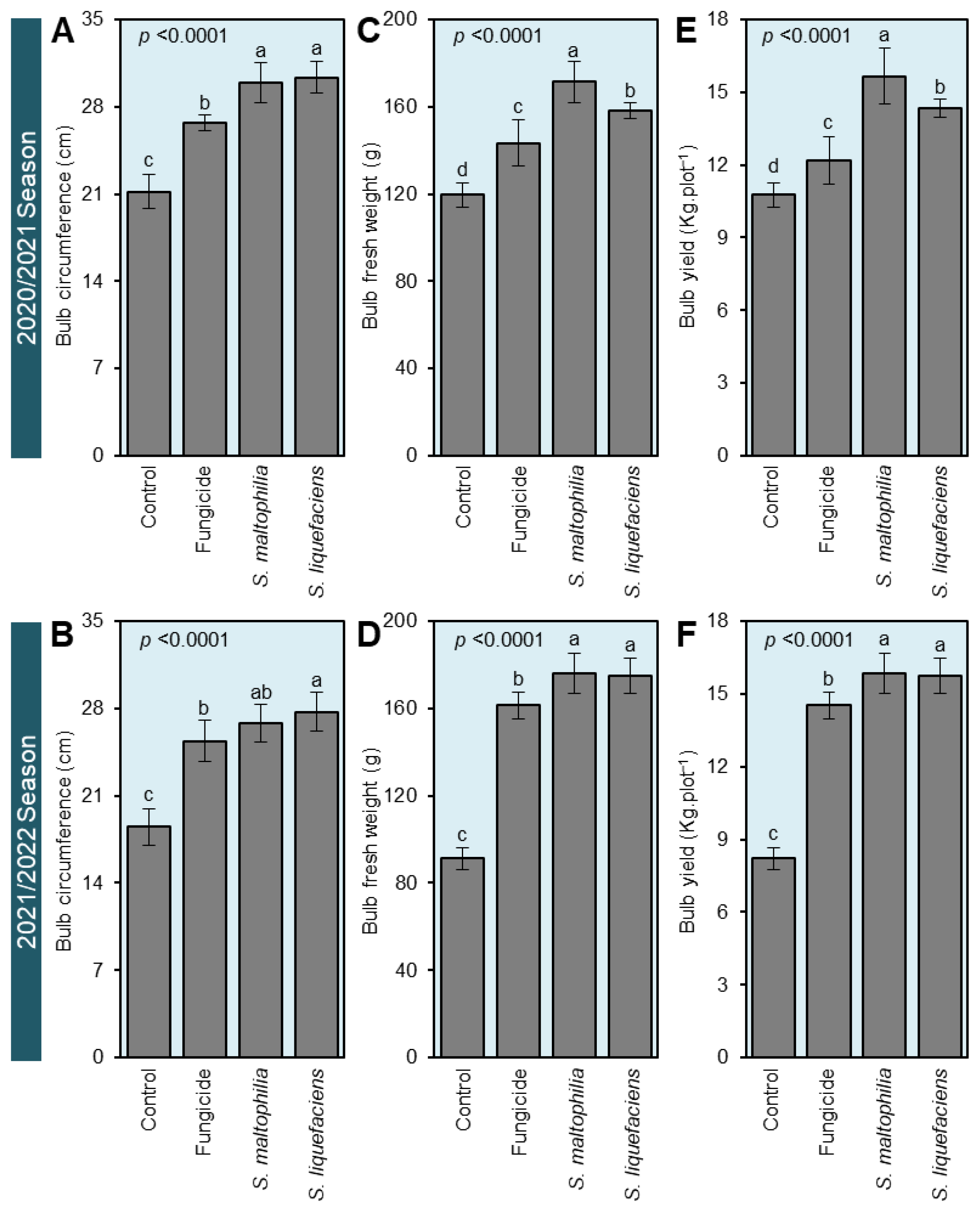
3.5. Cell-Free Culture Filtrates of S. maltophilia and S. liquefaciens Stimulated the Growth of S. cepivora-Infected Onion Plants
3.6. Culture Filtrates of S. maltophilia and S. liquefaciens Enhanced the Yield Components of S. cepivora-Infected Onion Plants
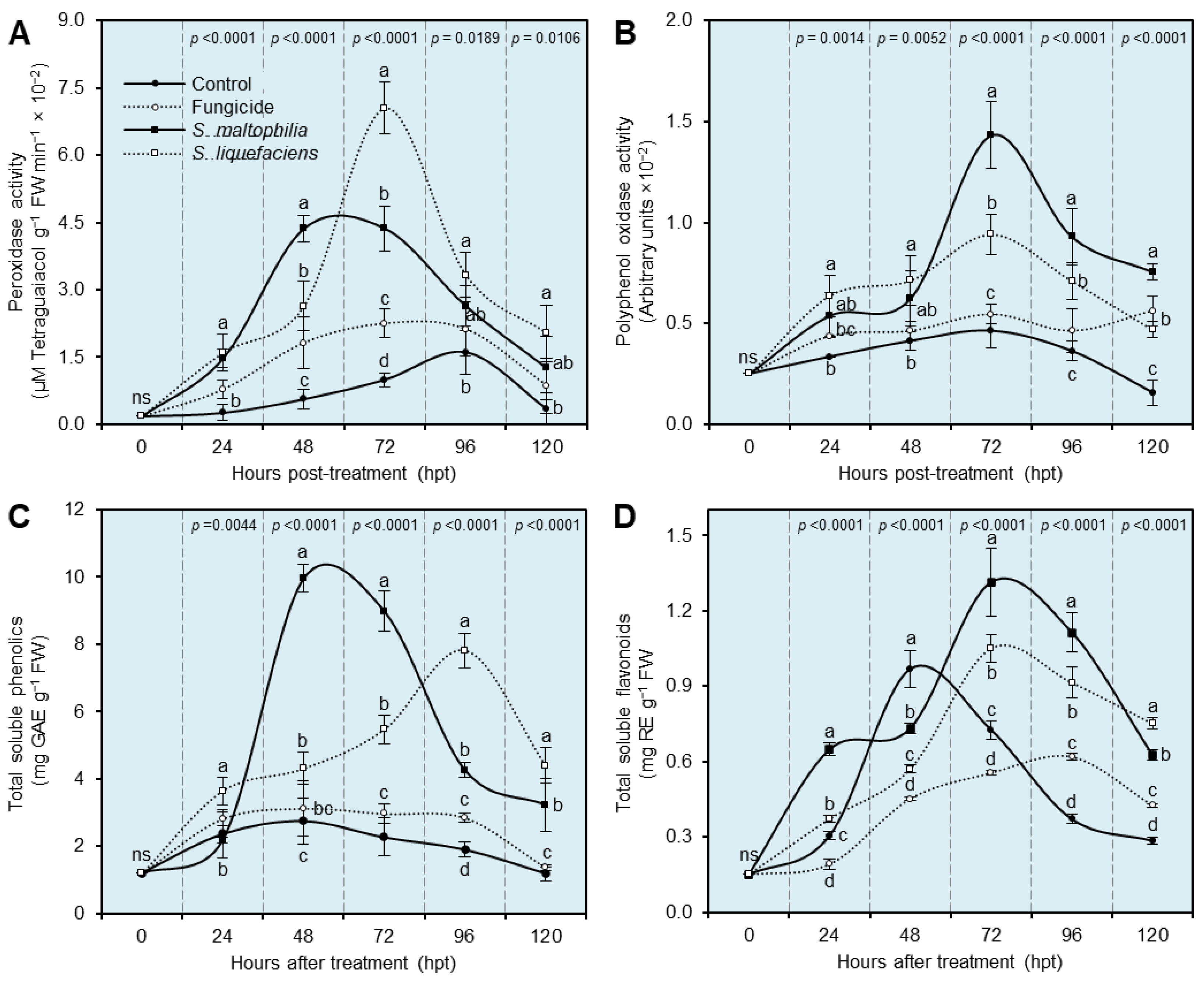
