Genome-Wide Identification of the RsSWEET Gene Family and Functional Analysis of RsSWEET17 in Root Growth and Development in Radish
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of SWEET Genes in Radish
2.2. Phylogenetic Analysis and Gene Structure Analyses of the RsSWEET Genes
2.3. Promoter Cis-Element Analysis of RsSWEETs
2.4. Plant Materials and Treatments
2.5. Expression Profile Analysis of RsSWEET Genes
2.6. Functional Complementation Analyses in Yeast
2.7. Soluble Sugar Determination
2.8. RNA Isolation and RT-qPCR
2.9. Statistical Analysis
3. Results
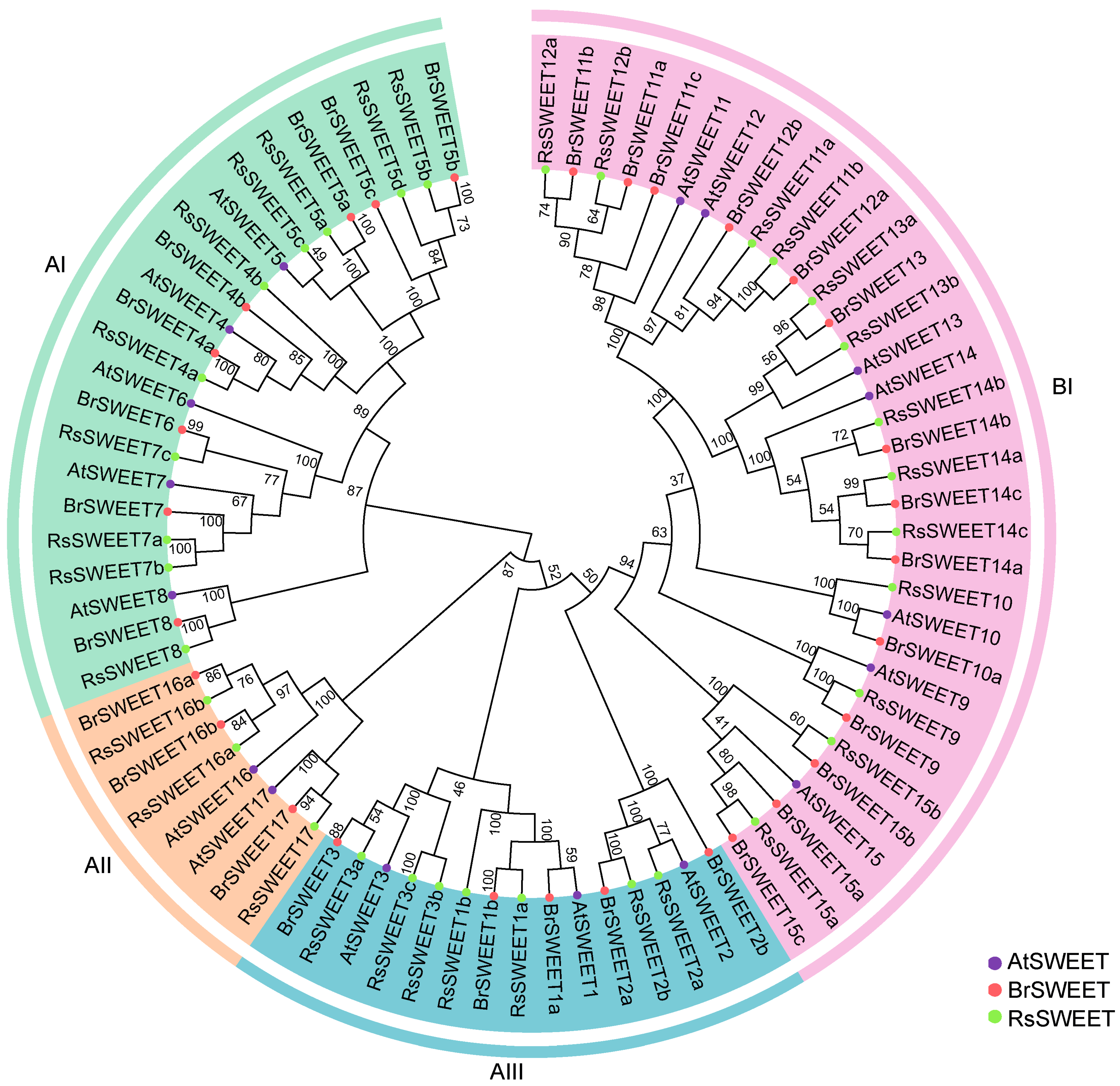
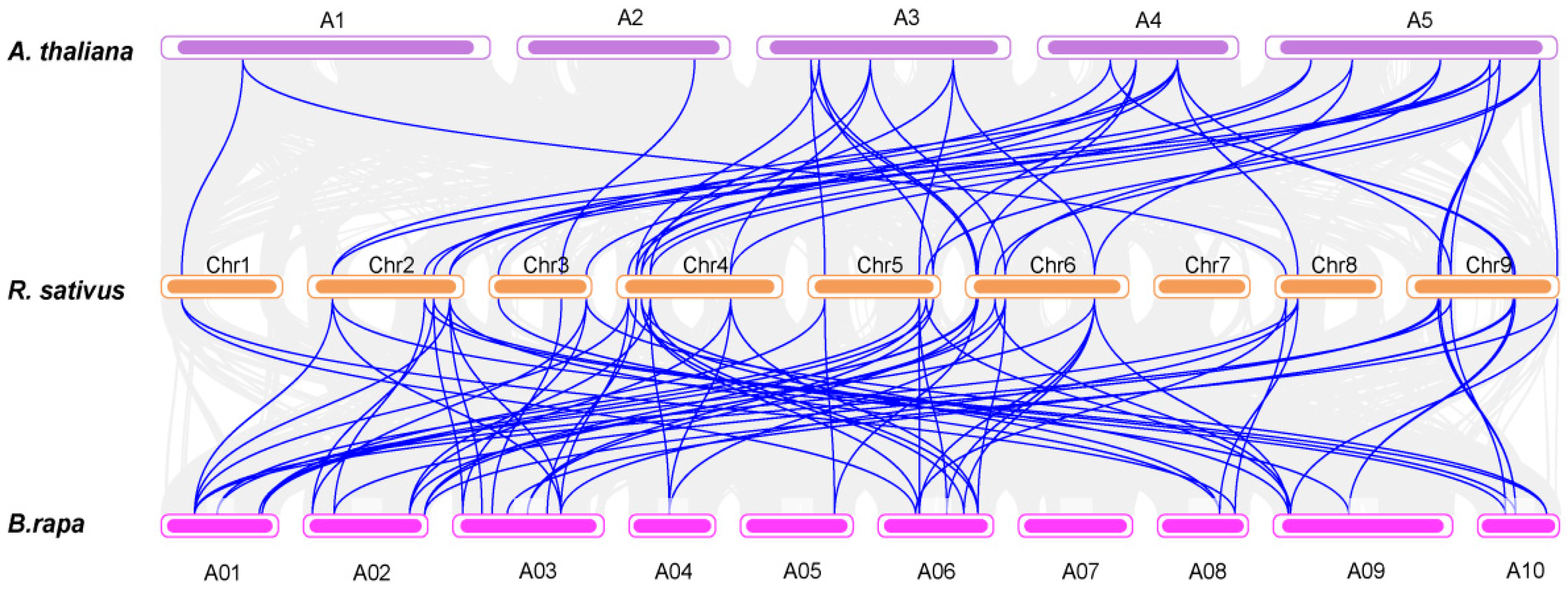
3.1. Identification, Phylogenetic and Collinearity Analysis of RsSWEETs in Radish
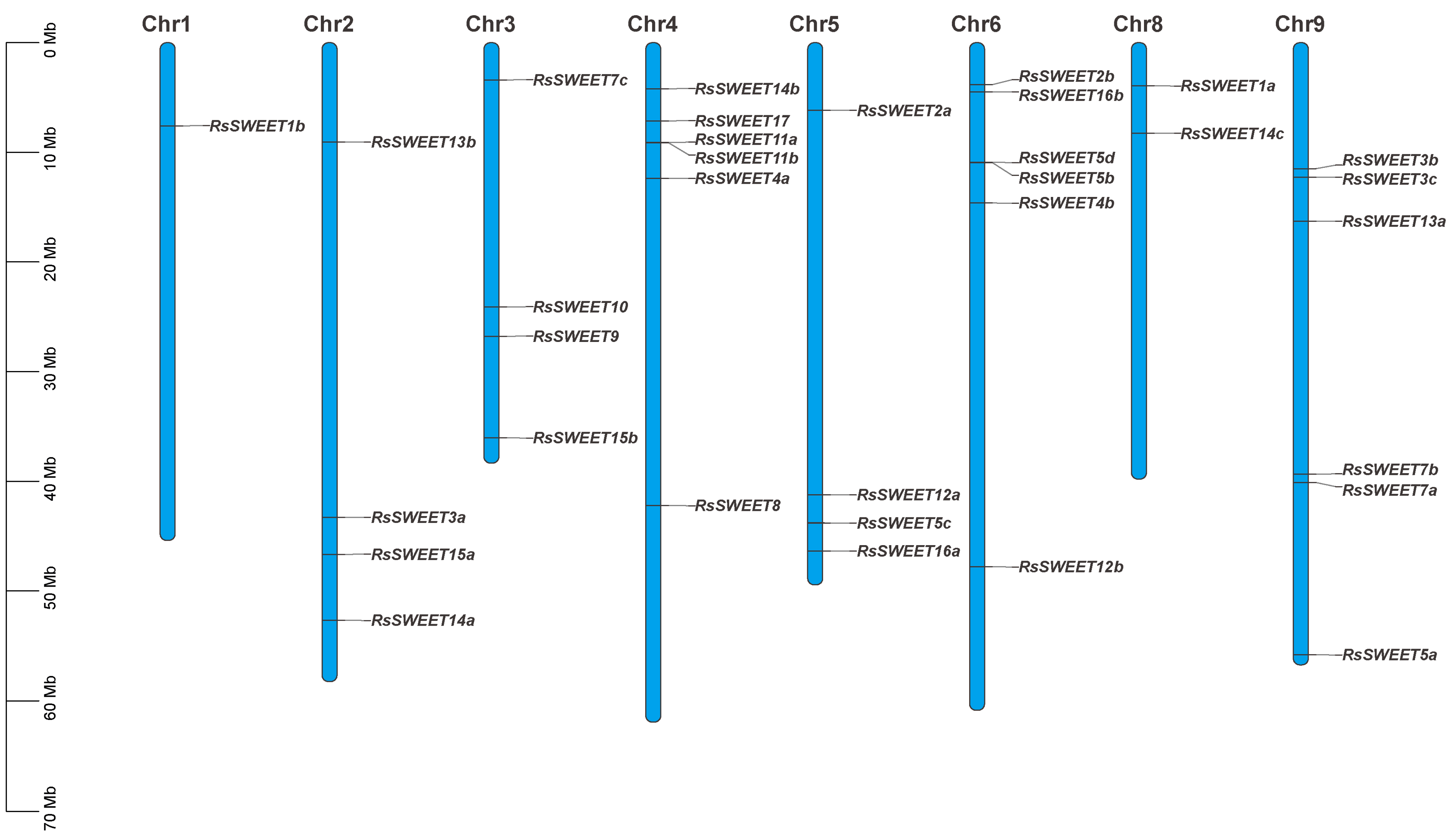
3.2. Chromosomal Distribution of RsSWEET Proteins in Radish
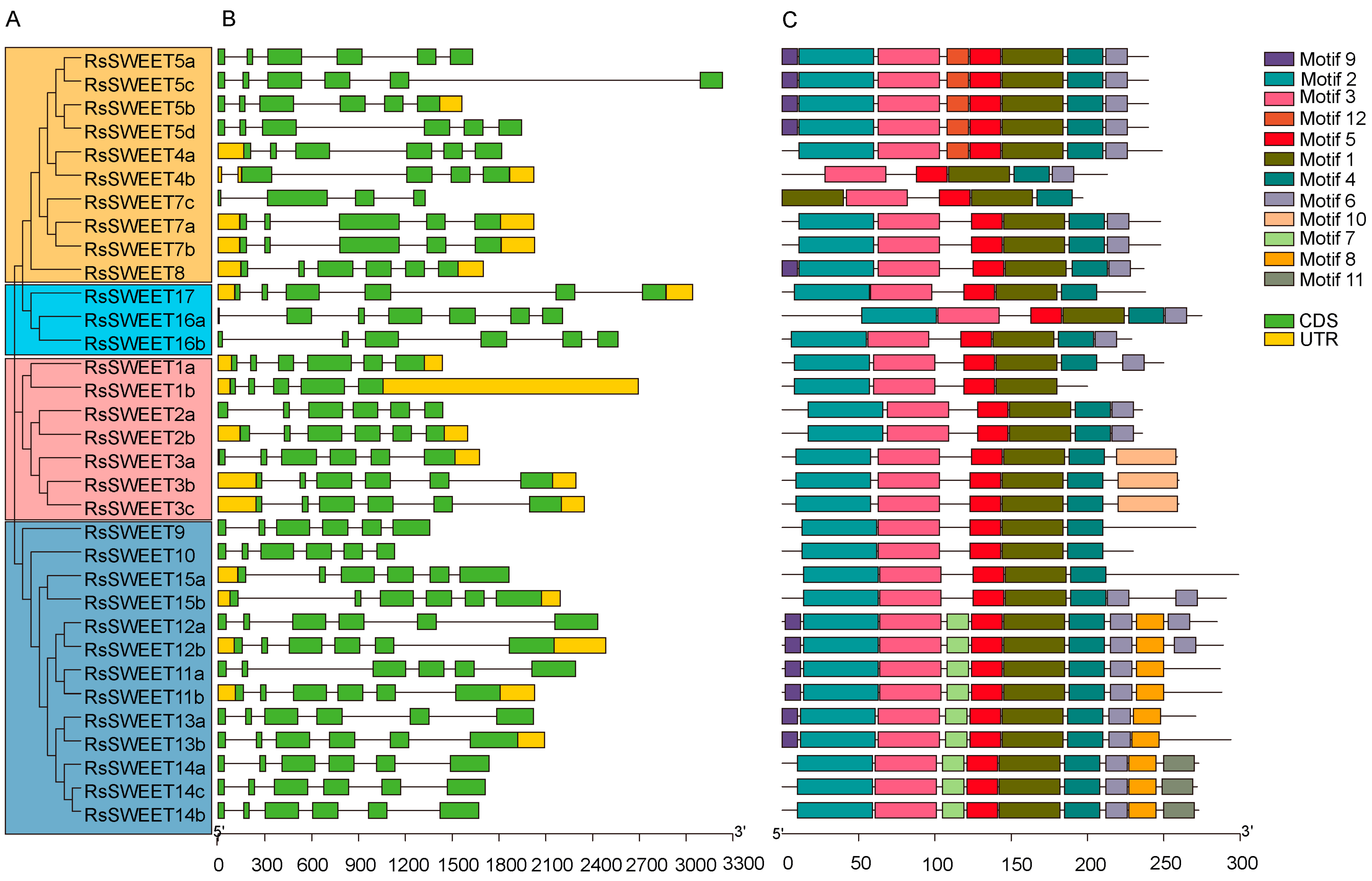
3.3. Gene Structure and Conserved Motif Distribution of RsSWEETs
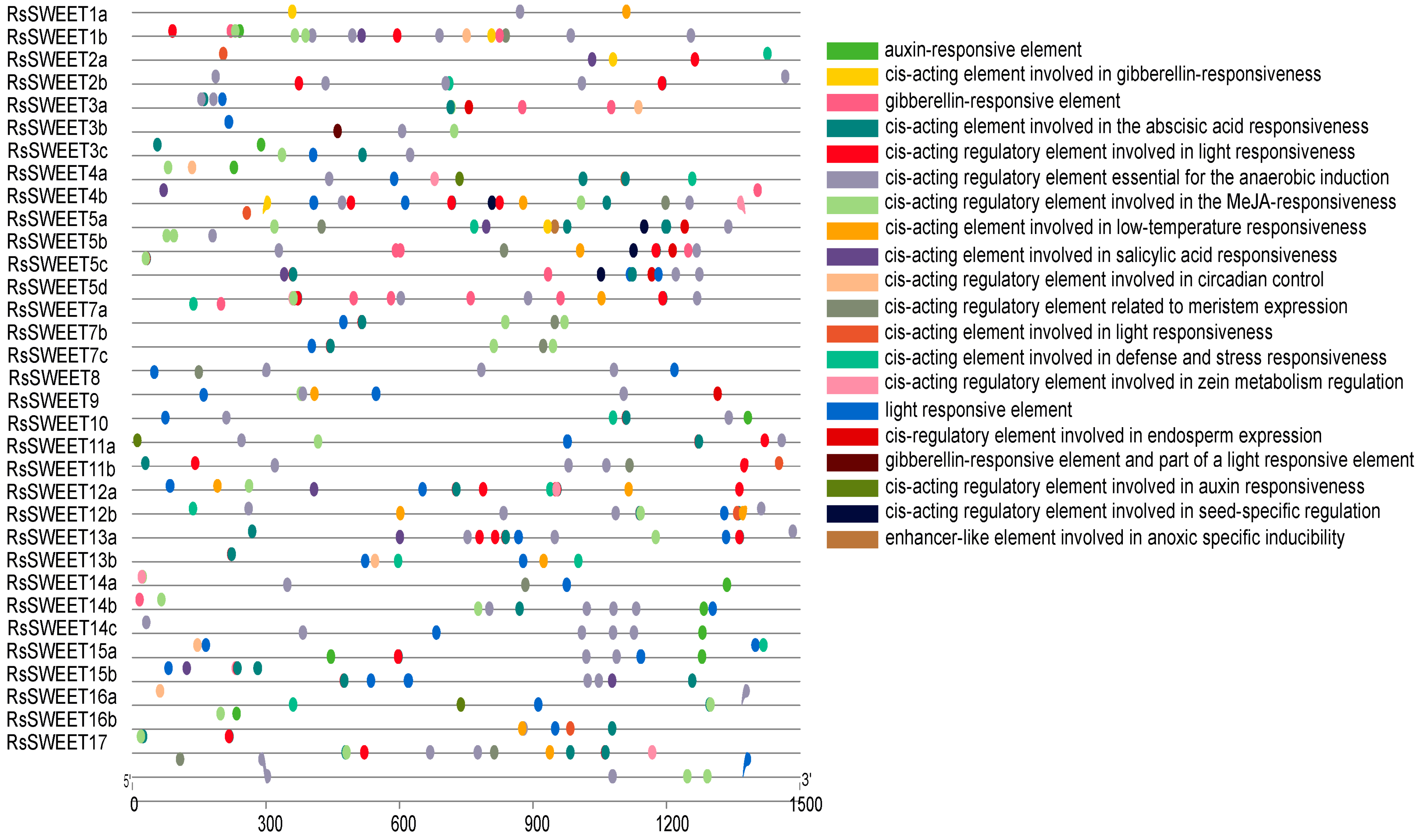
3.4. The Cis-Acting Regulatory Elements in RsSWEET Gene Promoters
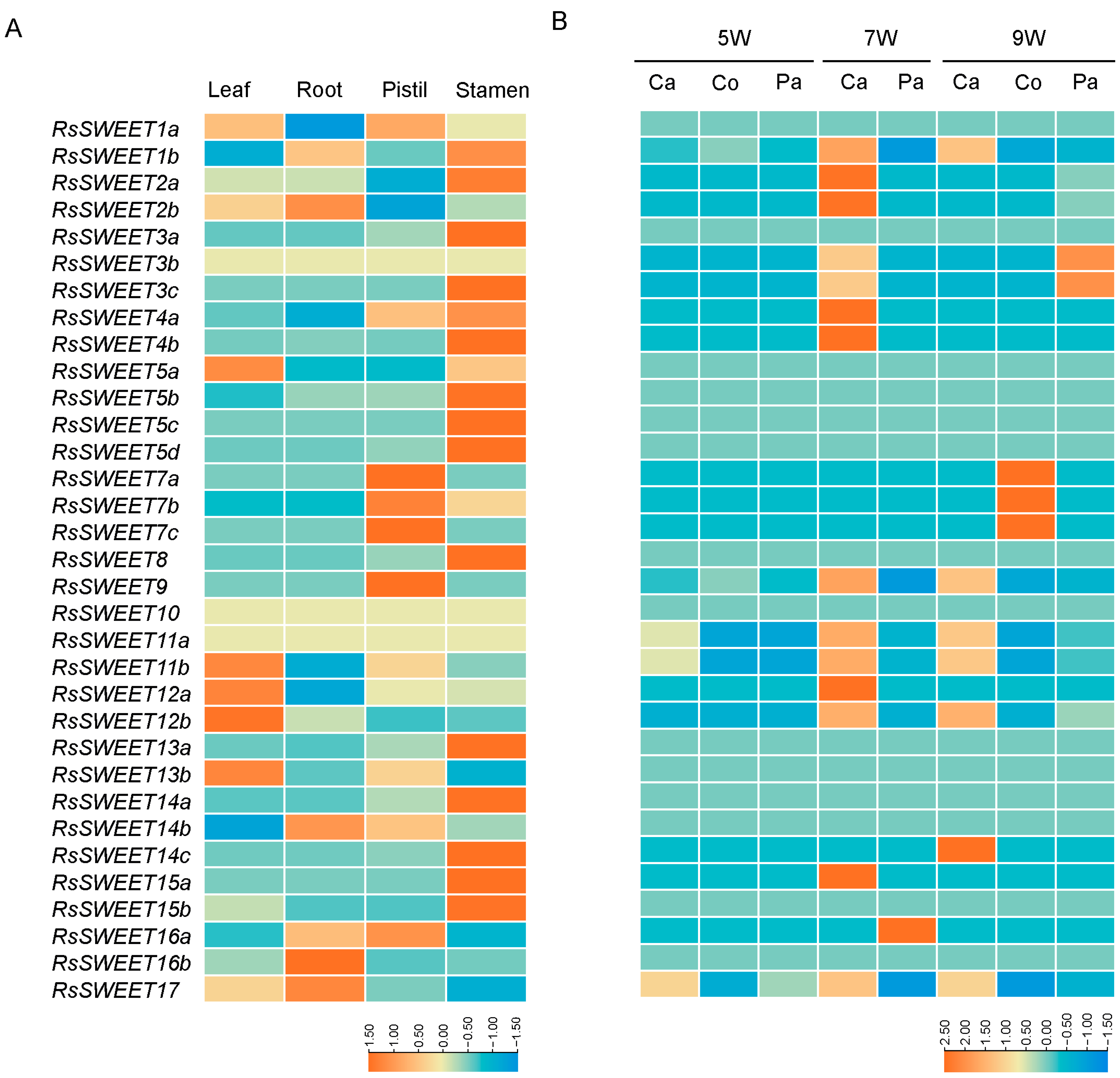
3.5. Differential Expression Analysis of RsSWEET Genes
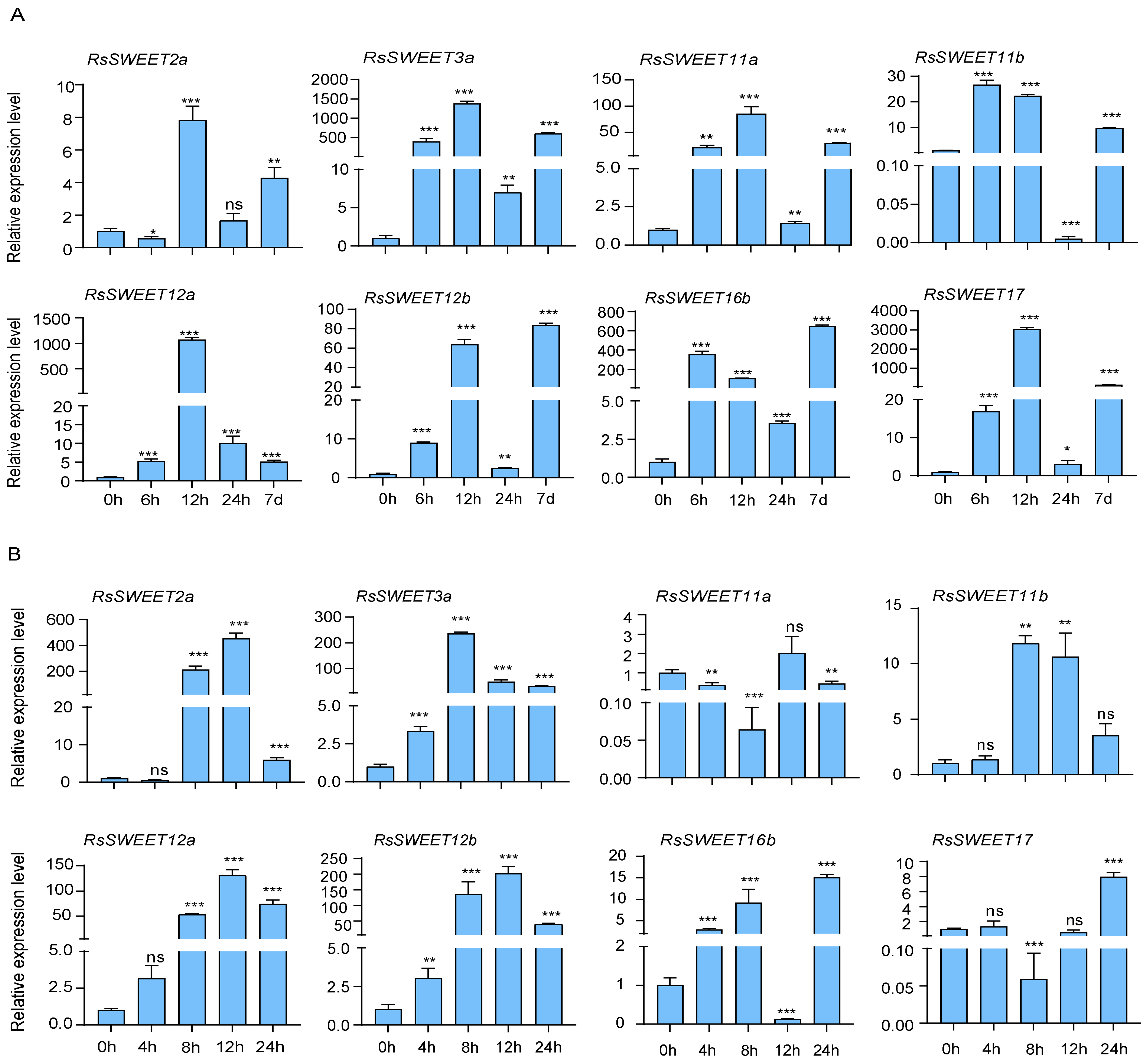
3.6. Expression Profiles of RsSWEET Genes under Abiotic Stress in Radish
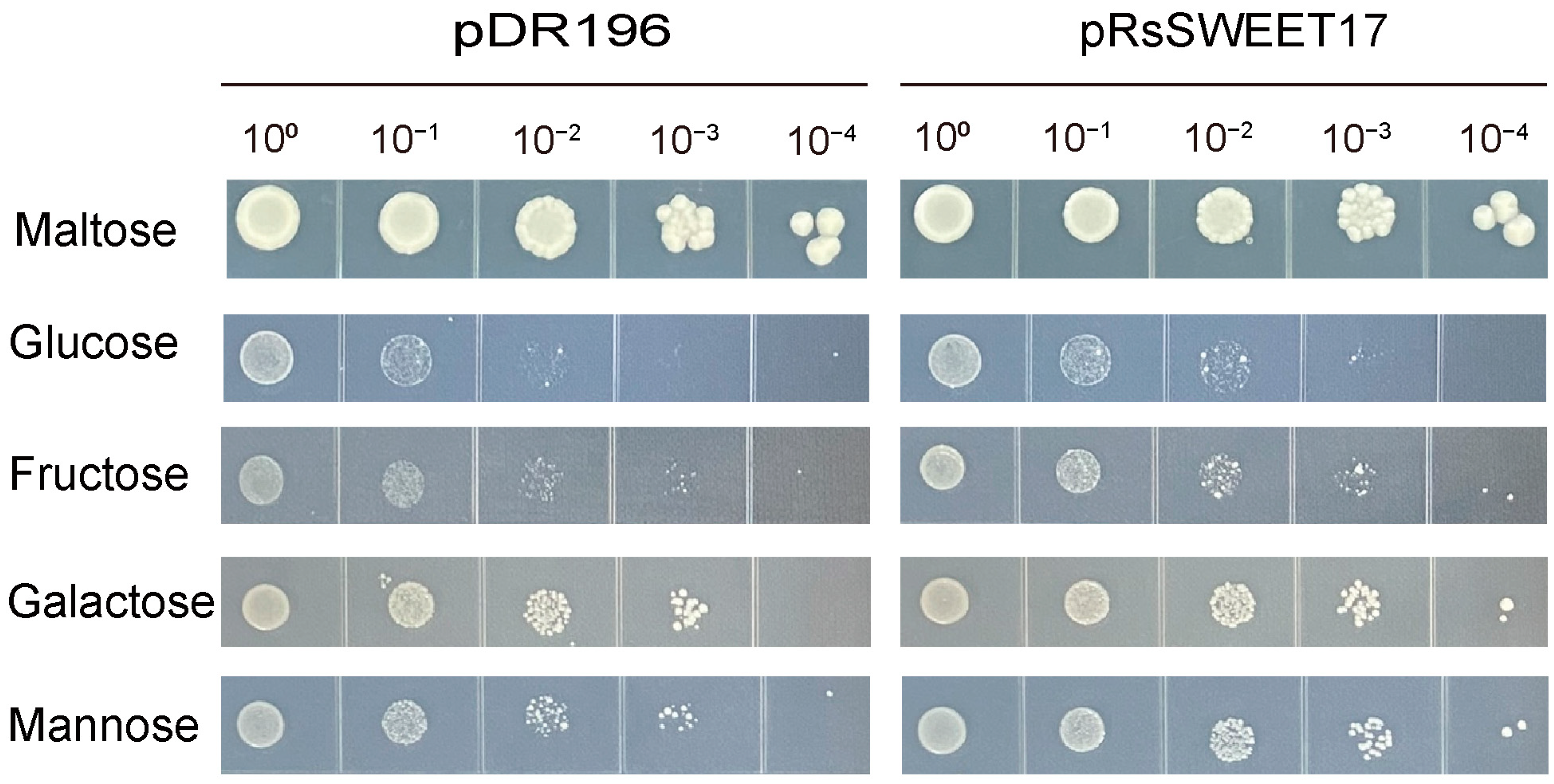
3.7. Sugar Transport Activity of RsSWEET7 in Yeast
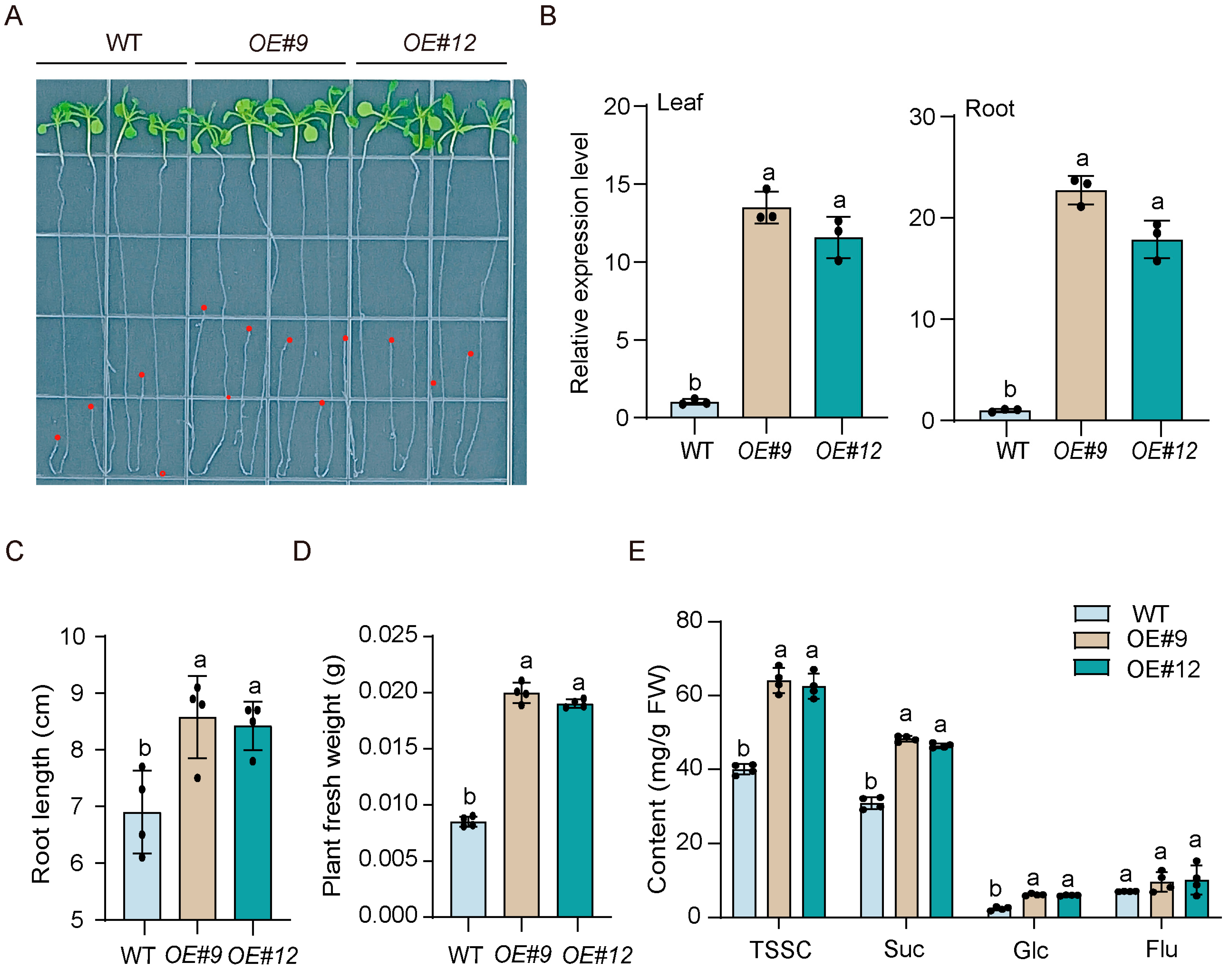
3.8. Overexpression of RsSWEET17 Improves Root Sugar Accumulation and Root Growth
4. Discussion
4.1. Characterization of the RsSWEET Gene Family in Radish
4.2. Potential Roles of RsSWEET Genes in Various Stress Responses and Plant Growth and Development
4.3. RsSWEET17 Positively Regulates Sugar Accumulation and Root Growth in Radish
