Abstract
The availability of adequate information about the documentation and characterization of germplasm is fundamental for any crop improvement program. The importance of cumin as a medicinal plant yet the lack of information about its genetic variability encouraged us to initiate the current study aiming at assessing the genetic variability among 17 cumin genotypes from different geographical regions using four molecular markers (RAPD, ISSR, SRAP and SCoT). Further, the potential of six accessions to induce callus was studied under in vitro conditions on MS and B5 basal media supplemented with various combinations between 2,4-D and kinetin. Our findings showed that combining 87 primers, including 42, 15, 7 and 23 primers of RAPD, ISSR, SCoT and SRAP, respectively, facilitated detecting the relationship among the assessed cumin accessions. A total number of 765 bands were analyzed, among which only 74 bands were polymorphic. The polymorphism was low (9.67%) and varied among and within markers. The SCoT markers exposed the highest average values of polymorphism information content (0.06), resolving power (0.91) and diversity index (0.08), while ISSR induced the highest expected heterozygosity (0.06) and marker index (0.08). The UPGMA dendrogram based on data from all the molecular markers separated the genotypes into three main clusters, with a partial geographic-based relationship among the genotypes. Out of the six accessions evaluated for callus induction in vitro, five were potent to induce callus, with a frequency ranging from 90.4 to 97.5% and no significant differences among the five accessions tested using ANOVA. Two medium combinations showed superior results: MS amended with 2,4-D (4.44 mg/L) + Kin (0.22 mg/L) and B5 with 2,4-D (8.88 mg/L) + Kin (0.22 mg/L). Statistically significant variations in the relative growth rate of the produced callus were detected among accessions, where EG-4 accessions induced the highest values, followed by EG-5. All medium combinations, including 2,4-D alone, exhibited significant superiority compared with those including both 2,4-D and Kin. Our findings exposed low variability among the studied cumin accessions, implying the real need for more effort to assess wider populations from different geographic regions together with the need for reliable diversification programs.
1. Introduction
The importance of medicinal plants is as old as mankind, either in their crude forms or after the isolation of active constituents. The vast diversity in the plant kingdom developed by nature has great importance for producing a wide variety of phytochemicals [1]. High genetic diversity also represents a prerequisite for the development of breeding programs for important crops, including medicinal plants [2,3]. A lack of adequate information about genetic diversity and intraspecific relatedness hinders the development of breeding programs for many medicinal plants [4]. A good example of this is cumin, despite its importance as a medicinal and aromatic plant. Cumin (Cuminum cyminum L.) belongs to the family Apiaceae and is widely cultivated in its origin countries of India, Iran and the Middle East [5]. It is consumed as a powder of its seeds (fruits) as a flavor in foods and beverages. The valuable essential oil contained in the seeds has high antioxidant, carminative and antiflatulent activity, which qualifies it for many applications in foods, medicines and perfumery [5,6,7,8,9].
To fill the gap in the characterization of cumin germplasm, great effort is necessary from specialists in different regions with widespread cultivation of cumin in order to obtain reliable documentation and characterization of cumin germplasm [5]. Assessments of genetic variability have been reported for several cumin populations using the traditional phenotypic methods, yet they expose environmental interactions with genotypes together with being costly and time-consuming [4,10]. Thus, dominant markers such as RAPD, ISSR and AFLP and simple sequence repeats (SSRs) or microsatellites have been employed by several authors looking for a more reliable, reproducible, informative and effective way of assessing genetic variability [10,11]. Countless plant species have been subjected to fingerprinting using different DNA markers. Examples of this include utilizing molecular markers to facilitate the early and precise discovery of pathogens in cotton (Gossypium sp.) [12]. RAPD, ISSR and SSR genotyping have been widely applied for assessing genetic variability and, thus, clonal fidelity analyses in several important cultivated plants, such as bananas (Musa spp.) [13,14]. In addition, microsatellite and RAPD primers were also employed to identify duplicates/misnomers and to differentiate grapevines clones [15]. Nevertheless, the available published work on the characterization of cumin germplasm is limited. Information about the development of SSRs was stated by Kumar et al. [11]. Bhatt et al. [16] evaluated genetic diversity among 16 cumin genotypes using 10 sequence-related amplified polymorphism (SRAP) markers. Their results revealed a moderate level of genetic diversity confirmed by the obtained Jaccard similarity coefficient (0.59), polymorphism information content (PIC) value (0.34) and marker index (MI) value (2.43). Accordingly, they suggested more efforts to increase cumin genetic diversity through different approaches, such as mutagenesis and somaclonal variation. A higher level of similarity (46%) was reported using RAPD by Baghizadeh et al. [17] among 32 Iranian cumin genotypes categorized into six groups using the UPGMA method. Likewise, Bahraminejad et al. [18] reported a 43% similarity level using RAPD markers among 49 Iranian genotypes, showing five different categories based on UPGMA. Thus, they deduced a high potential for variability in Iranian cumin populations. Based on RAPD and ISSR markers, respectively, the percentages of polymorphism were 62% and 70%, and the means of the PIC were 0.26 and 0.3 [19]. The start-codon-targeted (SCoT) marker was successfully utilized to determine genetic diversity among 12 cumin genotypes, showing 79.80% polymorphism, as reported by Parashar and Malik [8].
Callus culture plays a special role in the plant biotechnology area for producing medicinal compounds on a large scale [20]. Moreover, callus masses derived from plant tissues can sometimes produce high amounts of secondary metabolites [21]. The formation of callus involves the process of cell dedifferentiation, where cells lose their differentiated characters. This leads to a phenomenon known as somaclonal variation, where genetic variability is observed in the derived progeny. In the commercial production of crops, where true-to-type progeny is targeted, these variations are obstructive. On the contrary, somaclonal variation is considered very helpful in the field of crop improvement to introduce new cultivars with specific characteristics or to produce secondary metabolites [22]. Callus production from roots, shoots and leaves is generally applied to determine the conditions required for explants to survive and grow, to exploit products coming from primary and secondary metabolism, to study cell development and to obtain cell suspension in propagation [20]. Jha et al. [23] published the first study on the callus induction of cumin. They used hypocotyl and leaf explants in B5 medium complemented with various plant growth regulators (PGRs). Beiki et al. [24] studied callus induction in three Iranian landraces with the use of embryo explants. Gupta et al. [25] produced embryogenic and nonembryogenic callus in cumin on Murashige and Skoog (MS) supplemented with 0.5 mg/L of 2,4- dichlorophenoxyacetic acid (2,4-D) after 35 days of inoculation from all four explants, viz., root, hypocotyl, cotyledon and shoot apex, as well as organogenic and nonorganogenic callus on MS medium supplemented with 0.1 mg/L of TDZ after 40 days of inoculation from hypocotyl explants. In contrast to cytokinin-induced callus, the utilization of 2,4-D can establish a long-term culture of embryogenic callus that may be used for the induction and screening of somaclonal cell lines [26].
