Abstract
Plants in their natural habitat frequently face different biotic and abiotic stresses, which lead to the production of reactive oxygen species (ROS) that can damage cell membranes, cause peroxidation and deterioration of macromolecules, and ultimately result in cell death. Superoxide dismutase (SOD), a class of metalloenzymes, is primarily found in living organisms and serves as the principal line of defense against ROS. The SOD gene family has not yet been characterized in any species of water lily from the genus Nymphaea. The present study aims to conduct a genome-wide study to discover SOD genes in four representative water lily species. In our present comparative study, we discovered 43 SOD genes in the genomes of four water lily species. The phylogenetic investigation results revealed that SOD genes from water lily and closely related plant species formed two distinct groups, as determined by their binding domains with high bootstrap values. Enzymatic ion-binding classified the SOD gene family into three groups, FeSOD, Cu/ZnSOD, and MnSOD. The analysis of gene structure indicated that the SOD gene family exhibited a relatively conserved organization of exons and introns, as well as motif configuration. Moreover, we discovered that the promoters of water lily SODs contained five phytohormones, four stress-responsive elements, and numerous light-responsive cis-elements. The predicted 3D protein structures revealed water lily SODs form conserved protein dimer signatures that were comparable to each other. Finally, the RT-qPCR gene expression analysis of nine NcSOD genes revealed their responsiveness to heat, saline, cold, cadmium chloride, and copper sulphate stress. These findings establish a basis for further investigation into the role of the SOD gene family in Nymphaea colorata and offer potential avenues for genetic enhancement of water lily aquaculture.
1. Introduction
Plants residing in their native environments often encounter a multitude of stress factors, including elevated salinity levels, prolonged drought periods, high temperatures, and the presence of heavy metals. These stressors exert notable influences on the plants’ overall growth, development, and productivity [1,2]. Under stress, plants adapt their homeostatic mechanisms by generating an excess of reactive oxygen species (ROS) within their cells. ROS are primarily generated in various parts of the plant cell, including the plasma membrane, peroxisomes, apoplast, cell walls, endoplasmic reticulum, mitochondria, and chloroplasts [3]. These ROS are toxic free radicals that can oxidize proteins, damage cell membranes, and cause harm to DNA when formed in excessive amounts [4,5]. The occurrence of stresses in plants inevitably leads to the production of ROS, such as peroxide radicals (HOO−), hydrogen peroxide (H2O2), and singlet oxygen (1O2). For instance, several potent scavengers of active oxygen have the ability to mitigate environmental stresses by regulating the expression of genes belonging to enzyme reaction families, such as superoxide dismutase (SOD), catalase (CAT), peroxidase (POD), glutathione peroxidase (GPX), and peroxidase (PrxR) [6,7,8,9]. Plants have developed effective and intricate antioxidant defense mechanisms comprising a variety of enzymatic and non-enzymatic antioxidants to manage the harmful effects of ROS. Among various antioxidant enzymes, SOD, a group of metalloenzymes, is predominantly present in living creatures. In managing environmental cues, SODs show a vital part in the physio-biochemical processes of plants via acting as the primary defense against ROS [3]. In plants, SOD enzymes are encoded by a family of genes that are classified based on their metal cofactors: (FeSOD), (Cu/ZnSOD), (MnSOD), and (NiSOD) [10,11,12]. Among them NiSOD is predominantly found in cyanobacteria, streptomyces, and marine organisms, but has not yet been documented in plants [13,14]. Iron and manganese superoxide dismutase are mainly present in lower plants, whereas copper and zinc are found in higher plants [15]. Such SODs are usually dispersed in different cell parts [16]. In the main, Cu/ZnSODs are localized in the cytosol, peroxisomes, and chloroplasts. MnSODs are found inside mitochondria and FeSODs are usually found in the peroxisomes and chloroplasts [17,18].
SODs have been demonstrated in recent studies to secure plants against abiotic stress factors including cold, drought, heat, salinity, ethylene and abscisic acid [19,20,21,22]. Various findings have demonstrated that SOD genes might be transcribed and induced in many plants in different stress circumstances [23,24]. In recently published articles, the SOD gene family under various abiotic and hormones stress situations in Brassica napus [25], Zostera marina [26], Salvia miltiorrhiza [27], and Hordeum vulgare [28] were reported. Furthermore, different stress conditions can result in varied expression patterns of diverse forms of SOD genes. For example, tomatoes (Solanum lycopersicum) exhibit specific patterns of regulation in their SOD genes; for instance, under salt stress, SlSOD1 is a single gene among the nine SlSOD genes that shows significant upregulation, while SlSOD2, SlSOD5, SlSOD6, and SlSOD8 are also regulated. However, in drought conditions, the expression levels of four genes among the nine, namely “SlSOD2, SlSOD5, SlSOD6, and SlSOD8,” are observed to be high [23]. Moreover, the expression profiles of the identical SOD gene type varied in the presence of stress. For instance, the studies revealed that the expression of MnSODs in Arabidopsis remained unchanged during oxidative stress, while scientists observed a considerable alteration in the expression of MnSODs in Zostera marina, peas (Pisum sativum), and wheat (Triticum aestivum) during salinity stress [26,29,30,31]. The findings imply that diverse SOD genes unveil distinct expression patterns in reaction to varying environmental stresses. In addition, scientists have revealed that the regulation of SOD expression may involve various miRNAs and alternative splicing mechanisms [32,33].
Water lilies are the most significant ornamental waterscape plants in the world. It is a perennial aquatic plant of the order Nymphaeales, genus Nymphaea in the family Nymphaeaceae. Nymphaea (Nymphaeaceae), also called flowering plants, are angiosperms with large and showy flowers. There are more than 60 species in the world, mostly distributed in tropical, subtropical, and temperate regions. They have curved or rounded and variously notched waxy-coated leaves on long stalks, usually grow on the water, and surround flowers. Each plant can grow approximately 70 to 80 flowers. The aquaculture of water lily, flowers can also be used as fresh cut flowers, in tea, in dried flower crafts, and in textile production. Water lilies have garnered significant attention from scientists, researchers, and entrepreneurs worldwide due to their immense economic, medicinal, and cultural value. While these plants hold great importance in phylogenetic research, the accessibility to comprehensive genetic and genomic information remains somewhat limited [34]. Since we released the first water lily (Nymphaea colorata) genome sequence in 2020 [35], the SOD gene family has not yet been discovered in any species of water lily.
To fill in this gap, the present study aims to conduct a comparative genome-wide study to discover SOD genes in representative water lily species genomes. Our analysis included the characterization of their phylogenetic connections, conserved motifs, cis-elements, gene structure, expression analysis, protein-protein interaction and 3D structures, in order to decode its structural characteristics and functions under stresses.