3.7. Cell-Free Culture Filtrates of S. maltophilia and S. liquefaciens Stimulate the Antioxidant Defense Machinery of S. cepivora-Infected Onion Plants
3.8. Culture Filtrates of S. maltophilia and S. liquefaciens and the Endogenous Auxin Content of S. cepivora-Infected Onion Plants
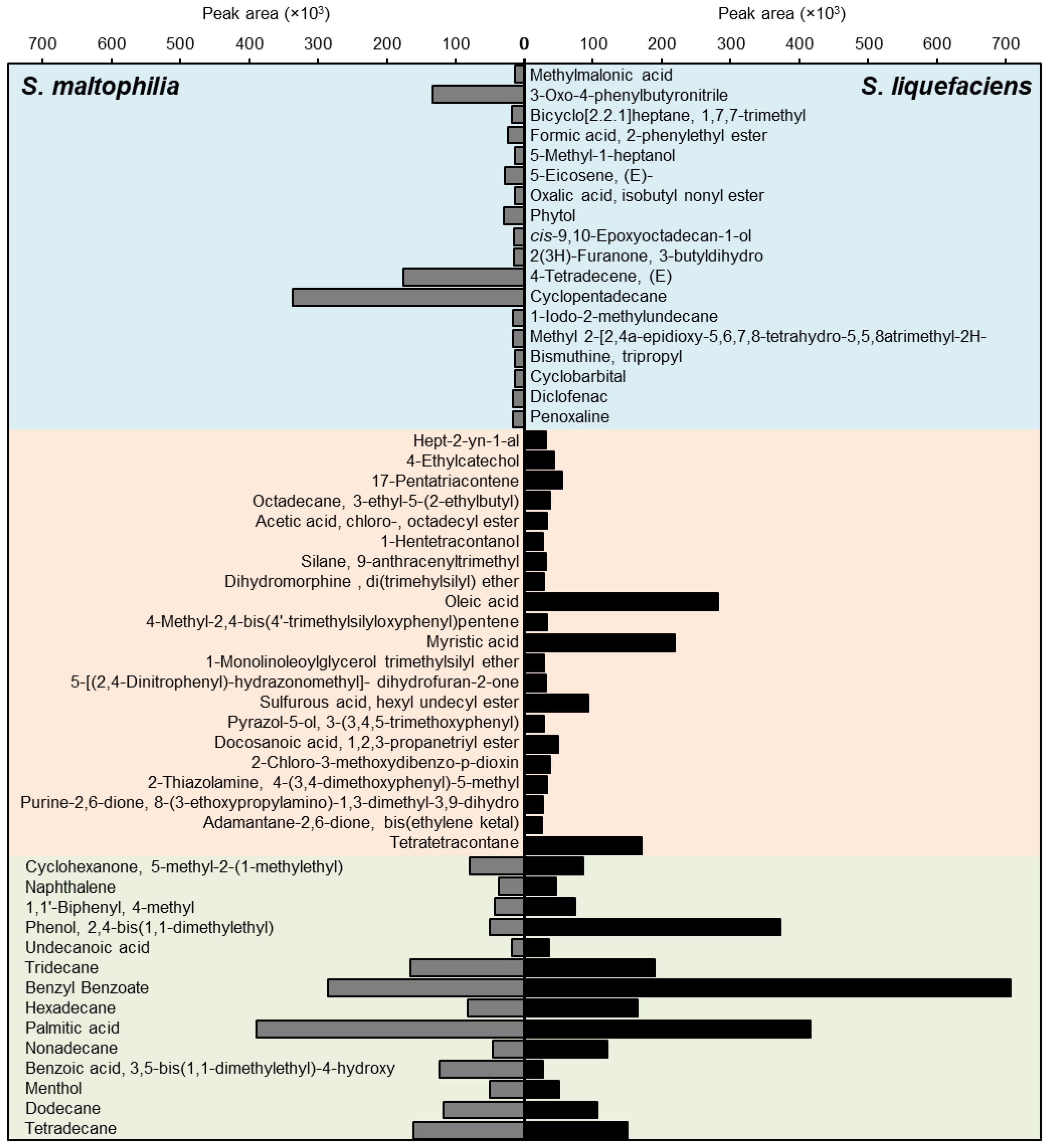
3.9. Chemical Composition of Culture Filtrates of S. maltophilia and S. liquefaciens
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Shultz, S. Onions. J. Agric. Food Inf. 2010, 11, 8–15. [Google Scholar]
- Javadzadeh, A.; Ghorbanihaghjo, A.; Bonyadi, S.; Rashidi, M.R.; Mesgari, M.; Rashtchizadeh, N.; Argani, H. Preventive Effect of Onion Juice on Selenite-Induced Experimental Cataract. Indian J. Ophthalmol. 2009, 57, 185. [Google Scholar] [PubMed]
- FAOSTAT. Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 2 June 2023).