The aim of the current study, therefore, is to assess the genetic variability among seventeen cumin genotypes from different geographical regions using different molecular markers (RAPD, ISSR, SRAP and SCoT) and to study the variability of in vitro callus induced therefrom.
2. Materials and Methods
2.1. Plant Material
The current study involved seventeen genotypes of cumin (Cuminum cyminum L.) from different origins, as indicated in Table 1. A total of ten accessions were brought from the Genebank of the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) in Gatersleben, Germany, which were collected from different origins: Pakistan (7 accessions), Iran (1 accession), Iraq (1 accession) and Colombia (1 accession). In addition, seven genotypes were collected from cumin fields as follows: 3 landraces from Assiut region, Egypt; 3 landraces from El-Menia region, Egypt; and one landrace from Germany, kindly donated by Prof. Dr. Evelyn Klocke, JKI, Quedlinburg, Germany.

Table 1.
List of cumin genotypes involved in the current study and their sources. Accession names and IDs are given for seeds brought from the Genebank, and accession codes are given for seeds collected from cultivated fields.
2.2. Molecular Analysis
2.2.1. DNA Extraction and Purification
DNA was extracted from cumin seeds of the 17 tested genotypes according to the protocol suggested by [27]. Briefly, 30 mg of dry seeds were homogenized in a mortar with 1.5 mL of extraction buffer consisting of 100 mM Tris-HCl (pH 8), 2.0 M NaCl, 20 mM EDTA (pH 8), 2% (w/v) CTAB, 1% (w/v) PVP and 0.5% (w/v) activated charcoal. After 30 min of incubation at 55 °C with frequent agitation, the mixture was cooled down and incubated for 2 h at room temperature (RT). The mixture was then centrifuged at 16,000× g for 10 min at RT. The supernatant was thoroughly vortex with one vol chloroform-isoamyl alcohol (4% v/v isoamyl alcohol in chloroform). The supernatant aqueous layer was mixed with 0.45 vol of isopropanol and incubated at 25 °C for 1 h before being centrifuged at 700× g for 10 min at RT. The supernatant was discarded, and the pellet was washed with 1 mL of wash buffer (15 mM of ammonium acetate in 75% ethanol), vortexed and then centrifuged at 900× g for 10 min at RT. After removing the supernatant, the pellet was air-dried and then dissolved in 25 µL of TE (10 mM of Tris-HCL, 1 mM of EDTA, pH 8). The DNA solution was quantified using a NanoDrop 8000 UV-Vis spectrophotometer (Thermo Scientific, Waltham, MA, USA) and then stored at 4 °C. The chemicals used for DNA extraction were bought from Thermo Scientific, USA.
2.2.2. Molecular Markers
Four types of molecular markers were used, i.e., random amplified polymorphic DNA (RAPD), inter simple sequence repeats (ISSR), start-codon-targeted polymorphism (SCoT) and sequence-related amplified polymorphism (SRAP). Primers used for each marker are shown in Table S1 in the Supplementary Materials, including 42, 15, 7 and 23 primers of RAPD, ISSR, SCoT and SRAP, respectively, with combinations of 5 forward with 6 reverse primers. The PCR mixture and program conditions were performed according to Williams et al. [28] for RAPD, Gupta et al. [29] for ISSR, Collard and Mackill [30] for SCoT and Li and Quiros [31] for SRAP, where a GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA, USA) was utilized. PCR products were separated on 1.5% agarose gel (Cleaver Scientific, Rugby, UK) for RAPD and ISSR and 2% for SCoT and SRAP. All gels were stained with ethidium bromide (Thermo Fisher Scientific, USA) and visualized with a UV transilluminator imaging system. MyTaqTM mix was bought from Bioline GmbH, Luckenwalde, Germany, and all primers were bought from Eurogentec, Belgium.
2.2.3. Data Analysis
Forty-two primers for RAPD, fifteen primers for ISSR, seven primers for SCoT and twenty-three primer combinations for SRAP gave clear and strong band patterns and, subsequently, were selected for data analysis. Bands were scored and a binary matrix of the presence (1) or absence (0) of bands was made for all primers of all markers. A cluster analysis based on RAPD, ISSR, SCoT and SRAP data was performed separately using NTsys PC v.2.21q software, and an unweighted pair-group method with arithmetic averages (UPGMA) dendrogram was made using Jaccard’s coefficient [32]. Additionally, a dendrogram exhibiting combined data for all the markers was also obtained. Moreover, some diversity measures were calculated from the molecular data of all markers, including the following: the percentage of polymorphism (%P), as ; the polymorphism information content (PIC), using the formula , where f is the frequency of band presence among the samples [33]; the primer resolving power (Rp), using , where Ib is the informativeness of a band and is calculated as , where p is the proportion of a band among the samples [34]; the diversity index (DI) using , where fi is the frequency of band presence among the samples [35]; the expected heterozygosity (He), as , where f is the frequency of band presence among the samples [36]; and marker index (MI) as , where EMR is the effective multiplex ratio that equals the number of polymorphic bands × polymorphism ratio, and PIC is the polymorphism information content [37].
2.3. In Vitro Seed Germination and Callus Induction
Seeds of the 6 cumin genotypes originated from Egypt (EG-1, EG-2, EG-3, EG-4, EG5 and EG-6) were disinfected by rinsing with running water for 30 min and then soaking in 5% Ca (ClO)2 for 15 min with a few drops of Tween-20, followed by washing three times with sterile distilled water for 10 min each. The disinfested seeds were cultured in baby-food jars containing 30 mL/jar of MS basal salt medium [38] with vitamins solidified with 7 g/L agar but free from hormones and sucrose. The pH was adjusted to 5.7 and samples were then autoclaved at 120 °C and 1.2 kg/cm for 20 min. The cultures were incubated at 23 °C until germination.
