Molecular Characterization, Population Structure Analysis, and Association Mapping of Turkish Parsley Genotypes Using iPBS Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Phytochemical Analysis
2.3. Genetic Analysis
3. Results
3.1. Phytochemical Variations
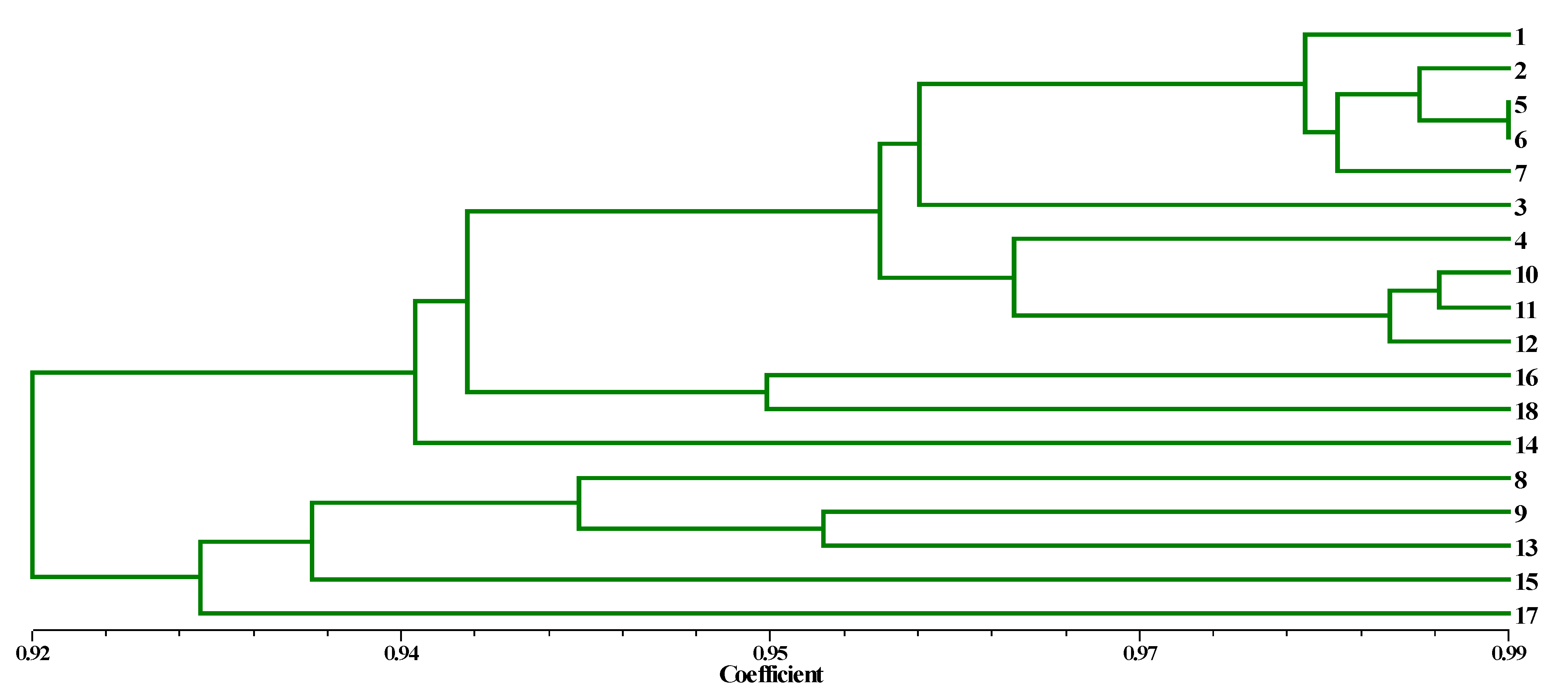
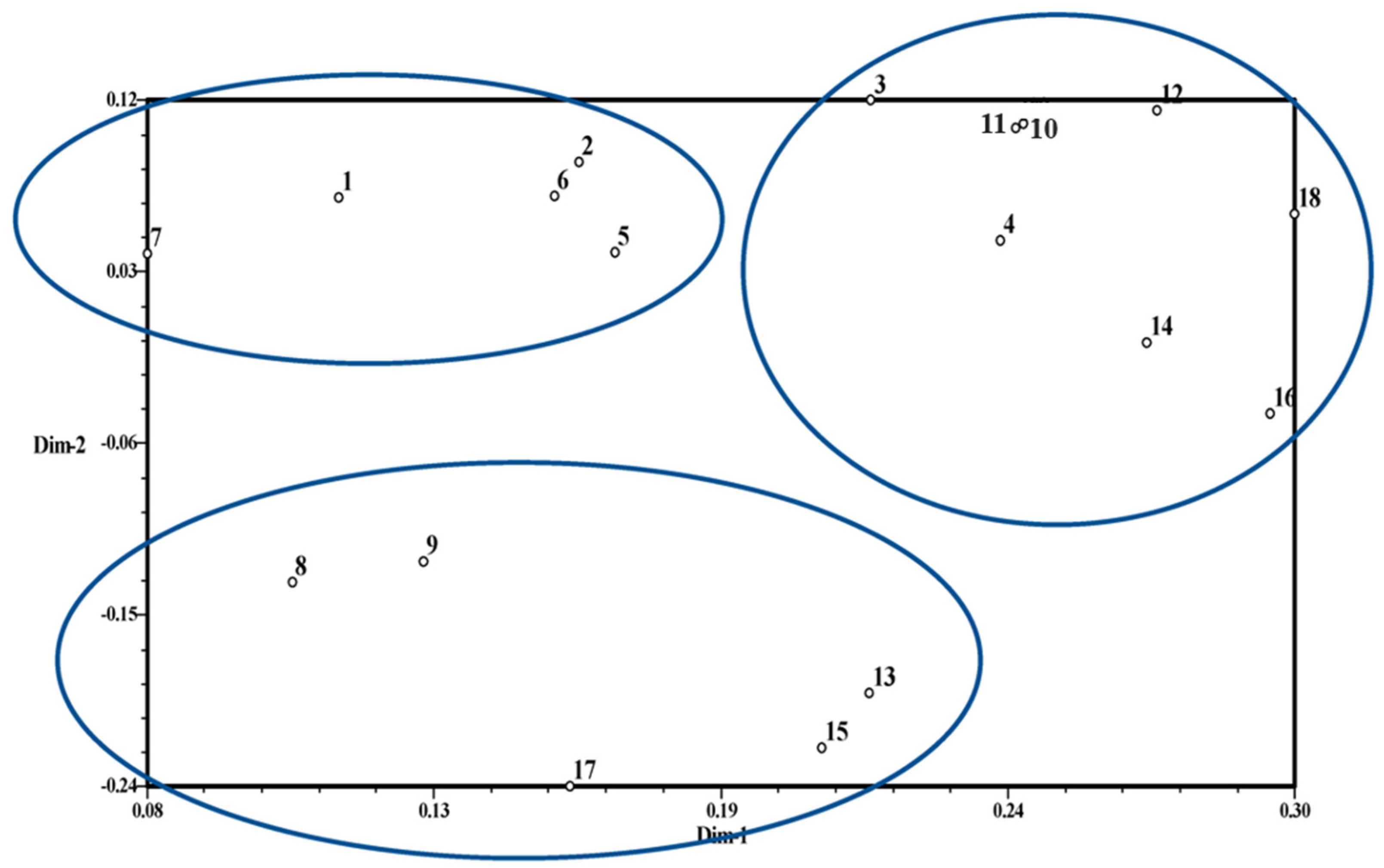
3.2. Genetic Diversity Analysis
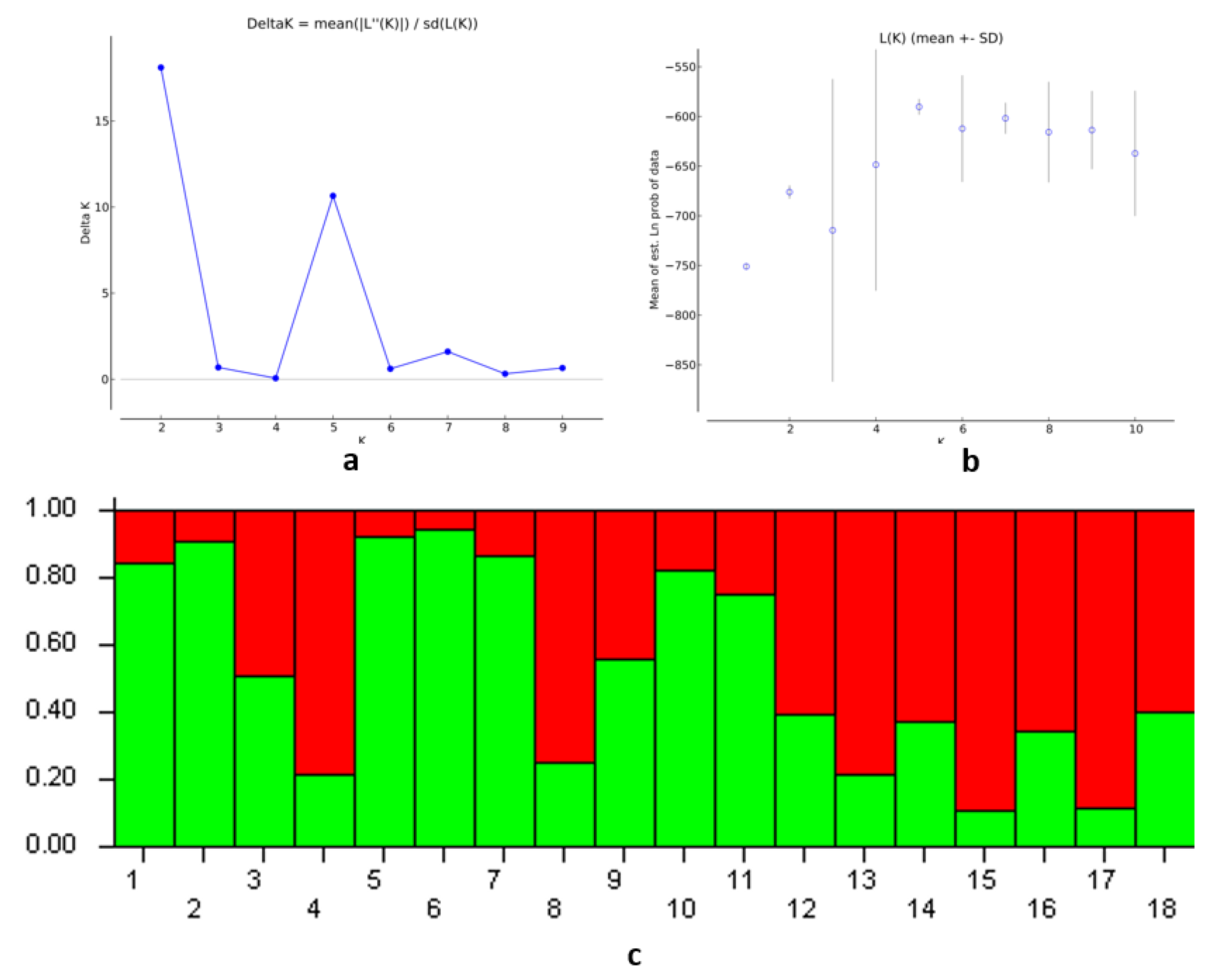
3.3. Population Structure
3.4. Associating Mapping
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dadan, M.; Rybak, K.; Wiktor, A.; Nowacka, M.; Zubernik, J.; Witrowa–Rajchert, D. Selected chemical composition changes in microwave–convective dried parsley leaves affected by ultrasound and steaming pre–treatments–An optimization approach. Food Chem. 2018, 239, 242–251. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, V.S.; Chávez, D.W.H.; Paiva, P.R.F.; Gamallo, O.D.; Castro, R.N.; Sawaya, A.C.H.F.; Sampaio, G.R.; Torres, E.A.F.D.S.; Saldanha, T. Parsley (Petroselinum crispum Mill.): A source of bioactive compounds as a domestic strategy to minimize cholesterol oxidation during the thermal preparation of omelets. Food Res. Int. 2022, 156, 111199. [Google Scholar] [CrossRef] [PubMed]
- Dziki, D.; Hassoon, W.H.; Biernacka, B.; Gawlik–Dziki, U. Dried and powdered leaves of parsley as a functional additive to wheat bread. Appl. Sci. 2022, 12, 7930. [Google Scholar] [CrossRef]
- Michalaki, A.; Karantonis, H.C.; Kritikou, A.S.; Thomaidis, N.S.; Dasenaki, M.E. Ultrasound–assisted extraction of specific phenolic compounds from Petroselinum crispum leaves using response surface methodology and HPLC–PDA and Q–TOF–MS/MS identification. Appl. Sci. 2023, 13, 798. [Google Scholar] [CrossRef]
- Dobričević, N.; Šic Žlabur, J.; Voća, S.; Pliestić, S.; Galić, A.; Delić, A.; Fabek Uher, S. Bioactive compounds content and nutritional potential of different parsley parts (Petroselinum crispum Mill.). J. Cent. Eur. Agr. 2019, 20, 900–910. [Google Scholar] [CrossRef]
- Mouhoubi, K.; Boulekbache–Makhlouf, L.; Madani, K.; Palatzidi, A.; Perez–Jimenez, J.; Mateos–Aparicio, I.; Garcia–Alonso, A. Phenolic compounds and antioxidant activity are differentially affected by drying processes in celery, coriander and parsley leaves. Int. J. Food Sci. Technol. 2022, 57, 3467–3476. [Google Scholar] [CrossRef]