- Dutta, R.; Nadig, S.M.; Manjunathagowda, D.C.; Gurav, V.S.; Singh, M. Anthracnose of Onion (Allium cepa L.): A Twister Disease. Pathogens 2022, 11, 884. [Google Scholar] [PubMed]
- Darwesh, O.M.; Elshahawy, I.E. Silver Nanoparticles Inactivate Sclerotial Formation in Controlling White Rot Disease in Onion and Garlic Caused by the Soil Borne Fungus Stromatinia cepivora. Eur. J. Plant Pathol. 2021, 160, 917–934. [Google Scholar]
- Elshahawy, I.E. Reduced Sclerotial Viability of Stromatinia cepivora and Control of White Rot Disease of Onion and Garlic by Means of Soil Bio-Solarization. Eur. J. Plant Pathol. 2021, 160, 519–540. [Google Scholar]
- Valdés-Santiago, L.; Vargas-Bernal, R.; Herrera-Pérez, G.; Colli-Mull, J.G.; Ordaz-Arias, A. Application of Two-Photon Microscopy to Study Sclerotium cepivorum Berk Sclerotia Isolated from Naturally Infested Soil and Produced In Vitro. Curr. Microbiol. 2021, 78, 749–755. [Google Scholar]
- Coley-Smith, J.R.; Mitchell, C.M.; Sansford, C.E. Long-term Survival of Sclerotia of Sclerotium cepivorum and Stromatinia gladioli. Plant Pathol. 1990, 39, 58–69. [Google Scholar]
- Amin, M.; Tadele, S.; Selvaraj, T. White Rot (Sclerotium cepivorum Berk)—An Aggressive Pest of Onion and Garlic in Ethiopia: An Overview. J. Agric. Biotechnol. Sustain. Dev. 2014, 6, 6–15. [Google Scholar] [CrossRef]
- Melero-Vara, J.M.; Basallote-Ureba, A.M.P.-L.; Basallote-Ureba, M.J. Comparison of Physical, Chemical and Biological Methods of Controlling Garlic White Rot. Eur. J. Plant Pathol. 2000, 106, 581–588. [Google Scholar]
- Coventry, E.; Noble, R.; Mead, A.; Whipps, J.M. Suppression of Allium White Rot (Sclerotium cepivorum) in Different Soils Using Vegetable Wastes. Eur. J. Plant Pathol. 2005, 111, 101–112. [Google Scholar]
- Clarkson, J.P.; Payne, T.; Mead, A.; Whipps, J.M. Selection of Fungal Biological Control Agents of Sclerotium cepivorum for Control of White Rot by Sclerotial Degradation in a UK Soil. Plant Pathol. 2002, 516, 735–745. [Google Scholar]
- Shalaby, M.E.; Ghoniem, K.E. Biological and Fungicidal Antagonism of Sclerotium cepivorum for Controlling Onion White Rot Disease. Ann. Microbiol. 2013, 63, 1579–1589. [Google Scholar] [CrossRef]
- Coley-Smith, J.R. White Rot Disease of Allium: Problems of Soil-Borne Diseases in Microcosm. Plant Pathol. 1990, 39, 214–222. [Google Scholar]
- Zubrod, J.P.; Bundschuh, M.; Arts, G.; Brühl, C.A.; Imfeld, G.; Knäbel, A.; Payraudeau, S.; Rasmussen, J.J.; Rohr, J.; Scharmüller, A.; et al. Fungicides: An Overlooked Pesticide Class? Environ. Sci. Technol. 2019, 53, 3347–3365. [Google Scholar] [PubMed]
- Deresa, E.M.; Diriba, T.F. Phytochemicals as Alternative Fungicides for Controlling Plant Diseases: A Comprehensive Review of Their Efficacy, Commercial Representatives, Advantages, Challenges for Adoption, and Possible Solutions. Heliyon 2023, 9, e13810. [Google Scholar] [CrossRef] [PubMed]
- Chet, I.; Inbar, J. Biological Control of Fungal Pathogens. Appl. Biochem. Biotechnol. 1994, 48, 37–43. [Google Scholar] [CrossRef]
- Sharma, I. Phytopathogenic Fungi and Their Biocontrol Applications. In Fungi Bio-Prospects in Sustainable Agriculture, Environment and Nano-Technology, Volume 1: Fungal Diversity of Sustainable Agriculture; Sharma, V.K., Shah, M.P., Parmar, S., Kumar, A., Eds.; Academic Press: Cambridge, MA, USA, 2021; Volume 1, pp. 155–188. ISBN 9780128213940. [Google Scholar]
- Aliye, N.; Fininsa, C.; Hiskias, Y. Evaluation of Rhizosphere Bacterial Antagonists for Their Potential to Bioprotect Potato (Solanum Tuberosum) against Bacterial Wilt (Ralstonia solanacearum). Biol. Control 2008, 47, 282–288. [Google Scholar]
- Messiha, N.A.S.; van Diepeningen, A.D.; Farag, N.S.; Abdallah, S.A.; Janse, J.D.; van Bruggen, A.H.C. Stenotrophomonas Maltophilia: A New Potential Biocontrol Agent of Ralstonia solanacearum, Causal Agent of Potato Brown Rot. Eur. J. Plant Pathol. 2007, 118, 211–225. [Google Scholar] [CrossRef]