Hypocotyl segments from germinated seeds (≈5 mm long) were utilized as explants for callus induction on eight combinations of growth media. These consisted of two basal salt media, i.e., MS medium with vitamins and Gamborg’s B5 with vitamins [39], each of which was supplemented with four combinations of dichlorophenoxyacetic acid (2,4-D) at 0.44 or 0.88 mg/L, either alone or plus kinetin (Kin) at 0.22 mg/L. All media contained 40 g/L of sucrose plus 7 g/L of agar, and the pH was adjusted to 5.7 before autoclaving at 120 °C and 1.2 kg/cm for 20 min. Hypocotyl segments were cultured in Petri dishes (10 segments/dish) containing 30 mL of medium/plate of the correspondent growth media. For each treatment, 15 Petri dishes, divided into three replicates, were assigned. The cultures were incubated at 23 °C under 30 µmol/m2/s of fluorescent light (16 h/day) for 5 weeks. Data were recorded on the percentage of callus induction frequency, calculated as the percentage of explants inducing callus relative to the total cultured explants. Callus proliferation was further studied by transferring uniform pieces of healthy, friable primary callus into the same eight fresh growth media combinations. Of the six tested cumin accessions, one accession (EG-3) failed to induce callus and, therefore, was excluded from the callus proliferation experiment. After 5 weeks, data on callus growth were recorded by the estimation of callus fresh weight and calculation of the relative growth rate (RGR) according to the following formula: . The basal salt media, MS and B5, were bought from Duchefa Biochemie B.V, Haarlem, The Netherlands. All other chemicals, including phytohormones, were purchased from Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany. Both experiments were arranged as two-way factorial experiments in a completely randomized design (CRD) and were statistically analyzed for differences among treatments using ANOVA, according to Gomez and Gomez [40]. Means were compared using LSD at p = 0.05. Statistix software (ver. 8.1, Analytical Software, Tallahassee, FL, USA) was used to perform the statistical analysis.
3. Results
3.1. Molecular Analysis
Four types of molecular markers, i.e., RAPD, ISSR, SCoT and SRAP, were used to perform a molecular analysis via the assessment of genetic diversity and relationship among seventeen cumin genotypes. Products of agarose gel electrophoresis of PCR amplification of different markers are presented in Figures S1–S14 in the Supplementary Materials. The results clearly show that all the molecular markers were able to expose the relationship among the tested genotypes. However, the amount of information and genetic diversity revealed by each was different. In this regard, a total number of 765 bands were analyzed, which were obtained using 87 primers, including 42, 15, 7 and 23 primers of RAPD, ISSR, SCoT and SRAP, respectively. Among the 765 bands, only 74 bands were polymorphic, with 9.67% polymorphism. However, the percentage of polymorphism varied among the markers; thus, the lowest average of %P was 5.46% for the RAPD marker, while the highest was 21.39% caused by the SCoT marker. Furthermore, the %P values within each marker generated by individual primers were different. In this regard, the highest %P for each marker was 53.85, 60.00, 42.86 and 78.57 for RAPD, ISSR, SCoT and SRAP, respectively.
In addition, some diversity measures, i.e., polymorphism information content (PIC), primer resolving power (Rp), diversity index (DI), expected heterozygosity (He) and marker index (MI), were calculated based on the data of the markers (Table 2). The averaged values of PIC obtained by the four markers were 0.01, 0.04, 0.06 and 0.01 for RAPD, ISSR, SCoT and SRAP, respectively. Moreover, the highest value of PIC was 0.17, as obtained by the B and UBC-112 ISSR primers. For primer resolving power, markers generated Rp values of 0.14, 0.64, 0.91 and 0.10 for RAPD, ISSR, SCoT and SRAP, respectively. The highest Rp was 2.59, as obtained by the UBC-112 ISSR primer. Additionally, averaged DI values were 0.01, 0.04, 0.08 and 0.01, while He averages were 0.01, 0.06, 0.05 and 0.02, as obtained by RAPD, ISSR, SCoT and SRAP, respectively. The highest value of DI was 0.17 and was generated by the B-ISSR primers and the SCT-06 and SCT-17 SCoT primers, while the highest value of He was 0.25, as generated by the UBC-112 ISSR primer. The marker index was calculated for each marker; its averages were 0.01, 0.08, 0.06 and 0.04 for RAPD, ISSR, SCoT and SRAP, respectively. Among marker primers, the highest MI was 0.81, as generated by the Me3/Em3 SRAP primer.

Table 2.
Some diversity measures and relationships among seventeen genotypes of cumin calculated based on RAPD, ISSR, SCoT and SRAP markers. TNB—total number of bands, NPB—number of polymorphic bands, %P—percentage of polymorphism, PIC—polymorphism information content, Rp—primer resolving power, DI—diversity index, He—expected heterozygosity, MI—marker index, RAPD—random amplified polymorphic DNA, ISSR—inter simple sequence repeats, SCoT—start-codon-targeted polymorphism, SRAP—sequence-related amplified polymorphism.
To figure out the genetic relationship among the tested genotypes, molecular data of the four different markers were used to perform a cluster analysis. Four dendrograms were obtained based on Jaccard’s coefficient of similarity (Figure 1). The RAPD-based dendrogram separated the genotypes into three main clusters (Figure 1A). The first cluster included all genotypes collected from Egypt and grouped them into three subclusters (EG-01 and EG-06, EG-02 and EG-03, EG-04 and EG-05). The second cluster gathered all the IPK genotypes, while the third cluster contained the genotype GRM-17 alone at a similarity value of 0.97. Some genotypes showed 100% similarity: these were IPK-10 and IPK-13 and IPK-12, IPK-14, IPK-15 and IPK-16. The ISSR-based dendrogram was different from that based on RAPD, in which the genotypes were separated into four main clusters (Figure 1B). All genotypes collected from Egypt were placed in the first main cluster, along with IPK-07, IPK-08 and PK-09. The second main cluster, interestingly, gathered IPK-10 with GRM-17, while the third main cluster consisted of the rest of the genotypes (IPK-12, IPK-16, IPK-13, IPK-15 and IPK-14). In addition, the genotype IPK-11 was separated alone in the fourth main cluster at a similarity value of 0.93. The dendrogram of SCoT data was more similar to that of RAPD than that of ISSR. However, a wider range of similarities was exposed on the SCoT-based dendrogram scale than on both others. All the genotypes collected from Egypt were grouped in the first main cluster except EG-06. The other genotypes of IPK with EG-06 formed the second main cluster, in which three subclusters were generated (Figure 1C). Meanwhile, the genotype GRM-17 was placed in the third main cluster alone at a similarity value of 0.82. The fourth dendrogram was made based on SRAP data, which was highly different from the previous ones. Out of the seventeen genotypes, twelve showed 100% similarity and were placed in one cluster at 1.00 similarity. The other five genotypes (i.e., IPK-13, EG-03, EG-06, IPK-12 and GRM-17) were placed in sub-branches separately, with the lowest similarity of 0.94 for GRM-17 (Figure 1D). This difference among the dendrograms might be due to the different regions of the genome that each molecular marker covered and the number of primers used.