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Walmsley, A.R.; Barrett, M.P.; Bringaud, F.; Gould, G.W. Sugar transporters from bacteria, parasites and mammals: Structure-activity relationships. Trends Biochem. Sci. 1988, 23, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Ding, C.; Hao, X.; Zeng, J.; Yang, Y.; Wang, X.; Wang, L. CsSWEET1a and CsSWEET17 mediate growth and freezing tolerance by promoting sugar transport across the plasma membrane. Plant. Cell Physiol. 2020, 61, 1669–1682. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant. Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef]
- Ko, H.Y.; Ho, L.H.; Neuhaus, H.E.; Guo, W.J. Transporter SlSWEET15 unloads sucrose from phloem and seed coat for fruit and seed development in tomato. Plant Physiol. 2021, 187, 2230–2245. [Google Scholar] [CrossRef] [PubMed]
- Turgeon, R. The role of phloem loading reconsidered. Plant Physiol. 2010, 152, 1817–1823. [Google Scholar] [CrossRef]
- Chen, L.; Qu, X.; Hou, B.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef]
- Chen, L.Q.; Lin, I.W.; Qu, X.Q.; Sosso, D.; McFarlane, H.E.; Londoño, A.; Samuels, A.L.; Frommer, W.B. A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 2015, 27, 607–619. [Google Scholar] [CrossRef]
- Ji, J.; Yang, L.; Fang, Z.; Zhang, Y.; Zhuang, M.; Lv, H.; Wang, Y. Plant SWEET family of sugar transporters: Structure, evolution and biological functions. Biomolecules 2022, 12, 205. [Google Scholar] [CrossRef]
- Abelenda, J.A.; Bergonzi, S.; Oortwijn, M.; Sonnewald, S.; Du, M.; Visser, R.G.F.; Sonnewald, U.; Bachem, C.W.B. Source-Sink regulation is mediated by interaction of an FT homolog with a SWEET Protein in Potato. Curr. Biol. 2019, 29, 1178–1186. [Google Scholar] [CrossRef]
- Liesche, J. Sucrose transporters and plasmodesmal regulation in passive phloem loading. J. Integr. Plant Biol. 2017, 59, 311–321. [Google Scholar] [CrossRef]
- Sun, L.; Sui, X.; Lucas, W.J.; Li, Y.; Feng, S.; Ma, S.; Fan, J.; Gao, L.; Zhang, Z. Down-regulation of the sucrose transporter CsSUT1 causes male sterility by altering carbohydrate supply. Plant Physiol. 2019, 180, 986–997. [Google Scholar] [CrossRef]