2. Materials and Methods
2.1. Retrieval of SOD Gene Family in Water Lily Species
To investigate the SOD gene family in water lilies, the Blastp search method was employed, utilizing the Arabidopsis SOD sequence as a query to examine the entire genome of each water lily species individually [36]. In our study, we used two methodologies, protein blast and the hidden Markov model (HMM), to detect SOD genes in four water lily species in which the genome sequences of two species Nymphaea colorata, and Nymphaea thermarum, are available online; while the other two, i.e., Nymphaea minuta, and Nymphaea mexicana, have unpublished genome sequences. For BLASTP, we utilized eight A. thaliana SOD amino acid sequences (AT1G08830.1/AtCSD1, AT2G28190.1/AtCSD2, AT5G18100.1/AtCSD3, AT4G25100.1/AtFSD1, AT5G51100.1/AtFSD2, AT5G23310.1/AtFSD3, AT3G10920.1/AtMSD1, and AT3G56350.1/At00MSD2) as the query, with an e-value set to 1 × 10−5. We obtained these eight AtSODs amino acid sequences from the Arabidopsis genome database TAIR (http://www.arabidopsis.org/ accessed on 5 January 2023). To identify conserved domains SOD_Cu (PF00080), SOD_Fe_C (PF00081) and SOD _Mn (PF02777), we performed scans of specific amino acid sequences using the web resources Pfam (Pfamv34.0-19178pSSMs) protein domain database (http://pfam.xfam.org/ accessed on 8 January 2023), and SMART (http://smart.embl-heidelberg.de/ accessed on 9 January 2023) [37].
2.2. Analysis of Physicochemical Features and Subcellular Localization
In order to anticipate the physicochemical characteristics of SOD proteins in water lily species, such as amino acid count (A.A), theoretical isoelectric point (pI), molecular weight (kDa), Pfam Domains, Functional annotations, and grand average of hydropathicity (GRAVY), we employed the ProtParam website accessible at http://web.expasy.org/protparam/ accessed on 13 January 2023 [38]. In order to predict the subcellular localization of SOD proteins, we employed the WoLF PSORT (https://wolfpsort.hgc.jp/ accessed on 17 January 2023) [39] and ProtComp 9.0 server (http://linux1.softberry.com/ accessed on 18 January 2023) [27].
2.3. Phylogenetic Analysis and Conserved Motifs
We generated a phylogenetic tree to investigate the evolutionary connections of the water lily SOD gene family. This was done using protein sequences from N. colorata, N. mexicana, N. minuta, N. thermarum, and A. trichopoda (Amborella trichopoda). First, we aligned the protein sequences using MUSCLE with default parameters [39]. Then, we used the MEGA11 software (https://megasoftware.net/home accessed on 27 January 2023) to make a phylogenetic tree using the Neighbor-Joining algorithms. We assigned confidence levels to each branch of the tree using bootstrap tests (1000). We used the MEME server (https://meme-suite.org/meme/db/motifs accessed on 2 February 2023) with default settings to detect conserved motifs in the protein sequences of water lily SODs [40].
2.4. Prediction of the Cis-Regulatory Elements in the Promoter
To examine the potential cis-elements in the promoters of water lily SODs, we obtained the 2 Kb sequence upstream of start codons from each species’ genome. Then we used the PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ accessed on 6 February 2023) [41], to examine the promoter sequence of each gene, and then we created figure using TBtools (V 1.068).
2.5. Examination of the 3D Structures of Water Lily SOD Proteins
The comprehension of a protein’s functions requires a detailed understanding of its 3D structure. To this end, we analyzed the predicted 3D structures of four water lily species, using the online servers SOPMA and SWISS MODEL, both available through ExPASy at https://www.expasy.org/ accessed on 15 February 2023. Finally, we applied the UCSF Chimera visualization tool to visualize the 3D structures [42].
2.6. Analysis of SOD Gene Structure of Water Lily
The Gene Structure Display Server (GSDS; http://gsds.cbi.pku.edu.cn/index.php accessed on 19 February 2023) program was used to visualize the organization of exons and introns in the SOD genes of water lilies [27].
2.7. Analysis of Potential Protein Interaction
To make the SOD protein interaction network, we utilized STRING 11.0 (https://string-db.org/cgi/input.pl accessed on 4 March 2023) tool for this purpose [43].
2.8. Expression Profiling of NcSOD Genes in Pollen and Ovule
The expression pattern of the NcSOD gene family was obtained from our own RNA-seq raw data (unpublished). All 9 NcSOD genes’ expression levels were explored in 1 day mature pollen, and 0, 1, 2, and 3 days mature ovule. The expression heatmap was constructed using TBtools (V 1.068, https://github.com/CJ-Chen/TBtools/ accessed on 9 March 2023), in which the color bar from light yellow to a dark red exhibited less to high levels of expression, and light blue to dark blue shows less or no expression of NcSOD genes.
2.9. Plant Materials and Abiotic Stresses
To examine how NcSOD members respond to different abiotic stresses, we grew N. colorata mature plants in water tub filled with tap water under an open environment. The plants were then subjected to various stress treatments, including 250 mM NaCl, 200 μM CuSO4, and 2.5 mM CdCl2, as well as being treated with cold stress at 8 °C and heat stress at 42 °C. Each treatment was performed with three independent biological replicates, and each sample was collected from at least five individual plants. Leaves from the plantlets were collected at 0, 2, 4, and 6 h for the salt, heat, cold, and heavy metal stress experiments. After collection, all samples were instantly frozen in liquid nitrogen and preserved at −80 °C until total RNA isolation.
2.10. RNA Isolation and Real-Time Quantitative PCR Expression Analysis
RNA extraction was performed using the RNAprep Pure Plant Kit (TIANGEN, Beijing, China). The concentration of the samples was determined using a NanoDrop 2000 C spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The genomic DNA, was removed by DNase I treatment, followed by cDNA synthesis using the QuantiTect Reverse Transcription Kit (Qiagen, Shanghai, China). The RT-qPCR expression was performed on the Roche LightCycler 96 PCR system, following the recommended guidelines for the ChamQTM SYBR RT-qPCR Master Mix (Vazyme Biotech Co., Ltd., Sanya, China). For each RT-qPCR, the expression level of the actin gene in N. colorata was employed to standardize the RNA samples. Three biological replicates for each sample were employed for RT-qPCR, analysis with actin as internal control. Gene-specific primers for NcSODs and Nc-actin in the RT-qPCR system were designed using the online NCBI Primer-BLAST Program and their specificity was confirmed using the Oligo Calculator online tool (http://mcb.berkeley.edu/labs/krantz/tools/oligocalc.html accessed on 5 April 2023). The primers were synthesized by NANSHAN BIOTECH, (Sanya), and listed in (Table S1). The 2−ΔΔCT method was used to analyze the RT-qPCR gene expression data [44].
3. Results
3.1. Genome-Wide Analysis of SOD Gene Family in Four Water Lily Species
In this comparative study, we discovered 43 SOD genes in the genomes of four water lily species. Protein sequences of eight A. thaliana (AtSODs) were used as queries and removed the repetitive redundant sequences (Table S2), and 9–15 genes were obtained for each species, for example three diploid water lilies N. colorata (9 SODs), N. thermarum (10 SODs), N. minuta (9 SODs), and a tetraploid water lily N. mexicana (15 SODs) (Table 1; Table S3). After conducting domain scrutiny, we identified 15 proteins with a Cu/Zn-SODs domain (Pfam; 00080), 19 with a Fe-SODs domain (Pfam; 00081), and 9 with a Mn-SODs domain (Pfam; 02777) in water lily species. These results are consistent among all species and contain all SOD genes and domains with sub-families.