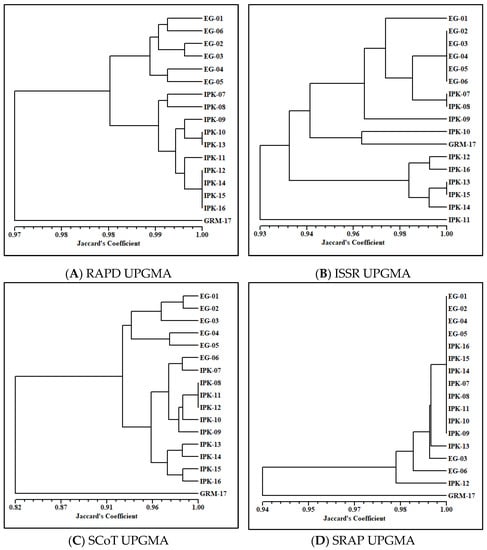
Figure 1.
UPGMA dendrogram using Jaccard’s similarity coefficient [32] illustrating relationships among 17 cumin accessions. (A) UPGMA dendrogram based on random amplified polymorphic DNA (RAPD) marker; (B) UPGMA dendrogram based on inter simple sequence repeats (ISSR) marker; (C) UPGMA dendrogram based on start-codon-targeted polymorphism (SCoT); (D) UPGMA dendrogram based on sequence-related amplified polymorphism (SRAP). A total of six accessions (EG-01 to EG-06) originated from Egypt; ten accessions (from IPK-07 to IPK-16) were brought from the Genebank of the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) in Gatersleben, Germany; and one accession (GRM-17) was from Germany. More details about the accessions are presented in Table 1.
For a better conclusion and to comprehend the relationship among the tested genotypes, a dendrogram was made based on data from all the molecular markers (Figure 2). The dendrogram separated the genotypes into three main clusters in a way somewhat similar to that based on RAPD data. The first main cluster consisted of all genotypes collected from Egypt. The second main cluster included all genotypes of IPK, while GRM-17 was separated alone in the third main cluster at a similarity value of 0.94. Based on the data of all the molecular markers, the IPK-14 and IPK-16 genotypes showed 100% similarity.
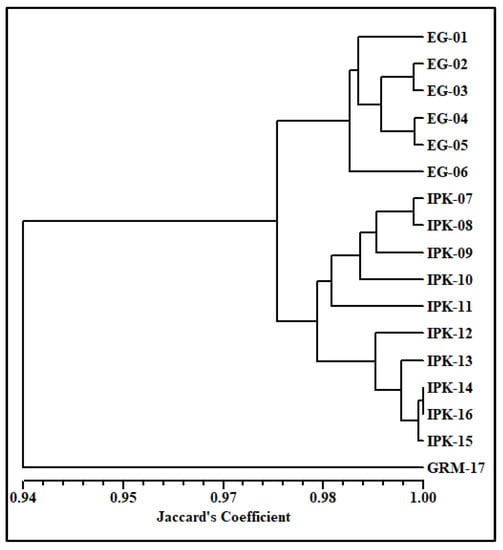
Figure 2.
UPGMA dendrogram using Jaccard’s similarity coefficient (Jaccard 1908) illustrating relationships among 17 cumin accessions based on combined data from random amplified polymorphic DNA (RAPD), inter simple sequence repeats (ISSR), start-codon-targeted polymorphism (SCoT) and sequence-related amplified polymorphism (SRAP). A total of six accessions (EG-01 to EG-06) originated from Egypt; ten accessions (from IPK-07 to IPK-16) were brought from the Genebank of the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) in Gatersleben, Germany; and one accession (GRM-17) was from Germany. More details about the accessions are presented in Table 1.
The correlation between each two of the four molecular markers was performed using the Mantel test. The highest correlation (r = 0.84) was between the RAPD and SCoT similarities, followed by the correlation between the SCoT and SRAP data (r = 0.78). In addition, the correlation between RAPD and SRAP was positively significant (r = 0.70). However, there was no correlation between ISSR and both RAPD (r = 0.33) and SCoT (r = 0.25).
3.2. Callus Induction and Growth
Variability in the callus induction of six cumin accessions was studied on different growth media combinations. The ability to induce callus significantly varied among accessions, as indicated by the callus induction frequency percentage illustrated in Table 3. EG-3 failed to induce callus in almost any of the tested media. The callus induction frequency induced by the other accessions ranged from 90.4 to 97.5%, with no significant differences among the five accessions. The employed combinations of growth media also showed nonsignificant potential to promote callus induction from various accessions. The two superior combinations were MS amended with 2,4-D (4.44 mg/L) + Kin (0.22 mg/L) and B5 with 2,4-D (8.88 mg/L) + Kin (0.22 mg/L). In general, it was noticed that the lower concentration of 2,4-D was more efficient when combined with the MS medium, unlike the B5 medium, which showed better results with a higher concentration. It is worth mentioning that the frequency of callus induction reached 100% in many cases (Table 3).

Table 3.
Callus induction frequency percentages of six cumin accessions originated from Egypt in response to eight combinations of growth media. Data are presented as means ± SD (n = 3).
Even though callus frequency showed a narrow range, indicating low variation among the five accessions, which showed the potential to induce callus, the RGR of the produced callus greatly varied (Table 4). The RGR (mg/g/day) based on callus fresh weight reflected this variation. All the media combinations, including the 2,4-D alone, exhibited significant superiority compared with those including both 2,4-D and Kin. Accessions also showed significant variability, where EG-4 induced the highest RGR, followed by EG-5, when cultured on either MS or B5 media supplemented with 2,4-D alone. Furthermore, the characteristics of the induced callus on different media combinations slightly varied (Figure S15 in the Supplementary Materials). Callus color was generally yellow, with a distinguished dark brown–yellow color of callus forming on the media supplemented with 2,4-D alone. Callus formed on these media was more compact and less friable than that formed on media amended with 2,4-D and Kin.