- Debette, J.; Blondeau, R. Presence of Pseudomonas maltophilia in the Rhizosphere of Several Cultivated Plants. Can. J. Microbiol. 1980, 26, 460–463. [Google Scholar]
- Garbeva, P.; Van Overbeek, L.S.; Van Vuurde, J.W.L.; Van Elsas, J.D. Analysis of Endophytic Bacterial Communities of Potato by Plating and Denaturing Gradient Gel Electrophoresis (DGGE) of 16S rDNA Based PCR Fragments. Microb. Ecol. 2001, 41, 369–383. [Google Scholar] [PubMed]
- Berg, G.; Marten, P.; Ballin, G. Stenotrophomonas maltophilia in the Rhizosphere of Oilseed Rape—Occurrence, Characterization and Interaction with Phytopathogenic Fungi. Microbiol. Res. 1996, 151, 19–27. [Google Scholar] [CrossRef]
- Sultana, F.; Hossain, M.M. Assessing the Potentials of Bacterial Antagonists for Plant Growth Promotion, Nutrient Acquisition, and Biological Control of Southern Blight Disease in Tomato. PLoS ONE 2022, 17, e0267253. [Google Scholar]
- Badri Fariman, A.; Abbasiliasi, S.; Akmar Abdullah, S.N.; Mohd Saud, H.; Wong, M.Y. Stenotrophomonas maltophilia Isolate UPMKH2 with the Abilities to Suppress Rice Blast Disease and Increase Yield a Promising Biocontrol Agent. Physiol. Mol. Plant Pathol. 2022, 121, 101872. [Google Scholar] [CrossRef]
- Lodewyckx, C.; Vangronsveld, J.; Porteous, F.; Edward, R.B.; Taghavi, S.; Mezgeay, M.; Van Der Lelie, D. Endophytic Bacteria and Their Potential Applications. Crit. Rev. Plant Sci. 2002, 21, 583–606. [Google Scholar]
- Santoyo, G.; Moreno-hagelsieb, G.; Orozco-mosqueda, C.; Glick, B.R. Plant Growth-Promoting Bacterial Endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, W.; Liang, Z. Endophytic Bacteria from Pinellia ternata, a New Source of Purine Alkaloids and Bacterial Manure. Pharm. Biol. 2015, 53, 1545–1548. [Google Scholar]
- Michail, G.; Reizopoulou, A.; Vagelas, I. Evaluation of The Biocontrol Efficacy of Serratia proteamaculans and S. liquefaciens Isolated From Bats Guano Pile From a Subterrestrial Cave (Greece). Agric. Biol. (Sel’skokhozyaistvennaya Biol.) 2022, 57, 566–578. [Google Scholar] [CrossRef]
- Tian, S.P.; Bertolini, P. Effects of Low Temperature on Mycelial Growth and Spore Germination of Botrytis allii in Culture and on Its Pathogenicity to Stored Garlic Bulbs. Plant Pathol. 1995, 44, 1008–1015. [Google Scholar] [CrossRef]
- Zewide, T.; Fininsa, C.; Sakhuja, P.K. Management of White Rot (Sclerotium cepivorum) of Garlic Using Fungicides in Ethiopia. Crop Prot. 2007, 26, 856–866. [Google Scholar] [CrossRef]
- El-Nagar, A.; Elzaawely, A.A.; Xuan, T.D.; Gaber, M.; El-Wakeil, N.; El-Sayed, Y.; Nehela, Y. Metal Complexation of Bis-Chalcone Derivatives Enhances Their Efficacy against Fusarium Wilt Disease, Caused by Fusarium equiseti, via Induction of Antioxidant Defense Machinery. Plants 2022, 11, 2418. [Google Scholar] [CrossRef]
- Nehela, Y.; Mazrou, Y.S.A.; Taha, N.A.; Elzaawely, A.A.; Xuan, T.D.; Makhlouf, A.H.; El-Nagar, A. Hydroxylated Cinnamates Enhance Tomato Resilience to Alternaria alternata, the Causal Agent of Early Blight Disease, and Stimulate Growth and Yield Traits. Plants 2023, 12, 1775. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the Number of Nucleotide Substitutions in the Control Region of Mitochondrial DNA in Humans and Chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.F.; Curl, E.A. Methods for Research on the Ecology of Soil-Borne Plant Pathogens. In Methods for Research on the Ecology of Soil-Borne Plant Pathogens; Burgess Publishing Co.: Clayton, NC, USA, 1972. [Google Scholar]