Table 1.
Characteristics of the SOD genes from four water lily species.
The biochemical and physiological characteristics of all SODs were investigated by calculating various parameters (Table 1). Protein length in the four representative water lily species ranged from 117 to 362 amino acids. Consequently, the molecular weight was found to be from 12.166 to 40.905 kDa. Most of the species investigated in this study displayed acidic properties, exhibiting pI values among 4.68 to 10.15. Furthermore, the predicted GRAVY values of SOD proteins were negative, showing that they are hydrophilic.
According to the subcellular localization prediction, the majority of water lily CSDs were found in the cytoplasm, while only a few localized in the chloroplast. Water lily FSDs and MnSDs were specifically located in the chloroplast and mitochondria, respectively (Table 1).
Furthermore, an NCBI domain search was conducted to perform domain-based analysis on all SOD proteins to acquire data and TBtools was then used to make the domain structures. As a result of this analysis, the presence of the SOD family was verified on all chosen protein sequences (Figure 1).
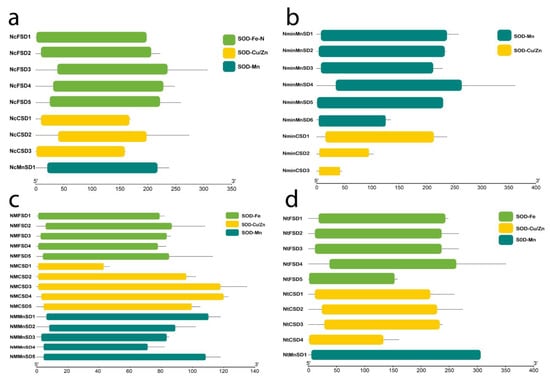
Figure 1.
Symbolic SOD domain structures of four water lily species: (a) Nymphaea colorata; (b) Nymphaea minuta; (c) Nymphaea mexicana; and (d) Nymphaea thermarum. Among four species only Nymphaea minuta contains Mn and Cu/Zn, while others constitute all three subfamilies of SOD.
3.2. Phylogenetic Relationships and Conserved Motif Analysis in Representative Water Lily Species
To uncover the evolutionary relations among SODs in various plant species, a phylogenetic tree was created using the complete protein sequences. The present research investigated the evolutionary relationships among genes of NcSODs, NtSODs, NminSODs, NMSODs, and AmtSODs. By considering the domains (Fe-SODs, Cu/Zn-SODs, and Mn-SODs) and analyzing a phylogenetic tree, a total of 50 SODs were classified into two main groups (Figure 2a). To assess the structural diversity of water lily SOD proteins and predict their functions, we utilized the MEME software to analyze their full-length protein sequences and identify conserved motifs. By examining conserved motifs in the SOD family, this analysis confirmed the categorization and evolutionary connections among SOD genes within the water lily species. The investigation revealed that 10 conserved motifs were present in all species (Figure 2b). The detailed information about the identified motifs, including their names, sequences, and widths is displayed in Table S4. The number of conserved motifs in SOD proteins ranged from two to six, and their distribution aligned with the groups. The Cu-SOD and Fe-SOD groups had only one motif in two genes (Figure 2b). Furthermore, MnSODs and FeSODs were classified in the similar group and subcluster, whereas Cu/ZnSODs were placed in a distinct group. Interestingly, motifs 1, 2, 4, and 9 were estimated to be particular to Cu/Zn-SOD whereas motifs 3, 5, 6, and 10 were exclusive to the MnSODs and FeSODs groups. In conclusion, the reliability of group arrangements was strongly supported by analyzing conserved motif patterns and phylogenetic relationships between water lily species. This suggests that water lily SODs proteins possess highly conserved amino acid residues within groups. Consequently, it is reasonable to infer that proteins with analogous structures and motifs may have similar efficient roles.

Figure 2.
Classification of SOD genes according to their subfamilies: (a) A neighbor-joining phylogenetic tree; (b) Conserved motif analysis. The motifs supported the two subfamilies which are mentioned in tree. Reliability of group arrangements was strongly supported by analyzing conserved motif patterns and phylogenetic relationships between water lily species. Different types of motifs represented by differently colored boxes.
3.3. Analyses of Cis-Elements in Water Lily SOD Gene Promoters
Retrieving cis-regulatory elements from the promoter regions of water lily SODs enabled the differentiation of gene functions and regulatory roles. By using the PlantCARE database, an analysis was conducted on the 2 kb upstream region from the start codon of individual SOD gene. Based on current findings, the cis-elements were classified into three categories: light related, stress related, and hormone response elements (Figure 3).
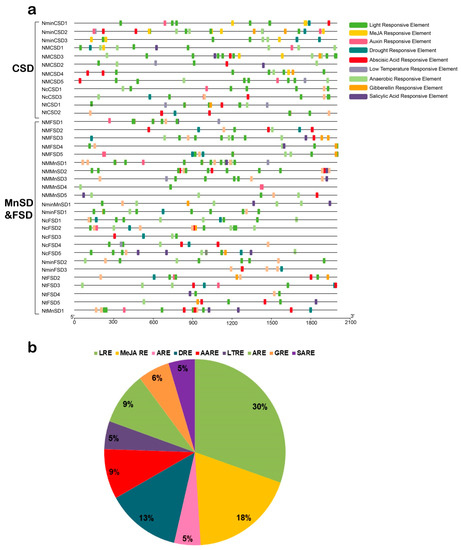
Figure 3.
Exploration of abiotic-stress related cis-regulatory components in the water lily SOD promoter regions. (a) Various hormone-associated and stress-responsive elements are explored. (b) The size of pie chart corresponds to the ratio of the respective promoter element. Cis-elements that share functional similarity are represented by the same colors.
In this study, five phytohormone-associated elements (Salicylic acid (SA), Gibberellin, Auxin, Abscisic acid (ABA), and Methyl jasmonate (MeJA), were detected, including ABRE, TCA-components, TGACG-and CGTCA-motif, TATC-box, P-box, etc. Moreover, four stress-responsive components (drought, low-temperature, light, and anaerobic) were recognized, such as LTR, ARE, TCT-motif, LAMP-element, MBS, etc (Figure 3). Generally many light reactive components were detected to be extensively dispersed in same group species, and demonstrating the important part of water lily SODs in response to light stress. Comparative analysis of the findings revealed that the SOD promoter cis-elements in water lily species can exhibit a significant response to abiotic stresses and can play a role in regulating plant growth and development and stress response.
3.4. 3D Structure Analysis of Water Lily SOD Proteins
The examination of a protein’s structure holds a great importance in understanding its function. We used SWISS MODEL and SOPMA online tools with the default search options, to predict the 3D structures of proteins. This study involved the prediction of three-dimensional models for four water lily proteins. The generated models were downloaded for the purpose of visualizing the 3D structures. The helices are represented by yellow, while the sheets or strands are represented by green (Figure 4). Proteins that fall under specific groups exhibit related structural evenness. The MnSD and FSD subfamilies share a nearly identical structure, with an equal amount of helices and sheets. Similarly, proteins in the CSD subfamily also possess an analogous structure.