Table 4.
Relative growth rate (RGR, mg/g/day) based on fresh weight of callus induced from five cumin accessions originated from Egypt in response to eight combinations of growth media. Data are presented as means ± SD (n = 3).
4. Discussion
The findings of the current study showed that combining 87 primers, including 42, 15, 7 and 23 primers of RAPD, ISSR, SCoT and SRAP, respectively, facilitated the detection of the relationship among 17 genotypes of cumin. Among the 765 bands, only 74 bands were polymorphic, with 9.67% polymorphism. However, the percentage of polymorphism varied among and within the markers; thus, the lowest average of %P was 5.46% for the RAPD marker, while the highest was 21.39% caused by the SCoT marker. In addition, the highest %P for each marker was 53.85, 60.00, 42.86 and 78.57 for RAPD, ISSR, SCoT and SRAP, respectively. The reason for incorporating several types of markers was to ensure the amplification of different regions of the genome and, thus, obtain a better analysis for genetic diversity, as reported by several authors such as Chen et al. [41]. Compared with previous studies conducted on the genetic diversity of cumin, our study exposed a lower rate of polymorphism, which could be ascribed to the larger sampling area with very different cumin accessions employed in previous studies. Using various RAPD primers, polymorphism percentages have reached about 86%, according to Baghizadeh et al. [17], among 32 cumin genotypes; 62% as reported by [19]; and 54% as reported by Rostami-Ahmadvani et al. [42]. ISSR markers have shown varying polymorphism percentages, attaining 25.74 [43], 67% [42], 70% [19] and a range from 56 to 89% [44]. SCoT markers have also exposed diverging polymorphism values ranging from 29.67% [43] to 79.80% [8,9].
Our findings revealed average values of the PIC of 0.01, 0.04, 0.06 and 0.01 obtained by the RAPD, ISSR, SCoT and SRAP markers, respectively. Moreover, the highest value of PIC was 0.17, as obtained by the B and UBC-112 ISSR primers. In previous studies, PIC values for RAPD markers have ranged from 0.18 [44] to 0.26 [19]; for ISSR, they have reached 0.3 [19], from 0.03 to 0.70 [5], and 0.37 [44] and, for SRAP, it was 0.34, with a range from 0.14 to 0.51 [16]. Regarding primer resolving power in the current study, markers generated Rp values of 0.14, 0.64, 0.91 and 0.10 for RAPD, ISSR, SCoT and SRAP, respectively. The highest Rp was 2.59, as obtained by the UBC-112 ISSR primer. These values are relatively lower than those previously reported by Bhatt et al. [16] based on SRAP primers. Additionally, average DI values were 0.01, 0.04, 0.08 and 0.01, while He averages were 0.01, 0.06, 0.05 and 0.02 obtained by RAPD, ISSR, SCoT and SRAP, respectively. The highest value of DI was 0.17, as generated by the B-ISSR primers and the SCT-06 and SCT-17 SCoT primers, while the highest value of He was 0.25, as generated by the UBC-112 ISSR primer. The MI was calculated for each marker, and its averages were 0.01, 0.08, 0.06 and 0.04 for RAPD, ISSR, SCoT and SRAP, respectively. Among marker primers, the highest MI was 0.81, as generated by the M3-E3 SRAP primer. Nevertheless, a higher MI (2.43) was reported by Bhatt et al. [16] for various cumin accessions based on a set of 10 SRAP primers.
The UPGMA dendrogram made based on data from all the molecular markers, i.e., RAPD, ISSR, SCoT and SRAP, employed in the current study illustrated the relationship among the seventeen cumin accessions studied. The dendrogram separated the genotypes into three main clusters. The first main cluster consisted of all genotypes collected from Egypt. The second main cluster included all genotypes of IPK, while GRM-17 was separated alone in the third main cluster at a similarity value of 0.94. Based on the data of all the molecular markers, genotypes IPK-14 and IPK-16, originating from Pakistan, showed 100% similarity. Both the RAPD and SCoT similarities showed the highest correlation (r = 0.84) performed using the Mantel test compared with the other markers. It is clear that the genotypes exhibited a partial geographic-based relationship. These results contradict those of Mohamamadizadeh et al. [45], indicating no influence of the increase in the geographical on genetic differentiation. The number of clusters exposed by UPGMA in previous studies has varied according to different cumin populations and the employed molecular markers. Bhatt et al. [16] reported three distinct clusters among 16 cumin genotypes based on a set of 10 SRAP. However, in other cumin populations, categories have reached six groups [17], as well as nine groups with a similarity level of 0.43 based on RAPD markers [18].
Regarding the potential of the tested accessions to induce callus under in vitro conditions, four accessions successfully induced callus with a frequency ranging from 85.0 to 93.8% and reached 100% in many cases, unlike the EG-3 accession, which failed to induce callus. The influence of the callus induction medium was evident with the superiority of two combinations: MS amended with 2,4-D (4.44 mg/L) + Kin (0.22 mg/L) and B5 with 2,4-D (8.88 mg/L) + Kin (0.22 mg/L). The RGR (mg/g/day) based on callus fresh weight reflected high variation among the accessions, where EG-4 induced the highest RGR followed by EG-5. All media combinations, including 2,4-D alone, exhibited significant superiority compared with those including both 2,4-D and Kin. The slight differences in callus color and texture could be attributed to the faster growth rate of the callus produced on the media supplemented with 2,4-D alone, indicating the need for a more frequent subculture to avoid color change. In the same context, the effect of subculture frequency on callus growth has been reported by several authors, where the subculture cycle improves callus characteristics and multiplication rate [46,47]. The callus induction frequency obtained in our study is relatively higher than those reported in several previous studies, such as Beiki et al. [24] (27%) on B5 medium supplemented with 0.2 NAA + 0.4 IAA + 0.1 mg/L of BAP. In line with our findings, some authors attributed the callus induction effect in cumin to the presence of 2,4-D, such as Soorni and Kahrizi [48]. Tawfik and Noga [26,49] confirmed the importance of 2,4-D together with kinetin for cumin callus induction and proliferation and recommended the use of hypocotyl segments as explants for callus induction on MS medium supplemented with 4 μM of 2,4-D alone or with Kin at 2 or 4 μM. Similar results were also reported by Suthar et al. [50], recommending MS basal medium with 1.0 μM of 2,4-D and 10 μM of kinetin as a callus induction medium for cumin.