- Sener, G.; Karakadıoglu, G.; Ozbeyli, D.; Ede, S.; Yanardag, R.; Sacan, O.; Aykac, A. Petroselinum crispum extract ameliorates scopolamine–induced cognitive dysfunction: Role on apoptosis, inflammation and oxidative stress. Food Sci. Hum. Wellness 2022, 11, 1290–1298. [Google Scholar] [CrossRef]
- Petropoulos, S.A.; Fernandes, Â.; Finimundy, T.C.; Polyzos, N.; Pinela, J.; Ivanov, M.; Soković, M.; Ferreira, I.C.F.R.; Barros, L. The bioactivities and chemical profile of turnip–rooted parsley germplasm. Horticulturae 2022, 8, 639. [Google Scholar] [CrossRef]
- Arsenov, D.; Župunski, M.; Pajević, S.; Nemeš, I.; Simin, N.; Alnuqaydan, A.M.; Watson, M.; Aloliqi, A.A.; Mimica–Dukić, N. Roots of Apium graveolens and Petroselinum crispum—Insight into phenolic status against toxicity level of trace elements. Plants 2021, 10, 1785. [Google Scholar] [CrossRef]
- Gadi, D.; Bnouham, M.; Aziz, M.; Ziyyat, A.; Legssyer, A.; Legrand, C.; Lafeve, F.F.; Mekhfi, H. Parsley extract inhibits in vitro and ex vivo platelet aggregation and prolongs bleeding time in rats. J. Ethnopharmacol. 2009, 125, 170–174. [Google Scholar] [CrossRef]
- Adsersen, A.; Gauguin, B.; Gudiksen, L.; Jäger, A.K. Screening of plants used in Danish folk medicine to treat memory dysfunction for acetylcholinesterase inhibitory activity. J. Ethnopharmacol. 2006, 104, 418–422. [Google Scholar] [CrossRef]
- Danciu, C.; Cioanca, O.; Watz–Farcaș, C.; Hancianu, M.; Racoviceanu, R.; Muntean, D.; Zupko, I.; Oprean, C.; Tatu, C.; Paunescu, V.; et al. Botanical Therapeutics (Part II): Antimicrobial and In vitro anticancer activity against mcf7 human breast cancer cells of chamomile, parsley and celery alcoholic extracts. Anticancer. Agents Med. Chem. 2020, 21, 187–200. [Google Scholar] [CrossRef]
- Ashry, M.; Atia, I.; Morsy, F. Elmashad, wael protective efficiency of parsley (Petroselinum crispum) against oxidative stress, DNA damage and nephrotoxicity induced with anti–tuberculosis drugs. Int. J. Cancer Biomed. Res. 2021, 5, 27–36. [Google Scholar] [CrossRef]