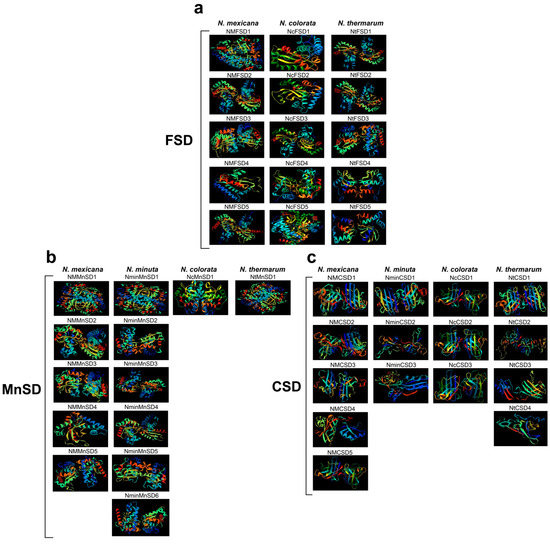
Figure 4.
The 3D structures of four water lilies’ (Nymphaea colorata, Nymphaea thermarum, Nymphaea minuta, and Nymphaea mexicana) SOD proteins categorized based on their sub families. (a) Represents the 3D structures of FSD subfamily; (b) Represents the 3D structures of MnSD subfamily; (c) Represents the 3D structures of CSD subfamily in all species. The final models are displayed, with diverse colors representing various sheets, domains, and helices. Note: In group (a) the Nymphaea minuta has no FSD subfamily.
3.5. Analysis of Exon-Intron Structure of NcSOD
The analysis of exon-intron structure of NcSOD genes was performed to elucidate the structural characteristics of species (Figure 5). NcSOD genes displayed varied exon-intron organizational patterns, with introns ranging from 5 (NcFSD5) to 9 (NcFSD4). The number of exons in NcSOD differs from 1 (NcFSD1) to 9 (NcFSD2). In one NcFSD1 gene, introns are absent, and there is only one exon. The gene structure investigation revealed that the SOD gene family displayed a relatively conserved exon/intron organization.
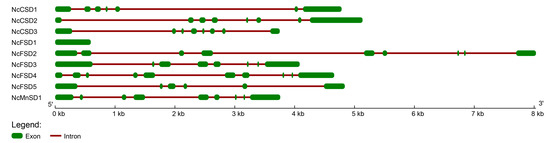
Figure 5.
Gene structure of NcSOD shows conserved exon/intron organization.
3.6. Expression Examination of NcSOD Genes in Reproductive Organs
The SOD gene family has a crucial role in plant growth, development, and response to stress. In order to investigate their specific biological functions in N. colorata, we observed the expression patterns of the 9 NcSOD genes in pollen and ovules using our own unpublished RNA-seq raw data. Under normal growth conditions, not all predicted genes in the N. colorata SOD family were expressed. Our analysis revealed that NcFSD3, NcFSD5, and NcMnSD1 were highly expressed in ovules at 0, 1, 2, and 3 days, while showing relatively lower expression in pollen on day 1 (Figure 6; Table S5). NcCSD1 and NcCSD2 were moderately expressed in ovules and pollen throughout all days. NcFSD2 and NcFSD4 showed a moderate expression in ovule but exhibited no expression in pollen. Both NcFSD1 and NcCSD3 showed no expression levels in both ovules and pollen. Generally, results exhibited that genes from all three subfamilies, i.e., Fe, Mn and Cu, play essential roles in N. colorata reproduction, growth, and development.
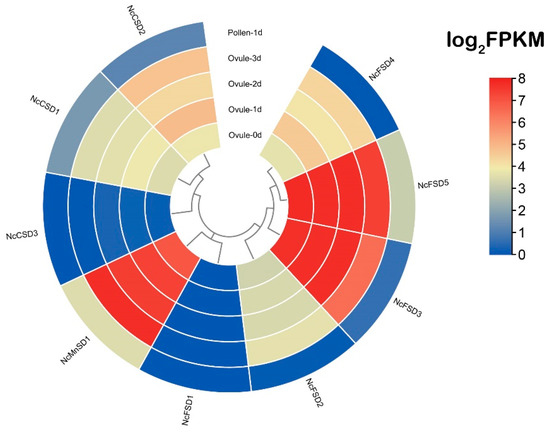
Figure 6.
Expression of the NcSOD genes was analyzed in pollen and ovule samples at four different time-points: 0 d, 1 d, 2 d, and 3 d. The expression bar from light blue to dark blue shows less or no expression of NcSOD genes. The light yellow to a dark red color exhibited less to high level of expression of these genes.
3.7. Potential NcSOD Protein–Protein Interaction
The potential NcSOD protein–protein interaction was analyzed via “STRING’’11.0 (https://string-db.org/cgi/input.pl accessed on 25 April 2023). As shown in Figure 7, among nine NcSOD genes, seven SOD proteins participate in strong interaction networks. Interestingly, we observed that different proteins co-regulate each other to respond to stress conditions. For example, NcCSD3, NcFSD1 and NcFSD4 are upregulated after 2 h under cold stress (Figure 8c). In water lilies, they potentially exert a regulatory function by forming protein complexes to improve cold tolerance and cope with various stresses.
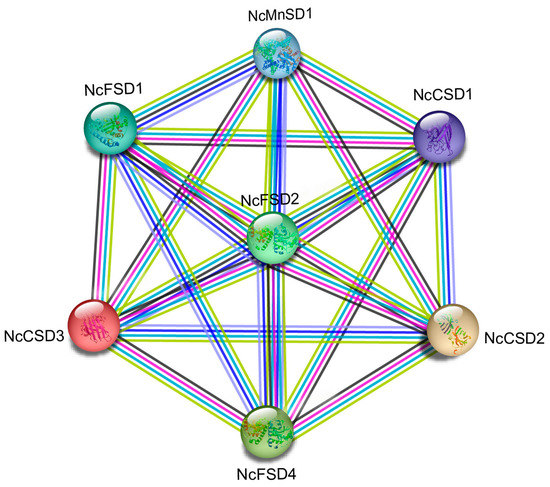
Figure 7.
Protein interaction linkage among the seven SOD genes from Nymphaea colorata. Different colored lines show the interaction of the genes.
3.8. Real-Time Quantitative PCR (RT-qPCR) Analysis of NcSOD Genes under Abiotic Stresses
In order to know the function of SODs, we employed RT-qPCR to examine the expression patterns of the SOD gene under various stress conditions like salinity, heat, cold, and heavy metals (copper sulphate and cadmium chloride). Substantial variations were perceived in the expression levels of the NcSOD genes across various treatments, indicating a complex and dynamic nature of their expression patterns.
Salt treatment strongly induced the expression of all NcCSDs, peaking at 2 and 4 h. Our study found high expression of NcCSDs at 6 h, suggesting its involvement in salt response in N. colorata. Additionally, NcMnSD1, NcFSD1, NcFSD2, and NcFSD5 were strongly induced and highly expressed under salt stress, implying their potential participation in the salt stress response (Figure 8a).