5. Conclusions
Aiming at assessing the genetic variability in cumin, 17 genotypes were assessed using four molecular markers (RAPD, ISSR, SRAP and SCoT). Low polymorphism was detected (9.67%) together with low values for the determined diversity measures, i.e., PIC (0.06), Rp (0.91), DI (0.08), He (0.06) and MI (0.08). The UPGMA dendrogram based on data from all the molecular markers separated the genotypes into three main clusters, with a partial geographic-based relationship among the genotypes. Out of the six accessions selected for the in vitro callus induction study, five were potent to induce callus, with a frequency ranging from 85.0 to 93.8%. Two media combinations showed superior results: MS amended with 2,4-D (4.44 mg/L) + Kin (0.22 mg/L) and B5 with 2,4-D (8.88 mg/L) + Kin (0.22 mg/L). The EG-4 accession was the superior accession regarding the frequency of callus induction and callus RGR. Additionally, 2,4-D alone exhibited better results than its combinations with Kin. Our findings exposed low variability among the studied cumin accessions, implying the real need for more effort to assess wider populations from different geographic regions together with the need for reliable diversification programs.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae9070742/s1, Table S1: List of RAPD, ISSR, SCoT and SRAP primers utilized to test variability with a collection of cumin genotypes; Figure S1: Products of agarose gel electrophoresis of RAPD-PCR amplification with OPA-01 primer for 17 cumin genotypes (lanes 2 to 18, in order from left to right: EG-01, EG-02, EG-03, EG-04, EG-05, EG-06, IPK-07, IPK-08, IPK-09, IPK-10, IPK-11, IPK-12, IPK-13, IPK-14, IPK-15, IPK-16 and GR-17 in addition to lane 19 for sweet fennel as a control ) using DNA size marker 100—10000 bp (lanes 1 and 20); Figure S2: Products of agarose gel electrophoresis of RAPD-PCR amplification with OPA-10 (upper image) and OPA-19 (lower image) primers for 17 cumin genotypes (lanes 2 to 18, in order from left to right: EG-01, EG-02, EG-03, EG-04, EG-05, EG-06, IPK-07, IPK-08, IPK-09, IPK-10, IPK-11, IPK-12, IPK-13, IPK-14, IPK-15, IPK-16 and GR-17 in addition to lane 19 for sweet fennel as a control) using DNA size marker 100—10000 bp (lane 1); Figure S3: Products of agarose gel electrophoresis of RAPD-PCR amplification with OPA-09 (upper image) and OPA-11 (lower image) primers for 17 cumin genotypes (lanes 2 to 18, in order from left to right: EG-01, EG-02, EG-03, EG-04, EG-05, EG-06, IPK-07, IPK-08, IPK-09, IPK-10, IPK-11, IPK-12, IPK-13, IPK-14, IPK-15, IPK-16 and GR-17 in addition to lane 19 for sweet fennel as a control) using DNA size marker 100—10000 bp (lane 1); Figure S4: Products of agarose gel electrophoresis of RAPD-PCR amplification with OPB-02 (upper image) and OPB-03 (lower image) primers for 17 cumin genotypes (lanes 2 to 18, in order from left to right: EG-01, EG-02, EG-03, EG-04, EG-05, EG-06, IPK-07, IPK-08, IPK-09, IPK-10, IPK-11, IPK-12, IPK-13, IPK-14, IPK-15, IPK-16 and GR-17 in addition to lane 19 for sweet fennel as a control) using DNA size marker 100—10000 bp (lane 1); Figure S5: Products of agarose gel electrophoresis of RAPD-PCR amplification with OPG-18 primer for 17 cumin genotypes (lanes 2 to 18, in order from left to right: EG-01, EG-02, EG-03, EG-04, EG-05, EG-06, IPK-07, IPK-08, IPK-09, IPK-10, IPK-11, IPK-12, IPK-13, IPK-14, IPK-15, IPK-16 and GR-17 in addition to lane 19 for sweet fennel as a control) using DNA size marker 100—10000 bp (lane 1); Figure S6: Products of agarose gel electrophoresis of RAPD-PCR amplification with OPL-16 (upper image) and OPL-07 (lower image) primers of 17 cumin genotypes (lanes 2 to 18, in order from left to right: EG-01, EG-02, EG-03, EG-04, EG-05, EG-06, IPK-07, IPK-08, IPK-09, IPK-10, IPK-11, IPK-12, IPK-13, IPK-14, IPK-15, IPK-16 and GR-17 in addition to lane 19 for sweet fennel as a control) using DNA size marker 100—10000 bp (lane 1); Figure S7: Products of agarose gel electrophoresis of ISSR-PCR amplification with A (upper image) and B (lower image) primers for 17 cumin genotypes (lanes 2 to 18, in order from left to right: EG-01, EG-02, EG-03, EG-04, EG-05, EG-06, IPK-07, IPK-08, IPK-09, IPK-10, IPK-11, IPK-12, IPK-13, IPK-14, IPK-15, IPK-16 and GR-17 in addition to lane 19 for sweet fennel as a control) using DNA size marker 100—10000 bp (lane 1); Figure S8: Products of agarose gel electrophoresis of ISSR-PCR amplification UBC-112 (upper image) and UBC -818 (lower image) primers for 17 cumin genotypes (lanes 2 to 18, in order from left to right: EG-01, EG-02, EG-03, EG-04, EG-05, EG-06, IPK-07, IPK-08, IPK-09, IPK-10, IPK-11, IPK-12, IPK-13, IPK-14, IPK-15, IPK-16 and GR-17 in addition to lane 19 for sweet fennel as a control) using DNA size marker 100—10000 bp (lane 1); Figure S9: Products of agarose gel electrophoresis of ISSR-PCR amplification UBC-841 (upper image) and UBC -854 (lower image) primers for 17 cumin genotypes (lanes 2 to 18, in order from left to right: EG-01, EG-02, EG-03, EG-04, EG-05, EG-06, IPK-07, IPK-08, IPK-09, IPK-10, IPK-11, IPK-12, IPK-13, IPK-14, IPK-15, IPK-16 and GR-17 in addition to lane 19 for sweet fennel as a control) using DNA size marker 100—10000 bp (lane 1); Figure S10: Products