- Petropoulos, S.A.; Daferera, D.; Akoumianakis, C.; Passam, H.; Polissiou, M. The effect of sowing date and growth stage on the essential oil composition of three types of parsley (Petroselinum crispum). J. Sci. Food Agric. 2004, 1610, 1606–1610. [Google Scholar] [CrossRef]
- Sarwar, S.; Ayyub, M.A.; Rezgui, M.; Nisar, S.; Jilani, I. Parsley: A review of habitat, phytochemistry, ethnopharmacology and biological activities. Int. J. Chem. Biochem. Sci. 2019, 9, 49–55. [Google Scholar]
- Turkish Statistical Institute (TUIK). 2022. Available online: https://data.tuik.gov.tr/Bulten/Index?p=Bitkisel–Uretim–Istatistikleri–2022–45504 (accessed on 10 January 2023).
- Simon, J.E.; Quinn, J. Characterization of essential oil of parsley. J. Agric. Food Chem. 1988, 36, 467–472. [Google Scholar] [CrossRef]
- Bekhradi, F.; Delshad, M.; Marín, A.; Luna, M.; Garrido, Y.; Kashi, A.; Babalar, M.; Gil, M. Effects of salt stress on physiological and postharvest quality characteristics of different Iranian genotypes of basil. Hortic. Environ. Biotechnol. 2015, 56, 777–785. [Google Scholar] [CrossRef]
- Jones, N.; Ougham, H.; Thomas, H.; Pasakinskiene, I. Markers and mapping revisited: Finding your gene. New Phytol. 2009, 183, 935–966. [Google Scholar] [CrossRef]
- Agarwal, M.; Shrivastava, N.; Padh, H. Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep. 2008, 27, 617–631. [Google Scholar] [CrossRef] [PubMed]
- Karaman, K.; Dalda–Sekerci, A.; Yetisir, H.; Gulsen, O.; Coskun, O.F. Molecular, morphological and biochemical characterization of some turkish bitter melon (Momordica charantia L.) genotypes. Ind. Crops Prod. 2008, 123, 93–99. [Google Scholar] [CrossRef]
- Kirac, H.; Dalda–Sekerci, A.; Coskun, O.F.; Gulsen, O. Morphological and molecular characterization of garlic (Allium sativum L.) genotypes sampled from Turkey. Gen. Res. Crop Evol. 2022, 69, 1833–1841. [Google Scholar] [CrossRef] [PubMed]
- Coskun, O.F. Determination of genetic diversity in some pumpkin genotypes using SSR marker technique. Erzincan Univ. J. Sci. Tech. 2022, 15, 942–952. [Google Scholar]
- Bennetzen, J.L. Transposable element contributions to plant gene and genome evolution. Plant Mol. Biol. 2000, 42, 251–269. [Google Scholar] [CrossRef] [PubMed]