- Bulgari, D.; Bozkurt, A.I.; Casati, P.; Çağlayan, K.; Quaglino, F.; Bianco, P.A. Endophytic Bacterial Community Living in Roots of Healthy and ‘Candidatus Phytoplasma Mali’-Infected Apple (Malus domestica, Borkh.) Trees. Antonie Van Leeuwenhoek 2012, 102, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Al-Hussini, H.S.; Al-Rawahi, A.Y.; Al-Marhoon, A.A.; Al-Abri, S.A.; Al-Mahmooli, I.H.; Al-Sadi, A.M.; Velazhahan, R. Biological Control of Damping-off of Tomato Caused by Pythium aphanidermatum by Using Native Antagonistic Rhizobacteria Isolated from Omani Soil. J. Plant Pathol. 2019, 101, 315–322. [Google Scholar]
- Amin, M.M.; Ahmed, M.F.A. Biological Control of Onion White Rot Disease Using Potential Bacillus subtilis Isolates. Egypt. J. Biol. Pest Control 2023, 33, 27. [Google Scholar] [CrossRef]
- Kähkönen, M.P.; Hopia, A.I.; Vuorela, H.J.; Rauha, J.P.; Pihlaja, K.; Kujala, T.S.; Heinonen, M. Antioxidant Activity of Plant Extracts Containing Phenolic Compounds. J. Agric. Food Chem. 1999, 47, 3954–3962. [Google Scholar] [CrossRef]
- Djeridane, A.; Yousfi, M.; Nadjemi, B.; Boutassouna, D.; Stocker, P.; Vidal, N. Antioxidant Activity of Some Algerian Medicinal Plants Extracts Containing Phenolic Compounds. Food Chem. 2006, 97, 654–660. [Google Scholar]
- Harrach, B.D.; Fodor, J.; Pogány, M.; Preuss, J.; Barna, B. Antioxidant, Ethylene and Membrane Leakage Responses to Powdery Mildew Infection of near-Isogenic Barley Lines with Various Types of Resistance. Eur. J. Plant Pathol. 2008, 121, 21–33. [Google Scholar] [CrossRef]
- Malik, C.P.; Singh, M.B. Plant Enzymology and Histo-Enzymology; Kalyani Publishers: New Delhi, India, 1980. [Google Scholar]
- Hijaz, F.; Nehela, Y.; Killiny, N. Possible Role of Plant Volatiles in Tolerance against Huanglongbing in Citrus. Plant Signal. Behav. 2016, 11, e1138193. [Google Scholar] [CrossRef]
- Nehela, Y.; Hijaz, F.; Elzaawely, A.A.; El-Zahaby, H.M.; Killiny, N. Phytohormone Profiling of the Sweet Orange (Citrus sinensis (L.) Osbeck) Leaves and Roots Using GC-MS-Based Method. J. Plant Physiol. 2016, 199, 12–17. [Google Scholar] [CrossRef]
- Killiny, N.; Nehela, Y. Metabolomic Response to Huanglongbing: Role of Carboxylic Compounds in Citrus sinensis Response to ‘Candidatus Liberibacter asiaticus’ and Its Vector. Diaphorina citri. Mol. Plant-Microbe Interact. 2017, 30, 666–678. [Google Scholar] [CrossRef] [PubMed]
- Nehela, Y.; Hijaz, F.; Elzaawely, A.A.; El-Zahaby, H.M.; Killiny, N. Citrus Phytohormonal Response to Candidatus Liberibacter Asiaticus and Its Vector. Diaphorina citri. Physiol. Mol. Plant Pathol. 2018, 102, 24–35. [Google Scholar] [CrossRef]
- Nehela, Y.; Killiny, N. Melatonin Is Involved in Citrus Response to the Pathogen Huanglongbing via Modulation of Phytohormonal Biosynthesis. Plant Physiol. 2020, 184, 2216–2239. [Google Scholar] [CrossRef] [PubMed]
- Nehela, Y.; Taha, N.A.; Elzaawely, A.A.; Xuan, T.D.; Amin, M.A.; Ahmed, M.E.; El-Nagar, A. Benzoic Acid and Its Hydroxylated Derivatives Suppress Early Blight of Tomato (Alternaria solani) via the Induction of Salicylic Acid Biosynthesis and Enzymatic and Nonenzymatic Antioxidant Defense Machinery. J. Fungi 2021, 7, 663. [Google Scholar] [CrossRef]
- Hussain, W.A.; Elzaawely, A.A.; El Sheery, N.I.; Ismail, A.A.; El-Zahaby, H.M. Biological Control of Onion White Rot Disease Caused by Sclerotium cepivorum. Environ. Biodivers. Soil Secur. 2017, 1, 101–107. [Google Scholar] [CrossRef]
- Rivera-Méndez, W.; Obregón, M.; Morán-Diez, M.E.; Hermosa, R.; Monte, E. Trichoderma asperellum Biocontrol Activity and Induction of Systemic Defenses against Sclerotium cepivorum in Onion Plants under Tropical Climate Conditions. Biol. Control 2020, 141, 104145. [Google Scholar] [CrossRef]
- Dunne, C.; Crowley, J.J.; Moënne-Loccoz, Y.; Dowling, D.N.; De Bruijn, F.J.; O’Gara, F. Biological Control of Pythium ultimum by Stenotrophomonas maltophilia W81 Is Mediated by an Extracellular Proteolytic Activity. Microbiology 1997, 143, 3921–3931. [Google Scholar] [CrossRef]