- Slewinski, T.L. Diverse functional roles of monosaccharide transporters and their homologs in vascular plants: A physiological perspective. Mol. Plant 2011, 4, 641–662. [Google Scholar] [CrossRef] [PubMed]
- Eom, J.S.; Chen, L.Q.; Sosso, D.; Julius, B.T.; Lin, I.W.; Qu, X.Q.; Braun, D.M.; Frommer, W.B. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015, 25, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Anjali, A.; Fatima, U.; Manu, M.S.; Ramasamy, S.; Senthil-Kumar, M. Structure and regulation of SWEET transporters in plants: An update. Plant Physiol. Biochem. 2020, 156, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Miao, L.; Lv, Y.; Kong, L.; Chen, Q.; Chen, C.; Li, J.; Zeng, F.; Wang, S.; Li, J.; Huang, L.; et al. Genome-wide identification, phylogeny, evolution, and expression patterns of MtN3/saliva/SWEET genes and functional analysis of BcNS in Brassica rapa. BMC Genom. 2018, 19, 174. [Google Scholar] [CrossRef]
- Feng, C.Y.; Han, J.X.; Han, X.X.; Jiang, J. Genome-wide identification, phylogeny, and expression analysis of the SWEET gene family in tomato. Gene 2015, 573, 261–272. [Google Scholar] [CrossRef]
- Sosso, D.; Luo, D.; Li, Q.; Sasse, J.; Yang, J.; Gendrot, G.; Suzuki, M.; Koch, K.E.; Mccarty, D.R.; Chourey, P.S.; et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015, 47, 1489–1493. [Google Scholar] [CrossRef]
- Huang, D.M.; Chen, Y.; Liu, X.; Ni, D.A.; Bai, L.; Qin, Q.P. Genome-wide identification and expression analysis of the SWEET gene family in daylily (Hemerocallis fulva) and functional analysis of HfSWEET17 in response to cold stress. BMC Plant Biol. 2022, 22, 211. [Google Scholar] [CrossRef]
- Li, X.Y.; Guo, W.; Li, J.C.; Yue, P.T.; Bu, H.D.; Jiang, J.; Liu, W.T.; Xu, Y.X.; Yuan, H.; Li, T.; et al. Histone Acetylation at the promoter for the transcription factor PuWRKY31 affects sucrose accumulation in Pear Fruit. Plant Physiol. 2020, 182, 2035–2046. [Google Scholar] [CrossRef]