- Nadeem, M.A. Deciphering the genetic diversity and population structure of Turkish bread wheat germplasm using iPBS–retrotransposons markers. Mol. Biol. Rep. 2021, 48, 6739–6748. [Google Scholar] [CrossRef]
- Öztürk, H.İ.; Dursun, A.; Hosseinpour, A.; Haliloğlu, K. Genetic diversity of pinto and fresh bean (Phaseolus vulgaris L.) germplasm collected from Erzincan province of Turkey by inter–primer binding site (iPBS) retrotransposon markers. Turk. J. Agric. For. 2020, 44, 417–427. [Google Scholar] [CrossRef]
- Domblides, A.S.; Domblides, E.A.; Kharchenko, V.A.; Potekhin, G.A. Study of genetic variation among parsley (Petroselinum crispum Mill. Nym.) samples using RAPD and ISSR markers. Moscow Univ. Biol. Sci. Bull. 2010, 65, 152–154. [Google Scholar] [CrossRef]
- Nasiri, K.; Shojaeiyan, A.; Yadollahi, A.; Mirshekari, A.; Ghanbari, K. Genetic diversity assessment of some Iranian parsley (Petroselinum crispum Mill.) accessions using SRAP molecular marker. J. Veg. Sci. 2015, 1, 1–10. [Google Scholar]
- Ibrahim, H.M.M.; El–Leel, O.F.A.; Emam, K.A. Molecular profiling for genetic variability in Petroselinum crispum based on ISSR and RAPD markers. Middle East J. Agric. Res. 2017, 6, 67–75. [Google Scholar]
- Boutsika, A.; Sarrou, E.; Cook, C.M.; Mellidou, I.; Avramidou, E.; Angeli, A.; Martens, S.; Ralli, P.; Letsiou, S.; Selini, A.; et al. Evaluation of parsley (Petroselinum crispum) germplasm diversity from the Greek Gene Bank using morphological, molecular and metabolic markers. Ind. Crops Prod. 2021, 170, 113767. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis, 16th ed.; Association of Official Analytical Chemists: Washington, DC, USA, 1995. [Google Scholar]
- Holm, G. Chlorophyll mutations in barley. Acta Agric. Scan. 1954, 4, 457–471. [Google Scholar] [CrossRef]
- Wettstein, D. Chlorophyll letale und der submikroskopische formwechsel der plastiden. Exp. Cell Res. 1957, 12, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population Structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software Structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Vigouroux, Y.; Glaubitz, J.C.; Matsuoka, Y.; Major, M.; Doebley, J. Population structure and genetic diversity of the new world maize races assessed by microsatellites. Am. J. Bot. 2008, 95, 1240–1253. [Google Scholar] [CrossRef]