of agarose gel electrophoresis of SCOT-PCR amplification with 06 (upper image) and 12 (lower image) primers for 17 cumin genotypes (lanes 2 to 18, in order from left to right: EG-01, EG-02, EG-03, EG-04, EG-05, EG-06, IPK-07, IPK-08, IPK-09, IPK-10, IPK-11, IPK-12, IPK-13, IPK-14, IPK-15, IPK-16 and GR-17 in addition to lane 19 for sweet fennel as a control) using DNA size marker 100—10000 bp (lane 1); Figure S11: Products of agarose gel electrophoresis of SCOT-PCR amplification with 17 (upper image) and 25 (lower image) primers for 17 cumin genotypes (lanes 2 to 18, in order from left to right: EG-01, EG-02, EG-03, EG-04, EG-05, EG-06, IPK-07, IPK-08, IPK-09, IPK-10, IPK-11, IPK-12, IPK-13, IPK-14, IPK-15, IPK-16 and GR-17 in addition to lane 19 for sweet fennel as a control) using DNA size marker 100—10000 bp (lane 1); Figure S12: Products of agarose gel electrophoresis of SCOT-PCR amplification with 30 (upper image) and 33 (lower image) primers for 17 cumin genotypes (lanes 2 to 18, in order from left to right: EG-01, EG-02, EG-03, EG-04, EG-05, EG-06, IPK-07, IPK-08, IPK-09, IPK-10, IPK-11, IPK-12, IPK-13, IPK-14, IPK-15, IPK-16 and GR-17 in addition to lane 19 for sweet fennel as a control) using DNA size marker 100—10000 bp (lane 1); Figure S13: Products of agarose gel electrophoresis of SRAP-PCR amplification with Me3/Em2 primers for 17 cumin genotypes (lanes 2 to 18, in order from left to right: EG-01, EG-02, EG-03, EG-04, EG-05, EG-06, IPK-07, IPK-08, IPK-09, IPK-10, IPK-11, IPK-12, IPK-13, IPK-14, IPK-15, IPK-16 and GR-17 in addition to lane 19 for sweet fennel as a control) using DNA size marker 100—10000 bp (lane 1); Figure S14: Products of agarose gel electrophoresis of SRAP-PCR amplification with Me3/Em3 (upper image) and Me3/Em4 (lower image) primers for 17 cumin genotypes (lanes 2 to 18, in order from left to right: EG-01, EG-02, EG-03, EG-04, EG-05, EG-06, IPK-07, IPK-08, IPK-09, IPK-10, IPK-11, IPK-12, IPK-13, IPK-14, IPK-15, IPK-16 and GR-17 in addition to lane 19 for sweet fennel as a control) using DNA size marker 100—10000 bp (lane 1) and Figure S15: Pictures of induced callus from five cumin genotypes originated from Egypt (EG-1 and EG-2 were collected from Assiut region and EG-4, EG-5 and EG-6 were collected from El-menia region) in response to various medium combinations. MS denotes Murashige and Skoog medium and B5 means Gamborg’s B5 medium; each of the two media combined with four hormonal combinations, namely, 2,4-D (4.44), 2,4-D (4.44)+ Kin (0.22), 2,4-D (8.88) and 2,4-D (8.88)+ Kin (0.22), respectively.
Author Contributions
Conceptualization, K.A.M.A.-E., M.A.A.M. and O.H.M.I.; Data curation, K.A.M.A.-E. and M.A.A.M.; Investigation, K.A.M.A.-E. and O.H.M.I.; Methodology, M.A.A.M.; Visualization, O.H.M.I.; Writing—original draft, K.A.M.A.-E., M.A.A.M. and O.H.M.I.; Writing—review and editing, M.A.A.M., K.A.M.A.-E. and O.H.M.I. All authors have read and agreed to the published version of the manuscript.
Funding
This research work was funded by the Ministry of Education and King Abdulaziz University, DSR, Jeddah, Saudi Arabia, under grant No. “IFPIP: 37-155-1443”.
Data Availability Statement
Data will be made available on request.
Acknowledgments
This research work was funded by Institutional Fund Project under grant No. “IFPIP: 37-155-1443”. The authors gratefully acknowledge technical and financial support provided by the Ministry of Education and King Abdulaziz University, DSR, Jeddah, Saudi Arabia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Süntar, I. Importance of ethnopharmacological studies in drug discovery: Role of medicinal plants. Phytochem. Rev. 2020, 19, 1199–1209. [Google Scholar] [CrossRef]
- Archangi, A.; Mohammadi-Nejad, G.; Heidari, B. Assessing genetic diversity and aggregate genotype selection in a collection of cumin (Cuminum cyminum L.) accessions under drought stress: Application of BLUP and BLUE. Sci. Hortic. 2022, 299, 111028. [Google Scholar] [CrossRef]
- Riasat, M.; Heidari, B.; Pakniyat, H.; Jafari, A.A. Assessment of variability in secondary metabolites and expected response to genotype selection in fenugreek (Trigonella spp.). Ind. Crops Prod. 2018, 123, 221–231. [Google Scholar] [CrossRef]
- Parashar, M.; Malik, C.P. Appraisal of genetic diversity in Cuminum cyminum L. using molecular markers. Int. J. Life Sci. 2014, 3, 143–156. [Google Scholar] [CrossRef]
- Bharti, R.; Kumar, S.; Parekh, M.J. Development of genomic simple sequence repeat (gSSR) markers in cumin and their application in diversity analyses and cross-transferability. Ind. Crops Prod. 2018, 111, 158–164. [Google Scholar] [CrossRef]
- Kumar, S.; Saxena, S.N.; Mistry, J.G.; Fougat, R.S.; Solanki, R.K.; Sharma, R. Understanding Cuminum cyminum: An important seed spice crop of arid and semi-arid regions. Int. J. Seed Spices 2015, 5, 1–19. [Google Scholar]
- Heidari, M.; Sadeghi, H. Germination and emergence of primed cumin (Cuminum cyminum L.) seeds with GA3 under different temperature regimes. Int. J. Biosci. 2014, 5, 266–272. [Google Scholar]
- Parashar, M.; Malik, C.P. Appraisal of genetic diversity in Cuminum cyminum Linn. using SCoT and CCMP markers. Phytomorphology 2015, 65, 31–38. [Google Scholar]