During the heat stress condition, the levels of expression of all NcSOD genes were upregulated at both 2 h and 4 h, with the exception of NcFSD4, which initially showed a decrease at 2 h and then an increase at 4 h. Following the 6 h treatment, the expression levels of the various genes showed variation (Figure 8b).
Under the cold treatment, distinct expression profiles were observed among all NcSODs. NcCSD3, NcFSD1, and NcFSD4 exhibited upregulated expression at almost all time points, and reached their maximum expression at 2 and at 4 h, while NcCSD1 and NcCSD2 expression was slightly low. On the other hand, the remaining members showed down-regulated expression (Figure 8c).
NcSOD genes showed a positive response against heavy metals. In response to the copper sulphate treatment, the expressions of NcSOD genes exhibited variations at different time points. NcCSD3 and NcFSD1 displayed high expression at 2, 4 and 6 h, while other genes were low-expressed and different levels of expression were recorded. Furthermore, the highest expression levels for all genes were observed at the 6 h treatment (Figure 8d). During the cadmium chloride treatment, all NcSOD genes were upregulated at 2 h. Notably; NcCSD3 exhibited consistently high expression levels across all time points, as shown in the figure. At the 4 h and 6 h treatment, all genes experienced a gradual increase and demonstrated robust expression levels (Figure 8e). These findings can enhance our comprehension of NcSOD genes across various environmental conditions.
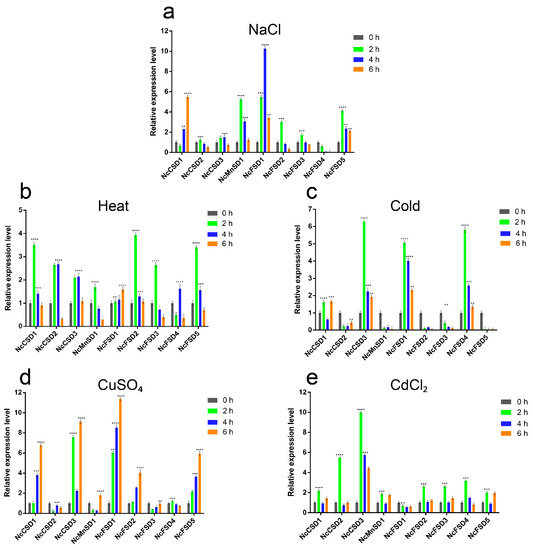
Figure 8.
RT-qPCR analysis of the expression patterns of NcSOD genes in the leaves under various abiotic stresses: (a) salt; (b) heat; (c) cold; (d) CuSO4; and (e) CdCl2 stress (0 (CK), 2, 4, and 6 h). Data presented as means, ± standard error, n = 3; statistically significant differences are exhibited by asterisks (p ≤ 0.05), according to the LSD test.
4. Discussion
Water lilies, with their significant ornamental, economic, medicinal, and cultural value, face challenges stemming from various abiotic stressors. However, through a combination of scientific research, technological innovations, and sustainable practices, we can optimize the growth and production of water lilies while preserving their aesthetic and functional benefits [45]. SODs have been demonstrated in recent studies to secure plants against abiotic stress factors including cold, drought, heat, salinity, ethylene, and abscisic acid [20,21,22,23,24,25]. In last several years, various plant species have been found to contain SOD family genes. For example, the aquatic sea grass (Zostera marina) has five SOD genes [26], Medicago truncatula [17] and barley (Hordeum vulgare) contain seven genes [28], sorghum (Sorghum bicolor) has eight genes [9], tomato (Solanum lycopersicum) has nine genes, and grapevine (Vitis vinifera) has ten genes [46]. Thus, we explored this family in four representative water lily species and checked the expression analysis in Nymphaea colorata.
In the present study, 43 SOD genes were identified in four water lily species, including 19 Fe-SODs, 15 Cu/Zn-SODs, and 9 Mn-SODs in all species (Table 1). The genes were classified into three major groups according to their binding domain (Figure 1). The number of genes in various water lily species was similar to that in cucumbers (Cucumis sativus) (9) and grapes (Vitis vinifera) (10), but fewer genes than the polyploidy crops cotton (Gossypium hirustum) (18) and wheat (Triticum aestivum) (26). However, the number of genes that encode Fe, Cu/Zn, and Mn-SOD differ among various species. For instance, N. colorata has three Cu/Zn-SODs, five Fe-SODs, and one Mn-SOD. The variation in SOD family member number could be due to the changes in genome sizes among species.
Previous research has shown that Cu/ZnSODs are consistently acidic, while Fe-MnSODs can be acidic or basic [42]. Most of the species investigated in this study displayed acidic properties. The results of subcellular localization of SOD proteins revealed that Cu/Zn-SODs are likely to be expressed in the cytoplasm, but Mn-SODs and Fe-SODs are expressed in mitochondria and chloroplasts, respectively, consistent with previous studies on SODs [18]. These distinct cellular locations enable Fe-SODs, Cu/Zn-SODs, and Mn-SODs to collaborate with one another to maintain the balance of free radicals in cells by functioning in different cellular parts.
Previous studies have indicated that the majority of cytoplasmic and chloroplast SODs comprise seven introns [13]. However, in our study it was revealed that NcSOD had a variable amount of exons ranging from 1 to 9. Furthermore, the number of introns in NcSOD varied from 5 to 9 (Figure 5). Notably, Figure 5 indicates that NcFSD1 comprises only one exon and lacks introns. The variability in the gene structure of SODs may arise from the mechanisms involving the insertion or deletion of exons and introns [47].
Various research studies have demonstrated that SOD genes from distinct species are divided into three subfamilies [12]. In our study, we examined the evolutionary connections of SOD proteins in N. colorata, N. thermarum, N. minuta, N. mexicana, and A. trichopoda which categorized within three subfamilies (Figure 2a): Fe, Cu/Zn, and Mn-SOD. Within the phylogenetic tree, the three subfamilies were classified into two distinct groups: Cu/ZnSODs and Fe-MnSODs. FeSODs and MnSODs were clustered together, and a high bootstrap value separated them. The water lily SODs exhibited a strong clustering relationship with closely related species, while showing less affinity with outgroup. This suggests that this gene family has undergone relatively conserved evolution. The presence of specific domains suggests a basis for classifying these genes and the possibility of shared ancestral genes. In cotton (Gossypium hirustum), the MSD and FSD families were found to have originated from a common ancestor, while the CSD subfamily developed independently. As a result, the two major groups expanded separately, as reported by Wang [48].