- Castillo, A.; Dorado, G.; Feuillet, C.; Sourdille, P.; Hernandez, P. Genetic structure and ecogeographical adaptation in wild barley (Hordeum chilense Roemer et Schultes) as revealed by microsatellite markers. BMC Plant Biol. 2010, 10, 266. [Google Scholar] [CrossRef]
- Parthasarathy, V.A.; Chempakam, B.; Zachariah, T.J. Chemistry of Spices; CAB International: London, UK, 2008. [Google Scholar]
- Karklelienė, R.; Dambrauskienė, E.; Juškevičienė, D.; Radzevičius, A.; Rubinskienė, M.; Viškelis, P. Productivity and nutritional value of dill and parsley. Hort. Sci. 2014, 41, 131–137. [Google Scholar] [CrossRef]
- Santos, J.; Herrero, M.; Mendiola, J.A.; Oliva–Teles, M.T.; Ibanez, E.; Delerue–Matos, C.; Oliveira, M.B.P.P. Fresh–cut aromatic herbs: Nutritional quality stability during shelf–life. LWT Food Sci. Tehnol. 2014, 59, 101–107. [Google Scholar] [CrossRef]
- Coskun, O.F.; Gunduz, Y.F.; Toprak, S.; Mavi, K. Molecular characterization of some parsley (Petroselinum crispum Mill.) genotypes. Mustafa Kemal Univ. J. Agri. Sci. 2023. Accepted. [Google Scholar]

| No | Code | Location | No | Code | Location |
|---|---|---|---|---|---|
| 1 | HMKU–MA1 | Hatay–Samandag | 10 | HMKU–MA10 | Hatay–Arsuz |
| 2 | HMKU–MA2 | Hatay–Samandag | 11 | HMKU–MA11 | Hatay–Antakya |
| 3 | HMKU–MA3 | Hatay–Samandag | 12 | HMKU–MA12 | Hatay–Antakya |
| 4 | HMKU–MA4 | Hatay–Samandag | 13 | HMKU–MA13 | Osmaniye |
| 5 | HMKU–MA5 | Hatay–Arsuz | 14 | HMKU–MA14 | Osmaniye |
| 6 | HMKU–MA6 | Hatay–Arsuz | 15 | HMKU–MA15 | Osmaniye |
| 7 | HMKU–MA7 | Hatay–Arsuz | 16 | HMKU–MA16 | Kilis |
| 8 | HMKU–MA8 | Hatay–Arsuz | 17 | HMKU–MA17 | Osmaniye |
| 9 | HMKU–MA9 | Hatay–Arsuz | 18 | HMKU–MA18 | Hatay–İskenderun |
| Genotype | Total Soluble Solids (%) | Total Acid Content (%) | pH–Value |
|---|---|---|---|
| 1 | 4.67 ± 0.34 ab | 2.59 ± 0.01 a | 5.86 ± 0.01 ab |
| 2 | 4.31 ± 0.31 a–c | 1.87 ± 0.01 f | 5.67 ± 0.02 h |
| 3 | 3.67 ± 0.32 b–d | 2.21 ± 0.03 c | 5.75 ± 0.01 ef |
| 4 | 4.32 ± 0.33 a–c | 2.11 ± 0.02 d | 5.74 ± 0.03 ef |