- Ren, Y.; Li, M.; Guo, S.; Sun, H.; Zhao, J.; Zhang, J.; Liu, G.; He, H.; Tian, S.; Yu, Y.; et al. evolutionary gain of oligosaccharide hydrolysis and sugar transport enhanced carbohydrate partitioning in sweet watermelon fruits. Plant Cell 2021, 33, 1554–1573. [Google Scholar] [CrossRef]
- Chardon, F.; Bedu, M.; Calenge, F.; Klemens, P.A.; Spinner, L.; Clement, G.; Chietera, G.; Léran, S.; Ferrand, M.; Lacombe, B.; et al. Leaf fructose content is controlled by the vacuolar transporter SWEET17 in Arabidopsis. Curr. Biol. 2013, 23, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Valifard, M.; Le Hir, R.; Müller, J.; Scheuring, D.; Neuhaus, H.E.; Pommerrenig, B. Vacuolar fructose transporter SWEET17 is critical for root development and drought tolerance. Plant Physiol. 2021, 187, 2716–2730. [Google Scholar] [CrossRef]
- Shammai, A.; Petreikov, M.; Yeselson, Y.; Faigenboim, A.; Moy-Komemi, M.; Cohen, S.; Cohen, D.; Besaulov, E.; Efrati, A.; Houminer, N.; et al. Natural genetic variation for expression of a SWEET transporter among wild species of Solanum lycopersicum (tomato) determines the hexose composition of ripening tomato fruit. Plant J. 2018, 96, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Asai, Y.; Kobayashi, Y.; Kobayashi, I. Increased expression of the tomato SlSWEET15 gene during grey mold infection and the possible involvement of the sugar effux to apoplasm in the disease susceptibility. J. Plant Pathol. 2016, 7, 329. [Google Scholar] [CrossRef]
- Zhou, A.; Ma, H.; Feng, S.; Gong, S.; Wang, J. DsSWEET17, a Tonoplast-Localized Sugar Transporter from dianthus spiculifolius, affects sugar metabolism and confers multiple stress tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 1564. [Google Scholar] [CrossRef]
- Zhang, R.; Niu, K.; Ma, H. Identification and expression analysis of the SWEET gene family from Poa pratensis under abiotic stresses. DNA Cell Biol. 2020, 39, 1606–1620. [Google Scholar] [CrossRef]