The analysis of promoters unveiled the existence of three main kinds of cis-components related to light, abiotic stress, and hormones response. Additionally, there were cis-elements associated with tissue-specific expression and developmental processes. Significant quantities of light-responsive cis-components were identified within SOD gene promoters, indicating the potential involvement of SODs in the abiotic stress response. Numerous investigations indicated the participation of SOD genes in the abiotic stress response across diverse plant species, including maize (Zea mays), Pennisetum glaucum, Dendrobium catenatum, and Arabidopsis [36,49,50,51]. Moreover, SOD gene promoters were found to contain a range of cis-elements linked to abiotic stress responses, including ARE, ABRE, MBS, ERE, Box-4, and TC-rich repeats. These cis-elements potentially contribute to the regulation of gene expression under diverse stress conditions. Among plant species like Arabidopsis, banana (Musa paradisiaca), rice (Oryza sativa), tomato (Solanum lycopersicum), poplar (Populus angusti-folia), and cotton (Gossypium herbaceum), the majority of SOD genes exhibit inducibility in response to various abiotic stresses [4,36,52,53,54,55]. Under various abiotic and hormones stress situations the SOD gene family were recently identified by many researchers in various different types of plants like in Brassica napus [25], Zostera marina [26], Salvia miltiorrhiza [27], and Hordeum vulgare [28].
The 3D structures of water lily SOD proteins remain relatively conserved, similar to conserved domains, gene structure, and phylogeny. The findings indicate that water lily SODs genes potentially perform diverse functions across various tissues and genotypes. These results supported earlier anticipated three dimensional structures of SODs in G. arboretum [54], sorghum (Sorghum bicolor) [9], rice (Oryza sative) [52], and in Gossypium raimondii. The preceding investigation demonstrated that metal ion binding active sites and the formation of conserved disulfide bonds within individual subunits contribute to protein stability, specificity, and dimerization [56].
To determine the specific expression profiles of NcSOD genes during various stages of development, we utilized RNA-seq data from ovules and pollen at various developmental stages. By analyzing the RNA-sequencing data from N. colorata, and examined the expressions of the 9 NcSOD genes in pollen and ovules at different days post-anthesis. Our analysis revealed that NcFSD3, NcFSD5, and NcMnSD1 were highly expressed in ovules at 0, 1, 2, and 3 days, while showing relatively lower expression in pollen on day 1 (Figure 6). While NcFSD1 and NcCSD3 showed no expression levels in both ovules and pollen that are in agreement with earlier findings [46].
The RT-qPCR analyses offer valuable insights into the potential role of NcSODs in reaction to diverse stresses. Our research revealed significant changes in the expression levels of nine NcSODs in varied stress environments, suggesting their crucial regulatory role in response to stress and possible functional interconnections. Overexpressing Cu/ZnSODs improved salinity stress resistance in Triticum aestivum, Oryza sativa, Puccinellia tenuiflora, and Arabidopsis [57,58]. Salt treatment strongly induced the expression of all NcCSDs, peaking at 2 h and 4 h. Our study found high expression of NcCSD at 6 h, suggesting its involvement in salt response in Nymphaea colorata. Additionally, NcMnSD1, NcFSD1, NcFSD2, and NcFSD5 were strongly induced and highly expressed under salt stress, implying their potential participation in the salt stress response, similar to NcCSDs, in N. colorata (Figure 8a). Particularly, most NcSOD genes exhibited upregulation throughout heat treatment, with some displaying analogous expression patterns (Figure 8b). During cold treatment, distinct expression profiles were observed among all NcSODs. NcCSD3, NcFSD1, and NcFSD4 exhibited upregulated expression at almost all time points, and reached their maximum expression at 2 h and at 4 h, while NcCSD1 and NcCSD2 were slightly expressed. On the other hand, the remaining members showed down-regulated expression (Figure 8c). These conclusions are consistent with previous findings, which reported a notable increase in SOD activity in rapeseed (Brassica napus) under cold stress conditions [59]. NcSOD genes showed a positive response against heavy metals. In response to the copper sulphate treatment, the expressions of NcSOD genes exhibited variations at different time points (Figure 8d). During the cadmium chloride treatment, all NcSOD genes were upregulated at 2 h. Notably, NcCSD3 exhibited consistently high expression levels across all time points, as showed in the (Figure 8e). Nevertheless, certain genes within the nine NcSODs exhibited a pattern of initially increasing and subsequently decreasing expressions in response to both heavy metal treatments. Similar results were also reported in reaction to heavy metals treatment in several plants [27]. However, experimental verification is still needed to fully elucidate the roles of NcSODs regulatory networks, and their interaction mechanism, under different abiotic stresses.
5. Conclusions
In conclusion, this study conducted a comprehensive genome-wide analysis of the SOD gene family in four representative water lily species, resulting in the identification of 43 water lily SODs. The gathered information, encompassing exon-intron structure, cis-components, protein features, phylogenetic relations, and expression profiles of N. colorata, has shed light on the significant roles played by NcSOD genes in responding to salt, heat, cold, and heavy metal stresses. Findings of this systematic investigation provide a valuable resource for future functional research on NcSOD proteins in biological processes and lay a solid foundation for stress-resistant breeding of N. colorata. To further deepen our understanding of NcSODs’ functions, our future studies will focus on gene engineering and comprehensive analysis, integrating genomics, transcriptomics, proteomics, and metabolomics.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/horticulturae9070781/s1, Table S1: the list of primer was used for gene expression analysis by RT-qPCR, Table S2: the protein sequences of SOD family genes in Arabidopsis thaliana, Table S3: the protein sequences of SOD family genes in four representative water lilies, Table S4: the information of identified 10 motifs in water lily SOD proteins, Table S5: The transcriptome data of Nymphaea colorata from ovules and pollen at various developmental stages.