| 5 | 4.67 ± 0.28 ab | 1.95 ± 0.01 ef | 5.84 ± 0.02 bc |
| 6 | 4.63 ± 0.41 ab | 1.94 ± 0.03 ef | 5.86 ± 0.01 ab |
| 7 | 4.28 ± 0.34 a–c | 1.97 ± 0.04 e | 5.85 ± 0.01 ab |
| 8 | 3.33 ± 0.30 c–e | 1.54 ± 0.02 hi | 5.87 ± 0.02 a |
| 9 | 3.67 ± 0.34 b–d | 1.62 ± 0.02 h | 5.77 ± 0.01 e |
| 10 | 5.34 ± 0.29 a | 1.88 ± 0.02 ef | 5.71 ± 0.02 g |
| 11 | 3.00 ± 0.01 de | 1.28 ± 0.02 jk | 5.81 ± 0.02 cd |
| 12 | 3.33 ± 0.32 c–e | 1.34 ± 0.03 j | 5.81 ± 0.02 cd |
| 13 | 2.32 ± 0.41 e | 1.53 ± 0.02 i | 5.85 ± 0.02 ab |
| 14 | 3.67 ± 0.27 b–d | 1.22 ± 0.01 k | 5.73 ± 0.02 fg |
| 15 | 2.33 ± 0.35 e | 1.04 ± 0.03 l | 5.75 ± 0.02 ef |
| 16 | 4.55 ± 0.53 ab | 2.44 ± 0.02 b | 5.80 ± 0.02 d |
| 17 | 4.31 ± 0.56 a–c | 1.74 ± 0.02 g | 5.80 ± 0.02 d |
| 18 | 4.01 ± 0.86 b–d | 1.33 ± 0.01 j | 5.81 ± 0.01 cd |
| Average | 4.43 ± 0.36 | 1.76 ± 0.02 | 5.79 ± 0.02 |
| Genotype | Chlorophyll a Content (mg/g) | Chlorophyll b Content (mg/g) | Total Chlorophyll Content (mg/g) | Total Carotenoid Content (mg/g) |
|---|---|---|---|---|
| 1 | 0.61 ± 0.01 a | 0.33 ± 0.02 b | 0.94 ± 0.03 ab | 0.18 ± 0.02 a |
| 2 | 0.42 ± 0.02 e–g | 0.22 ± 0.01 f | 0.65 ± 0.03 i | 0.12 ± 0.01 de |
| 3 | 0.31 ± 0.01 h | 0.18 ± 0.01 g | 0.50 ± 0.02 j | 0.10 ± 0.01 e |
| 4 | 0.57 ± 0.02 ab | 0.31 ± 0.02 bc | 0.88 ± 0.04 bc | 0.14 ± 0.02 b–d |
| 5 | 0.49 ± 0.03 cd | 0.29 ± 0.02 b–d | 0.78 ± 0.02 d–g | 0.14 ± 0.02 b–d |
| 6 | 0.37 ± 0.04 g | 0.29 ± 0.02 b–d | 0.66 ± 0.05 hi | 0.13 ± 0.01 b–d |
| 7 | 0.58 ± 0.03 ab | 0.24 ± 0.01 ef | 0.82 ± 0.04 c–f | 0.15 ± 0.01 b |
| 8 | 0.46 ± 0.02 de | 0.28 ± 0.01 cd | 0.74 ± 0.03 f–h | 0.14 ± 0.01 b–d |
| 9 | 0.54 ± 0.02 bc | 0.30 ± 0.01 bc | 0.84 ± 0.03 c–e | 0.14 ± 0.02 b–d |
| 10 | 0.40 ± 0.01 fg | 0.24 ± 0.01 ef | 0.64 ± 0.03 i | 0.13 ± 0.01 b–d |
| 11 | 0.61 ± 0.02 a | 0.37 ± 0.01 a | 0.98 ± 0.04 a | 0.18 ± 0.01 a |
| 12 | 0.45 ± 0.02 d–f | 0.26 ± 0.02 d–f | 0.71 ± 0.03 g–i | 0.13 ± 0.01 b–d |
| 13 | 0.45 ± 0.01 d–f | 0.25 ± 0.01 d–f | 0.71 ± 0.02 g–i | 0.12 ± 0.01 c–e |
| 14 | 0.54 ± 0.02 bc | 0.32 ± 0.02 b | 0.86 ± 0.02 cd | 0.15 ± 0.02 bc |
| 15 | 0.50 ± 0.03 cd | 0.30 ± 0.01 bc | 0.80 ± 0.03 c–f | 0.14 ± 0.01 b–d |
| 16 | 0.48 ± 0.02 cd | 0.26 ± 0.03 de | 0.75 ± 0.01 fg | 0.14 ± 0.01 b–d |