- Mathan, J.; Singh, A.; Ranjan, A. Sucrose transport in response to drought and salt stress involves ABA-mediated induction of OsSWEET13 and OsSWEET15 in rice. Physiol. Plantarum. 2020, 171, 620–637. [Google Scholar] [CrossRef]
- Yang, G.; Xu, H.; Zou, Q.; Zhang, J.; Jiang, S.; Fang, H.; Wang, Y.; Su, M.; Wang, N.; Chen, X. The vacuolar membrane sucrose transporter MdSWEET16 plays essential roles in the cold tolerance of apple. Plant Cell Tiss. Org. 2019, 140, 129–142. [Google Scholar] [CrossRef]
- Xu, L.; Wang, Y.; Dong, J.H.; Zhang, W.; Tang, M.J.; Zhang, W.; Wang, K.; Chen, Y.L.; Zhang, X.L.; He, Q.; et al. A chromosome-level genome assembly of radish (Raphanus sativus L.) reveals insights into genome adaptation and differential bolting regulation. Plant Biotechnol. J. 2023, 21, 990–1004. [Google Scholar] [CrossRef]
- Hoang, N.V.; Park, C.; Kamran, M.; Lee, J.Y. Gene regulatory network guided investigations and engineering of storage root development in root crops. Front. Plant Sci. 2020, 11, 762. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef]
- Zhu, X.; Zhang, X.; Cao, Y.; Xin, R.; Ma, Y.; Wang, L.; Xu, L.; Wang, Y.; Liu, R.; Liu, L. Genome-Wide identification of sucrose transporter genes and functional analysis of RsSUC1b in Radish (Raphanus sativus L.). Horticulturae 2022, 8, 1058. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, 39–49. [Google Scholar] [CrossRef]
- Jian, H.; Lu, K.; Yang, B.; Wang, T.; Zhang, L.; Zhang, A.; Wang, J.; Liu, L.; Qu, C.; Li, J. Genome-Wide analysis and expression profiling of the SUC and SWEET gene families of sucrose transporters in Oilseed Rape (Brassica napus L.). Front. Plant Sci. 2016, 7, 1464. [Google Scholar] [CrossRef]
- Hoang, N.V.; Choe, G.; Zheng, Y.; Aliaga Fandino, A.C.; Sung, I.; Hur, J.; Kamran, M.; Park, C.; Kim, H.; Ahn, H.; et al. Identification of conserved gene-regulatory networks that integrate environmental sensing and growth in the root cambium. Curr. Biol. 2020, 30, 2887–2900. [Google Scholar] [CrossRef]