Author Contributions
F.C. and W.U.K. conceived and designed this project. W.U.K. performed the analyses. L.U.K. and D.C. participated in the data analyses. W.U.K. carried out the experimental study and wrote the draft manuscript. F.C. and W.U.K. checked and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by a grant from National Natural Science Foundation of China (32172614), a grant from Hainan Province Science and Technology Special Fund (ZDYF2023XDNY050), a fund from collaborative Innovation Center of Nanfan and Tropical High Efficient Agriculture, XTCX2022NYB04, and a start-up fund from Hainan Institute of Zhejiang University.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
Acknowledgments
The authors thank the editor and anonymous reviewers for their valuable and insightful comments on this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mittler, R.; Blumwald, E. Genetic Engineering for Modern Agriculture: Challenges and Perspectives. Annu. Rev. Plant Biol. 2010, 61, 443–462. [Google Scholar] [CrossRef] [PubMed]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive Oxygen Species and Antioxidant Defense in Plants under Abiotic Stress: Revisiting the Crucial Role of a Universal Defense Regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Lee, S.-H.; Ahsan, N.; Lee, K.-W.; Kim, D.-H.; Lee, D.-G.; Kwak, S.-S.; Kwon, S.-Y.; Kim, T.-H.; Lee, B.-H. Simultaneous overexpression of both CuZn superoxide dismutase and ascorbate peroxidase in transgenic tall fescue plants confers increased tolerance to a wide range of abiotic stresses. J. Plant Physiol. 2007, 164, 1626–1638. [Google Scholar] [CrossRef]
- Karuppanapandian, T.; Wang, H.W.; Prabakaran, N.; Jeyalakshmi, K.; Kwon, M.; Manoharan, K.; Kim, W. 2,4-dichlorophenoxyacetic acid-induced leaf senescence in mung bean (Vigna radiata L. Wilczek) and senescence inhibition by co-treatment with silver nanoparticles. Plant Physiol. Biochem. 2011, 49, 168–177. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, P.; Umar, S.; Sharma, S. Mechanism of Free Radical Scavenging and Role of Phytohormones in Plants Under Abiotic Stresses. Plant Adapt. Phytoremediation 2010, 99–118. [Google Scholar] [CrossRef]
- Bafana, A.; Dutt, S.; Kumar, S.; Ahuja, P.S. Superoxide dismutase: An industrial perspective. Crit. Rev. Biotechnol. 2011, 31, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Filiz, E.; Tombuloğlu, H. Genome-Wide Distribution of Superoxide Dismutase (SOD) Gene Families in Sorghum Bicolor. Turkish J. Biol. 2015, 39, 49–59. [Google Scholar] [CrossRef]
- Hodgson, E.K.; Fridovich, I. Reversal of the superoxide dismutase reaction. Biochem. Biophys. Res. Commun. 1973, 54, 270–274. [Google Scholar] [CrossRef]
- Brawn, K.; Fridovich, I. Superoxide Radical and Superoxide Dismutases: Threat and Defense. Autoxid. Food Biol. Syst. 1980, 429–446. [Google Scholar] [CrossRef]
- Fink, R.C.; Scandalios, J.G. Molecular Evolution and Structure–Function Relationships of the Superoxide Dismutase Gene Families in Angiosperms and Their Relationship to Other Eukaryotic and Prokaryotic Superoxide Dismutases. Arch. Biochem. Biophys. 2002, 399, 19–36. [Google Scholar] [CrossRef] [PubMed]
- Abreu, I.A.; Cabelli, D.E. Superoxide dismutases—A review of the metal-associated mechanistic variations. Biochim. Biophys. Acta (BBA)—Proteins Proteom. 2010, 1804, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Dupont, C.L.; Neupane, K.; Shearer, J.; Palenik, B. Diversity, function and evolution of genes coding for putative Ni-containing superoxide dismutases. Environ. Microbiol. 2008, 10, 1831–1843. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.; Wang, W.; Yuan, R.; Den, F.; Shen, F.F. Superoxide Dismutase and Its Research in Plant Stress-Tolerance. Mol. Plant Breed. 2015, 13, 2633–2646. [Google Scholar]
- Zeng, X.-C.; Liu, Z.-G.; Shi, P.-H.; Xu, Y.-Z.; Sun, J.; Fang, Y.; Yang, G.; Wu, J.-Y.; Kong, D.-J.; Sun, W.-C. Cloning and Expression Analysis of Copper and Zinc Superoxide Dismutase (Cu/Zn-SOD) Gene from Brassica campestris L. Acta Agron. Sin. 2014, 40, 636–643. [Google Scholar] [CrossRef]
- Song, J.; Zeng, L.; Chen, R.; Wang, Y.; Zhou, Y. In Silico Identification and Expression Analysis of Superoxide Dismutase (SOD) Gene Family in Medicago Truncatula. 3 Biotech 2018, 8, 348. [Google Scholar] [CrossRef]
- Perry, J.J.P.; Shin, D.S.; Getzoff, E.D.; Tainer, J.A. The structural biochemistry of the superoxide dismutases. Biochim. Biophys. Acta (BBA)—Proteins Proteom. 2010, 1804, 245–262. [Google Scholar] [CrossRef]
- Wang, B.; Lüttge, U.; Ratajczak, R. Specific regulation of SOD isoforms by NaCl and osmotic stress in leaves of the C3 halophyte Suaeda salsa L. J. Plant Physiol. 2004, 161, 285–293. [Google Scholar] [CrossRef]
- Pilon, M.; Ravet, K.; Tapken, W. The Biogenesis and Physiological Function of Chloroplast Superoxide Dismutases. Biochim. Biophys. Acta (BBA)—Bioenerg. 2011, 1807, 989–998. [Google Scholar] [CrossRef]
- Krouma, A.; Drevon, J.-J.; Abdelly, C. Genotypic variation of N2-fixing common bean (Phaseolus vulgaris L.) in response to iron deficiency. J. Plant Physiol. 2006, 163, 1094–1100. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Lai, Z.; Lin, Y.; Lai, G.; Lian, C. Genome-wide identification and characterization of the superoxide dismutase gene family in Musa acuminata cv. Tianbaojiao (AAA group). BMC Genom. 2015, 16, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Yu, J.; Cheng, Y.; Ruan, M.; Wang, R.; Ye, Q.; Zhou, G.; Li, Z.; Yao, Z.; Yang, Y.; et al. The SOD Gene Family in Tomato: Identification, Phylogenetic Relationships, and Expression Patterns. Front. Plant Sci. 2016, 7, 1279. [Google Scholar] [CrossRef]
- Yan, J.-J.; Zhang, L.; Wang, R.-Q.; Xie, B.; Li, X.; Chen, R.-L.; Guo, L.-X.; Xie, B.-G. The Sequence Characteristics and Expression Models Reveal Superoxide Dismutase Involved in Cold Response and Fruiting Body Development in Volvariella volvacea. Int. J. Mol. Sci. 2015, 17, 34. [Google Scholar] [CrossRef]
- Su, W.; Raza, A.; Gao, A.; Jia, Z.; Zhang, Y.; Hussain, M.A.; Mehmood, S.S.; Cheng, Y.; Lv, Y.; Zou, X. Genome-Wide Analysis and Expression Profile of Superoxide Dismutase (SOD) Gene Family in Rapeseed (Brassica napus L.) under Different Hormones and Abiotic Stress Conditions. Antioxidants 2021, 10, 1182. [Google Scholar] [CrossRef] [PubMed]
- Zang, Y.; Chen, J.; Li, R.; Shang, S.; Tang, X. Genome-wide analysis of the superoxide dismutase (SOD) gene family in Zostera marina and expression profile analysis under temperature stress. PeerJ 2020, 8, e9063. [Google Scholar] [CrossRef] [PubMed]
- Han, L.-M.; Hua, W.-P.; Cao, X.-Y.; Yan, J.-A.; Chen, C.; Wang, Z.-Z. Genome-wide identification and expression analysis of the superoxide dismutase (SOD) gene family in Salvia miltiorrhiza. Gene 2020, 742, 144603. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, L.; Chen, Y.; Wang, S.; Fang, Y.; Zhang, X.; Wu, Y.; Xue, D. Genome-wide identification of the SOD gene family and expression analysis under drought and salt stress in barley. Plant Growth Regul. 2021, 94, 49–60. [Google Scholar] [CrossRef]