- Dunne, C.; Moenne-Loccoz, Y.; de Bruijn, F.J.; O’Gara, F. Overproduction of an Inducible Extracellular Serine Protease Improves Biological Control of Pythium ultimum by Stenotrophomonas maltophilia Strain W81. Microbiology 2000, 146, 2069–2078. [Google Scholar] [CrossRef]
- Jankiewicz, U.; Brzezinska, M.S.; Saks, E. Identification and Characterization of a Chitinase of Stenotrophomonas maltophilia, a Bacterium That Is Antagonistic towards Fungal Phytopathogens. J. Biosci. Bioeng. 2012, 113, 30–35. [Google Scholar] [CrossRef]
- Giesler, L.J.; Yuen, G.Y. Evaluation of Stenotrophomonas maltophilia Strain C3 for Biocontrol of Brown Patch Disease. Crop Prot. 1998, 17, 509–513. [Google Scholar] [CrossRef]
- Dewi, R.R.; Rahmah, S.M.; Taruna, A.; Aini, L.Q.; Fernando, I.; Abadi, A.L.; Syib’li, M.A. The Effectiveness Comparison Between Application of Indigenous Arbuscular Mycorrhizal Fungal Community and Stenotrophomonas maltophilia to Suppress Fusarium Wilt Incidence on Local Garlic Plant (Lumbu Hijau). AGRIVITA J. Agric. Sci. 2023, 45, 131–146. [Google Scholar] [CrossRef]
- Antonio Saministrado-Fudalan, P.; Sendaydiego, J.P.; Sinco, A.L. Serratia liquefaciens from IMO: Antibacterial and Chitinolytic Potentials. Int. J. Sci. Basic Appl. Res. 2017, 36, 306–317. [Google Scholar]
- Sneh, B.; Agami, O.; Baker, R. Biological Control of Fusarium-Wilt in Carnation with Serratia liquefaciens and Hafnia Alvei Isolated from Rhizosphere of Carnation. J. Phytopathol. 1985, 113, 271–276. [Google Scholar] [CrossRef]
- Kshetri, L.; Naseem, F.; Pandey, P. Role of Serratia sp. as Biocontrol Agent and Plant Growth Stimulator, with Prospects of Biotic Stress Management in Plant. In Plant Growth Promoting Rhizobacteria for Sustainable Stress Management: Volume 2: Rhizobacteria in Biotic Stress Management; Springer: Singapore, 2019; pp. 169–200. [Google Scholar] [CrossRef]
- Lucini, E.I.; Zunino, M.P.; López, M.L.; Zygadlo, J.A. Effect of Monoterpenes on Lipid Composition and Sclerotial Development of Sclerotium cepivorum Berk. J. Phytopathol. 2006, 154, 441–446. [Google Scholar] [CrossRef]
- Diastuti, H.; Chasani, M. Suwandri Antibacterial Activity of Benzyl Benzoate and Crotepoxide from Kaempferia rotunda L. Rhizome. Indones. J. Chem. 2020, 20, 9–15. [Google Scholar] [CrossRef]
- Walters, D.; Raynor, L.; Mitchell, A.; Walker, R.; Walker, K. Antifungal Activities of Four Fatty Acids against Plant Pathogenic Fungi. Mycopathologia 2004, 157, 87–90. [Google Scholar] [CrossRef]
- Liu, S.; Ruan, W.; Li, J.; Xu, H.; Wang, J.; Gao, Y.; Wang, J. Biological Control of Phytopathogenic Fungi by Fatty Acids. Mycopathologia 2008, 166, 93–102. [Google Scholar] [CrossRef]
- Casillas-Vargas, G.; Ocasio-Malavé, C.; Medina, S.; Morales-Guzmán, C.; Del Valle, R.G.; Carballeira, N.M.; Sanabria-Ríos, D.J. Antibacterial Fatty Acids: An Update of Possible Mechanisms of Action and Implications in the Development of the next-Generation of Antibacterial Agents. Prog. Lipid Res. 2021, 82, 101093. [Google Scholar] [CrossRef]
- Yogeswari, S.; Ramalakshmi, S.; Neelavathy, R.; Muthumary, J. Identification and Comparative Studies of Different Volatile Fractions from Monochaetia Kansensis by GCMS. Glob. J. Pharmacol. 2012, 6, 65–71. [Google Scholar]
- Faridha Begum, I.; Mohankumar, R.; Jeevan, M.; Ramani, K. GC–MS Analysis of Bio-Active Molecules Derived from Paracoccus pantotrophus FMR19 and the Antimicrobial Activity Against Bacterial Pathogens and MDROs. Indian J. Microbiol. 2016, 56, 426–432. [Google Scholar] [CrossRef]
- Wagay, N.A.; Mohiuddin, Y.G.; Khan, N.A. Phytochemical Evaluation and Identification of Bioactive Phytochemical Evaluation and Identification of Bioactive Compounds in Camel Thorn Alhagi Pseudalhagi (M. Bieb.). Int. J. Adv. Res. Sci. Eng. 2018, 7, 300–311. [Google Scholar]
- Zhang, Z.; Yuen, G.Y. The Role of Chitinase Production by Stenotrophomonas maltophilia Strain C3 in Biological Control of Bipolaris Sorokiniana. Phytopathology 2007, 90, 384–389. [Google Scholar] [CrossRef]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef]