- Wieczorke, R.; Krampe, S.; Weierstall, T.; Freidel, K.; Hollenberg, C.P.; Boles, E. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 1999, 464, 123–128. [Google Scholar] [CrossRef]
- Kang, J.N.; Kim, J.S.; Lee, S.M.; Won, S.Y.; Seo, M.S.; Kwon, S.J. Analysis of phenotypic characteristics and sucrose metabolism in the roots of Raphanus sativus L. Front. Plant Sci. 2021, 12, 716782. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, X.; Gong, Y.; Xu, L.; Wang, Y.; Liu, L. Evaluation of reference genes for gene expression studies in radish (Raphanus sativus L.) using quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2012, 424, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, S.; Lin, X.; Fang, H.; Shi, Y.; Grierson, D.; Chen, K. Transcription factor CitERF16 is involved in citrus fruit sucrose accumulation by activating CitSWEET11d. Front. Plant Sci. 2021, 12, 809619. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Qin, M.; Qiao, X.; Cheng, Y.; Li, X.; Zhang, H.; Wu, J. A newinsight into the evolution and functional divergence of SWEET transporters in Chinese White Pear (Pyrus bretschneideri). Plant Cell Physiol. 2017, 58, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Patil, G.; Valliyodan, B.; Deshmukh, R.; Prince, S.; Nicander, B.; Zhao, M.; Sonah, H.; Song, L.; Lin, L.; Chaudhary, J.; et al. Soybean (Glycine max) SWEET gene family: Insights through comparative genomics, transcriptome profiling and whole genome resequence analysis. BMC Genom. 2015, 16, 520. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, S.; Yu, F.; Tang, J.; Shan, X.; Bao, K.; Yu, L.; Wang, H.; Fei, Z.; Li, J. Genome-wide characterization and expression profiling of SWEET genes in cabbage (Brassica oleracea var. capitata L.) reveal their roles in chilling and clubroot disease responses. BMC Genom. 2019, 20, 93. [Google Scholar] [CrossRef]