- Gomez, J.M.; Hernandez, J.A.; Jimenez, A.; Del Rio, L.A.; Sevilla, F. Differential Response of Antioxidative Enzymes of Chloroplasts and Mitochondria to Long-term NaCl Stress of Pea Plants. Free. Radic. Res. 1999, 31, 11–18. [Google Scholar] [CrossRef]
- Wu, G.; Wilen, R.W.; Robertson, A.J.; Gusta, L.V. Isolation, Chromosomal Localization, and Differential Expression of Mitochondrial Manganese Superoxide Dismutase and Chloroplastic Copper/Zinc Superoxide Dismutase Genes in Wheat. Plant Physiol. 1999, 120, 513–520. [Google Scholar] [CrossRef]
- Baek, K.-H.; Skinner, D.Z. Alteration of antioxidant enzyme gene expression during cold acclimation of near-isogenic wheat lines. Plant Sci. 2003, 165, 1221–1227. [Google Scholar] [CrossRef]
- Srivastava, V.; Srivastava, M.K.; Chibani, K.; Nilsson, R.; Rouhier, N.; Melzer, M.; Wingsle, G. Alternative Splicing Studies of the Reactive Oxygen Species Gene Network in Populus Reveal Two Isoforms of High-Isoelectric-Point Superoxide Dismutase. Plant Physiol. 2009, 149, 1848–1859. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Feng, Z.; Bian, L.; Xie, H.; Liang, J. miR398 regulation in rice of the responses to abiotic and biotic stresses depends on CSD1 and CSD2 expression. Funct. Plant Biol. 2010, 38, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Diao, Y.; Chen, L.; Yang, G.; Zhou, M.; Song, Y.; Hu, Z.; Liu, J.Y. Nuclear DNA C-values in 12 species in Nymphaeales. Caryologia 2006, 59, 25–30. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, F.; Zhang, X.; Li, Z.; Zhao, Y.; Lohaus, R.; Chang, X.; Dong, W.; Ho, S.Y.W.; Liu, X.; et al. The water lily genome and the early evolution of flowering plants. Nature 2020, 577, 79–84. [Google Scholar] [CrossRef]
- Kliebenstein, D.J.; Monde, R.-A.; Last, R.L. Superoxide Dismutase in Arabidopsis: An Eclectic Enzyme Family with Disparate Regulation and Protein Localization. Plant Physiol. 1998, 118, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5857–5864. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The Proteomics Server for in-Depth Protein Knowledge and Analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Edgar, R. MUSCLE: Multiple sequence alignment with improved accuracy and speed. In Proceedings of the 2004 IEEE Computational Systems Bioinformatics Conference, CSB 2004, Stanford, CA, USA, 19 August 2004; pp. 728–729. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for Motif Discovery and Searching. Nucleic Acid Res. 2009, 37, 202–208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Zhou, Y.; Hu, L.; Wu, H.; Jiang, L.; Liu, S. Genome-Wide Identification and Transcriptional Expression Analysis of Cucumber Superoxide Dismutase (SOD) Family in Response to Various Abiotic Stresses. Int. J. Genom. 2017, 2017, 7243973. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P. STRING V11: Protein–Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Xiong, X.; Zhang, J.; Yang, Y.; Chen, Y.; Su, Q.; Zhao, Y.; Wang, J.; Xia, Z.; Wang, L.; Zhang, L.; et al. Water lily research: Past, present, and future. Trop. Plants 2023, 2, 1–8. [Google Scholar] [CrossRef]
- Hu, X.; Hao, C.; Cheng, Z.-M.; Zhong, Y. Genome-Wide Identification, Characterization, and Expression Analysis of the Grapevine Superoxide Dismutase (SOD) Family. Int. J. Genom. 2019, 2019, 7350414. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon–intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhang, X.; Deng, F.; Yuan, R.; Shen, F. Genome-wide characterization and expression analyses of superoxide dismutase (SOD) genes in Gossypium hirsutum. BMC Genom. 2017, 18, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Divya, K.; Kavi Kishor, P.B.; Bhatnagar-Mathur, P.; Singam, P.; Sharma, K.K.; Vadez, V.; Reddy, P.S. Isolation and functional characterization of three abiotic stress-inducible (Apx, Dhn and Hsc70) promoters from pearl millet (Pennisetum glaucum L.). Mol. Biol. Rep. 2019, 46, 6039–6052. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Wang, H.; Tong, Y.; Wang, Y. Insights into the Superoxide Dismutase Gene Family and Its Roles in Dendrobium catenatum under Abiotic Stresses. Plants 2020, 9, 1452. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, L.; Shang, J.; Hu, X.; Yu, H.; Wu, H.; Lv, W.; Zhao, Y. Genome-wide analysis of the maize superoxide dismutase (SOD) gene family reveals important roles in drought and salt responses. Genet. Mol. Biol. 2021, 44, 1–12. [Google Scholar] [CrossRef]
- Dehury, B.; Sarma, K.; Sarmah, R.; Sahu, J.; Sahoo, S.; Sahu, M.; Sen, P.; Modi, M.K.; Sharma, G.D.; Choudhury, M.D.; et al. In silico analyses of superoxide dismutases (SODs) of rice (Oryza sativa L.). J. Plant Biochem. Biotechnol. 2013, 22, 150–156. [Google Scholar] [CrossRef]
- Nath, K.; Kumar, S.; Poudyal, R.S.; Yang, Y.N.; Timilsina, R.; Park, Y.S.; Nath, J.; Chauhan, P.S.; Pant, B.; Lee, C.-H. Developmental stage-dependent differential gene expression of superoxide dismutase isoenzymes and their localization and physical interaction network in rice (Oryza sativa L.). Genes Genom. 2014, 36, 45–55. [Google Scholar] [CrossRef]
- Wang, W.; Xia, M.; Chen, J.; Deng, F.; Yuan, R.; Zhang, X.; Shen, F. Genome-wide analysis of superoxide dismutase gene family in Gossypium raimondii and G. arboreum. Plant Gene 2016, 6, 18–29. [Google Scholar] [CrossRef]
- Verma, D.; Lakhanpal, N.; Singh, K. Genome-wide identification and characterization of abiotic-stress responsive SOD (superoxide dismutase) gene family in Brassica juncea and B. rapa. BMC Genom. 2019, 20, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Fisher, C.L.; Cabelli, D.E.; Tainer, J.A.; Hallewell, R.A.; Getzoff, E.D. The role of arginine 143 in the electrostatics and mechanism of Cu, Zn superoxide dismutase: Computational and experimental evaluation by mutational analysis. Proteins Struct. Funct. Bioinform. 1994, 19, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, X.; Xiao, Z.; Yin, X.; Xing, T.; Xia, G. A wheat superoxide dismutase gene TaSOD2 enhances salt resistance through modulating redox homeostasis by promoting NADPH oxidase activity. Plant Mol. Biol. 2016, 91, 115–130. [Google Scholar] [CrossRef]
- Guan, Q.; Liao, X.; He, M.; Li, X.; Wang, Z.; Ma, H.; Yu, S.; Liu, S. Tolerance Analysis of Chloroplast OsCu/Zn-SOD Overexpressing Rice under NaCl and NaHCO3 Stress. PLoS ONE 2017, 12, e0186052. [Google Scholar] [CrossRef]
- He, H.; Lei, Y.; Yi, Z.; Raza, A.; Zeng, L.; Yan, L.; Xiaoyu, D.; Yong, C.; Xiling, Z. Study on the mechanism of exogenous serotonin improving cold tolerance of rapeseed (Brassica napus L.) seedlings. Plant Growth Regul. 2021, 94, 161–170. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).