- Rajput, V.D.; Harish; Singh, R.K.; Verma, K.K.; Sharma, L.; Quiroz-Figueroa, F.R.; Meena, M.; Gour, V.S.; Minkina, T.; Sushkova, S.; et al. Recent Developments in Enzymatic Antioxidant Defence Mechanism in Plants with Special Reference to Abiotic Stress. Biology 2021, 10, 267. [Google Scholar] [CrossRef] [PubMed]
- Racchi, M.L. Antioxidant Defenses in Plants with Attention to Prunus and Citrus spp. Antioxidants 2013, 2, 340–369. [Google Scholar] [PubMed]
- Granér, G.; Hamberg, M.; Meijer, J. Screening of Oxylipins for Control of Oilseed Rape (Brassica napus) Fungal Pathogens. Phytochemistry 2003, 63, 89–95. [Google Scholar] [CrossRef]
- Desai, M.D.; Suthar, K.P.; Singh, D.; Parekh, V.; Khunt, M.D.; Haidar, A. Molecular Evidence of Phytohormonal Modulatory Effect of Serratia liquefaciens on Rice. Appl. Biol. Res. 2020, 22, 126. [Google Scholar] [CrossRef]
- Hassan, T.U.; Bano, A. Comparative Effects of Wild Type Stenotrophomonas maltophilia and Its Indole Acetic Acid-Deficient Mutants on Wheat. Plant Biol. 2016, 18, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Ambawade, M.S.; Pathade, G.R. Production of Indole Acetic Acid (IAA) by Stenotrophomonas maltophilia BE25 Isolated from Roots of Banana (Musa spp). Int. J. Sci. Res. 2013, 4, 2644–2650. [Google Scholar]
- Singh, R.P.; Jha, P.N. The PGPR Stenotrophomonas maltophilia SBP-9 Augments Resistance against Biotic and Abiotic Stress in Wheat Plants. Front. Microbiol. 2017, 8, 1945. [Google Scholar] [CrossRef] [PubMed]
- Caneschi, W.L.; Sanchez, A.B.; Felestrino, É.B.; de Carvalho Lemes, C.G.; Cordeiro, I.F.; Fonseca, N.P.; Villa, M.M.; Vieira, I.T.; Moraes, L.Â.G.; de Almeida Barbosa Assis, R.; et al. Serratia liquefaciens FG3 Isolated from a Metallophyte Plant Sheds Light on the Evolution and Mechanisms of Adaptive Traits in Extreme Environments. Sci. Rep. 2019, 9, 18006. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Nie, Z.W.; He, L.Y.; Sheng, X.F. Rice-Derived Facultative Endophytic Serratia liquefaciens F2 Decreases Rice Grain Arsenic Accumulation in Arsenic-Polluted Soil. Environ. Pollut. 2020, 259, 113832. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Wang, Q.; He, L.Y.; Sheng, X.F. Increased Biomass and Reduced Rapeseed Cd Accumulation of Oilseed Rape in the Presence of Cd-Immobilizing and Polyamine-Producing Bacteria. J. Hazard. Mater. 2018, 353, 280–289. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osman, H.E.M.; Nehela, Y.; Elzaawely, A.A.; El-Morsy, M.H.; El-Nagar, A. Two Bacterial Bioagents Boost Onion Response to Stromatinia cepivora and Promote Growth and Yield via Enhancing the Antioxidant Defense System and Auxin Production. Horticulturae 2023, 9, 780. https://doi.org/10.3390/horticulturae9070780
Osman HEM, Nehela Y, Elzaawely AA, El-Morsy MH, El-Nagar A. Two Bacterial Bioagents Boost Onion Response to Stromatinia cepivora and Promote Growth and Yield via Enhancing the Antioxidant Defense System and Auxin Production. Horticulturae. 2023; 9(7):780. https://doi.org/10.3390/horticulturae9070780
Chicago/Turabian StyleOsman, Hanan E. M., Yasser Nehela, Abdelnaser A. Elzaawely, Mohamed H. El-Morsy, and Asmaa El-Nagar. 2023. "Two Bacterial Bioagents Boost Onion Response to Stromatinia cepivora and Promote Growth and Yield via Enhancing the Antioxidant Defense System and Auxin Production" Horticulturae 9, no. 7: 780. https://doi.org/10.3390/horticulturae9070780
APA StyleOsman, H. E. M., Nehela, Y., Elzaawely, A. A., El-Morsy, M. H., & El-Nagar, A. (2023). Two Bacterial Bioagents Boost Onion Response to Stromatinia cepivora and Promote Growth and Yield via Enhancing the Antioxidant Defense System and Auxin Production. Horticulturae, 9(7), 780. https://doi.org/10.3390/horticulturae9070780