- Xu, Y.; Tao, Y.; Cheung, L.S.; Fan, C.; Chen, L.Q.; Xu, S.; Perry, K.; Frommer, W.B.; Feng, L. Structures of bacterial homologues of SWEET transporters in two distinct conformations. Nature 2014, 515, 448–452. [Google Scholar] [CrossRef]
- Feng, L.; Frommer, W.B. Structure and function of SemiSWEET and SWEET sugar transporters. Trends Biochem. Sci. 2015, 40, 480–486. [Google Scholar] [CrossRef]
- Lemoine, R.; La Camera, S.; Atanassova, R.; Dédaldéchamp, F.; Allario, T.; Pourtau, N.; Bonnemain, J.L.; Laloi, M.; Coutos-Thévenot, P.; Maurousset, L.; et al. Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 2013, 4, 272. [Google Scholar] [CrossRef]
- Baker, R.F.; Leach, K.A.; Braun, D.M. SWEET as sugar: New sucrose effluxers in plants. Mol. Plant 2012, 5, 766–768. [Google Scholar] [CrossRef] [PubMed]
- Le Hir, R.; Spinner, L.; Klemens, P.A.W.; Chakraborti, D.; de Marco, F.; Vilaine, F.; Wolff, N.; Lemoine, R.; Porcheron, B.; Gery, C.; et al. Disruption of the sugar transporters AtSWEET11 and AtSWEET12 affects vascular development and freezing tolerance in Arabidopsis. Mol. Plant 2015, 8, 1687–1690. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, Z.Y.; Kumar, V.; Xu, X.F.; Yuan, P.; Zhu, X.F.; Li, T.Y.; Jia, B.; Xuan, Y.H. Genome-wide identification of the SWEET gene family in wheat. Gene 2018, 642, 284–292. [Google Scholar] [CrossRef]
- Chong, J.; Piron, M.C.; Meyer, S.; Merdinoglu, D.; Bertsch, C.; Mestre, P. The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J. Exp. Bot. 2014, 65, 6589–6601. [Google Scholar] [CrossRef]
- Zhou, A.; Liu, E.; Ma, H.; Feng, S.; Gong, S.; Wang, J. NaCl-induced expression of AtVHA-c5 gene in the roots plays a role in response of Arabidopsis to salt stress. Plant Cell Rep. 2018, 37, 443–452. [Google Scholar] [CrossRef]