| 17 | 0.47 ± 0.02 de | 0.30 ± 0.02 bc | 0.77 ± 0.02 e–g | 0.12 ± 0.01 de |
| 18 | 0.40 ± 0.03 fg | 0.23 ± 0.01 ef | 0.63 ± 0.03 i | 0.12 ± 0.01 de |
| Average | 0.48 ± 0.02 | 0.28 ± 0.01 | 0.76 ± 0.03 | 0.14 ± 0.01 |
| Primer Name | Primer Sequence 5′–3′ | Tm °C | Number of Bands | % Rate of Polym. | |
|---|---|---|---|---|---|
| Polym. | Total | ||||
| iPBS–2272 | GGCTCAGATGCCA | 46 | 11 | 17 | 64.7 |
| iPBS–2277 | GGCGATGATACCA | 48 | 4 | 12 | 33.3 |
| iPBS–2217 | ACTTGGATGTCGATACCA | 51 | 3 | 8 | 37.5 |
| iPBS–2219 | GAACTTATGCCGATACCA | 51 | 3 | 10 | 30 |
| iPBS–2222 | ACTTGGATGCCGATACCA | 54 | 4 | 11 | 36.4 |
| iPBS–2226 | CGGTGACCTTTGATACCA | 54 | 5 | 11 | 45.5 |
| iPBS–2228 | CATTGGCTCTTGATACCA | 51 | 4 | 12 | 33.3 |
| iPBS–2230 | TCTAGGCGTCTGATACCA | 54 | 3 | 13 | 23.1 |
| iPBS–2232 | AGAGAGGCTCGGATACCA | 56 | 3 | 10 | 30 |
| iPBS–2239 | ACCTAGGCTCGGATGCCA | 58 | 6 | 8 | 75 |
| iPBS–2243 | AGTCAGGCTCTGTTACCA | 54 | 2 | 14 | 14.3 |
| iPBS–2244 | GGAAGGCTCTGATTACCA | 54 | 6 | 9 | 66.7 |
| iPBS–2246 | ACTAGGCTCTGTATACCA | 51 | 1 | 10 | 10 |
| iPBS–2249 | AACCGACCTCTGATACCA | 54 | 4 | 17 | 23.5 |
| iPBS–2251 | GAACAGGCGATGATACCA | 54 | 2 | 7 | 28.6 |
| iPBS–2252 | TCATGGCTCATGATACCA | 51 | 4 | 12 | 33.3 |
| Total | 65 | 181 | 574.6 | ||
| Average | 4.1 | 11.3 | 31.9 | ||
| Primer | Band Freq. | p | q | Na | Ne | I | h | uh | PIC |
|---|---|---|---|---|---|---|---|---|---|
| iPBS–2272 | 0.71 | 0.71 | 0.29 | 1.71 | 1.39 | 0.34 | 0.22 | 0.24 | 0.41 |
| iPBS–2277 | 0.87 | 0.87 | 0.13 | 1.33 | 1.22 | 0.19 | 0.13 | 0.14 | 0.19 |
| iPBS–2217 | 0.92 | 0.92 | 0.08 | 1.38 | 1.15 | 0.14 | 0.09 | 0.09 | 0.13 |
| iPBS–2219 | 0.96 | 0.96 | 0.04 | 1.30 | 1.09 | 0.11 | 0.07 | 0.07 | 0.07 |
| iPBS–2222 | 0.91 | 0.91 | 0.09 | 1.36 | 1.20 | 0.20 | 0.13 | 0.14 | 0.15 |
| iPBS–2226 | 0.75 | 0.75 | 0.25 | 1.82 | 1.37 | 0.36 | 0.23 | 0.24 | 0.37 |
| iPBS–2228 | 0.93 | 0.93 | 0.07 | 1.75 | 1.15 | 0.22 | 0.12 | 0.13 | 0.13 |
| iPBS–2230 | 0.91 | 0.91 | 0.09 | 1.31 | 1.10 | 0.12 | 0.07 | 0.08 | 0.13 |
| iPBS–2232 | 0.88 | 0.88 | 0.12 | 1.80 | 1.28 | 0.29 | 0.18 | 0.19 | 0.21 |
| iPBS–2239 | 0.91 | 0.91 | 0.09 | 1.75 | 1.20 | 0.23 | 0.14 | 0.14 | 0.16 |