- Guo, W.J.; Nagy, R.; Chen, H.Y.; Pfrunder, S.; Yu, Y.C.; Santelia, D.; Frommer, W.B.; Martinoia, E. SWEET17, a facilitative transporter, mediates fructose transport across the tonoplast of Arabidopsis roots and leaves. Plant Physiol. 2014, 164, 777–789. [Google Scholar] [CrossRef]
- Li, Z.; Wei, X.; Tong, X.; Zhao, J.; Liu, X.; Wang, H.; Tang, L.; Shu, Y.; Li, G.; Wang, Y.; et al. The OsNAC23-Tre6P-SnRK1a feed-forward loop regulates sugar homeostasis and grain yield in rice. Mol. Plant 2022, 15, 706–722. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Wang, Y.; Xu, L.; Li, B.; Wang, K.; Ying, J.; He, Q.; Liu, L. RsCLE22a regulates taproot growth through an auxin signaling-related pathway in radish (Raphanus sativus L.). J. Exp. Bot. 2023, 74, 233–250. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Cao, Y.; Xin, R.; Xu, L.; Wang, Y.; Wang, L.; Ma, Y.; Liu, L. Genome-Wide Identification of the RsSWEET Gene Family and Functional Analysis of RsSWEET17 in Root Growth and Development in Radish. Horticulturae 2023, 9, 698. https://doi.org/10.3390/horticulturae9060698
Zhang X, Cao Y, Xin R, Xu L, Wang Y, Wang L, Ma Y, Liu L. Genome-Wide Identification of the RsSWEET Gene Family and Functional Analysis of RsSWEET17 in Root Growth and Development in Radish. Horticulturae. 2023; 9(6):698. https://doi.org/10.3390/horticulturae9060698
Chicago/Turabian StyleZhang, Xiaoli, Yang Cao, Ruixian Xin, Liang Xu, Yan Wang, Lun Wang, Yinbo Ma, and Liwang Liu. 2023. "Genome-Wide Identification of the RsSWEET Gene Family and Functional Analysis of RsSWEET17 in Root Growth and Development in Radish" Horticulturae 9, no. 6: 698. https://doi.org/10.3390/horticulturae9060698
APA StyleZhang, X., Cao, Y., Xin, R., Xu, L., Wang, Y., Wang, L., Ma, Y., & Liu, L. (2023). Genome-Wide Identification of the RsSWEET Gene Family and Functional Analysis of RsSWEET17 in Root Growth and Development in Radish. Horticulturae, 9(6), 698. https://doi.org/10.3390/horticulturae9060698