| iPBS–2243 | 0.97 | 0.97 | 0.03 | 1.14 | 1.08 | 0.08 | 0.05 | 0.05 | 0.06 |
| iPBS–2244 | 0.93 | 0.93 | 0.07 | 1.78 | 1.17 | 0.23 | 0.13 | 0.14 | 0.14 |
| iPBS–2246 | 0.98 | 0.98 | 0.02 | 1.10 | 1.04 | 0.05 | 0.03 | 0.03 | 0.03 |
| iPBS–2249 | 0.90 | 0.90 | 0.10 | 1.24 | 1.09 | 0.09 | 0.05 | 0.06 | 0.12 |
| iPBS–2251 | 0.90 | 0.90 | 0.10 | 1.57 | 1.22 | 0.23 | 0.14 | 0.15 | 0.17 |
| iPBS–2252 | 0.89 | 0.89 | 0.11 | 1.75 | 1.24 | 0.26 | 0.16 | 0.17 | 0.19 |
| Average | 0.89 | 0.89 | 0.11 | 1.51 | 1.19 | 0.20 | 0.12 | 0.13 | 0.17 |
| Traits | Marker | p | R |
|---|---|---|---|
| Total Soluble Solids | iPBS–2239–420 | 0.008964 | 0.26 |
| Total Soluble Solids | iPBS–2243–400 | 0.028865 | 0.19 |
| Total Soluble Solids | iPBS–2251–330 | 0.028865 | 0.19 |
| Total Acid | iPBS–2226–1140 | 0.008191 | 0.33 |
| pH | iPBS–2272–770 | 0.045347 | 0.24 |
| Chlorophyll a | iPBS–2228–450 | 0.037363 | 0.25 |
| Chlorophyll a | iPBS–2244–200 | 0.037363 | 0.26 |
| Chlorophyll b | iPBS–2228–450 | 0.029188 | 0.27 |
| Chlorophyll b | iPBS–2244–200 | 0.029188 | 0.28 |
| Total Carotenoid | iPBS–2249–1080 | 0.041217 | 0.23 |
| Total Carotenoid | iPBS–2244–540 | 0.04448 | 0.22 |
| Total Carotenoid | iPBS–2272–850 | 0.048688 | 0.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coşkun, Ö.F. Molecular Characterization, Population Structure Analysis, and Association Mapping of Turkish Parsley Genotypes Using iPBS Markers. Horticulturae 2023, 9, 336. https://doi.org/10.3390/horticulturae9030336
Coşkun ÖF. Molecular Characterization, Population Structure Analysis, and Association Mapping of Turkish Parsley Genotypes Using iPBS Markers. Horticulturae. 2023; 9(3):336. https://doi.org/10.3390/horticulturae9030336
Chicago/Turabian StyleCoşkun, Ömer Faruk. 2023. "Molecular Characterization, Population Structure Analysis, and Association Mapping of Turkish Parsley Genotypes Using iPBS Markers" Horticulturae 9, no. 3: 336. https://doi.org/10.3390/horticulturae9030336
APA StyleCoşkun, Ö. F. (2023). Molecular Characterization, Population Structure Analysis, and Association Mapping of Turkish Parsley Genotypes Using iPBS Markers. Horticulturae, 9(3), 336. https://doi.org/10.3390/horticulturae9030336