- Parashar, M.; Jakhar, M.L.; Malik, C.P. A review of biotechnology, genetic diversity in cumin (cuminum cyminum). Int. J. Life Sci. Pharma Res. 2014, 4, 17–34. [Google Scholar]
- Rukhsar; Patel, M.P.; Parmar, D.J.; Kalola, A.D.; Kumar, S. Morphological and molecular diversity patterns in castor germplasm accessions. Ind. Crops Prod. 2017, 97, 316–323. [Google Scholar] [CrossRef]
- Kumar, S.; Mahendi, H.A.; Fougat, R.S.; Sakure, A.A.; Mistry, J.G. Transferability of carrot (Daucus carota) microsatellite markers to cumin (Cuminum cyminum). Int. J. Seed Spices 2014, 4, 88–90. [Google Scholar]
- Chavhan, R.L.; Sable, S.; Narwade, A.V.; Hinge, V.R.; Kalbande, B.B.; Mukherjee, A.K.; Chakrabarty, P.K.; Kadam, U.S. Multiplex molecular marker-assisted analysis of significant pathogens of cotton (Gossypium sp.). Biocatal. Agric. Biotechnol. 2023, 47, 102557. [Google Scholar] [CrossRef]
- Hinge, V.R.; Shaikh, I.M.; Chavhan, R.L.; Deshmukh, A.S.; Shelake, R.M.; Ghuge, S.A.; Dethe, A.M.; Suprasanna, P.; Kadam, U.S. Assessment of genetic diversity and volatile content of commercially grown banana (Musa spp.) cultivars. Sci. Rep. 2022, 12, 7979. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.; Kadam, U.S.; Chacko, P.M.; Aher, L.; Karibasappa, G.S. Microsatellite analysis to differentiate clones of Thompson seedless grapevine. Indian J. Hortic. 2010, 67, 260–263. [Google Scholar]
- Upadhyay, A.; Kadam, U.S.; Chacko, P.; Karibasappa, G.S. Microsatellite and RAPD analysis of grape (Vitis spp.) accessions and identification of duplicates/misnomers in germplasm collection. Indian J. Hortic. 2010, 67, 8–15. [Google Scholar]
- Bhatt, J.; Kumar, S.; Patel, S.; Solanki, R. Sequence-related amplified polymorphism (SRAP) markers based genetic diversity analysis of cumin genotypes. Ann. Agrar. Sci. 2017, 15, 434–438. [Google Scholar] [CrossRef]
- Baghizadeh, A.; Salar Karimi, M.; Pourseyedi, S.H. Genetic diversity assessment of Iranian green cumin genotypes by RAPD molecular markers. Int. J. Plant Prod. 2013, 4, 472–479. [Google Scholar]
- Bahraminejad, A.; Mohammadi-Nejad, G.; Abdul Kadir, K.; Bin Yusop, M.R.; Samia, M.A. Molecular diversity of Cumin (’Cuminum cyminum’ L.) using RAPD markers. Aust. J. Crop Sci. 2012, 6, 194–199. [Google Scholar]
- Zabet, M.; Rahimi, A.; Izanlo, A.; Alizadeh, Z. Investigation of genetic variation in cumin (Cuminum cyminum) ecotypes of Khorasan Provinces using RAPD and ISSR markers. Agric. Biotechnol. J. 2019, 11, 75–98. [Google Scholar] [CrossRef]
- Sen, M.K.; Nasrin, S.; Rahman, S.; Jamal, A.H.M. In vitro callus induction and plantlet regeneration of Achyranthes aspera L., a high value medicinal plant. Asian Pac. J. Trop. Biomed. 2014, 4, 40–46. [Google Scholar] [CrossRef]
- Kirillova, N.V.; Smirnova, M.G.; Komov, V.P. Sequential Isolation of Superoxide Dismutase and Ajmaline from Tissue Cultures of Rauwolfia serpentina Benth. Appl. Biochem. Microbiol. 2001, 37, 160–163. [Google Scholar] [CrossRef]
- Bhatia, S. Application of Plant Biotechnology. In Modern Applications of Plant Biotechnology in Pharmaceutical Sciences; Bhatia, S., Sharma, K., Dahiya, R., Bera, T., Eds.; Academic Press: Boston, MA, USA, 2015; Chapter 5; pp. 157–207. [Google Scholar]
- Jha, T.B.; Roy, S.C.; Mitra, G.C. In vitro culture of Cuminum cyminum regeneration of flowering shoots from calli of hypocotyl and leaf explants. Plant Cell Tissue Organ Cult. 1982, 2, 11–14. [Google Scholar] [CrossRef]
- Beiki, A.H.; Mafavi-fard, M.R.; Ahmadi, J. Optimization of Two Different Morphogenesis Pathways in Three Iranian Cumin Landraces with the use of an Embryo. Biotechnol. Biotechnol. Equip. 2011, 25, 2228–2232. [Google Scholar] [CrossRef]
- Gupta, D. Studies on biochemical markers associated with regeneration potential in Cuminum cyminum L. Res. J. Chem. Environ. Sci. 2013, 1, 63–65. [Google Scholar]
- Tawfik, A.A.; Noga, G. Cumin regeneration from seedling derived embryogenic callus in response to amended kinetin. Plant Cell Tissue Organ Cult. 2002, 69, 35–40. [Google Scholar] [CrossRef]
- Križman, M.; Jakše, J.; Baričevič, D.; Javornik, B.; Prošek, M. Robust CTAB-activated charcoal protocol for plant DNA extraction. Acta Agric. Slov. 2006, 87, 427–433. [Google Scholar]
- Williams, J.G.K.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef]
- Gupta, M.; Chyi, Y.S.; Romero-Severson, J.; Owen, J.L. Amplification of DNA markers from evolutionarily diverse genomes using single primers of simple-sequence repeats. Theor. Appl. Genet. 1994, 89, 998–1006. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Mackill, D.J. Start Codon Targeted (SCoT) Polymorphism: A Simple, Novel DNA Marker Technique for Generating Gene-Targeted Markers in Plants. Plant Mol. Biol. Rep. 2009, 27, 86–93. [Google Scholar] [CrossRef]
- Li, G.; Quiros, C.F. Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: Its application to mapping and gene tagging in Brassica. Theor. Appl. Genet. 2001, 103, 455–461. [Google Scholar] [CrossRef]
- Jaccard, P. Nouvelles recherches sur la distribution florale. Bull. Soc. Vaud. Sci. Nat. 1908, 44, 223–270. [Google Scholar]
- Roldán-Ruiz, I.; Dendauw, J.; Van Bockstaele, E.; Depicker, A.; De Loose, M. AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol. Breed. 2000, 6, 125–134. [Google Scholar] [CrossRef]
- Prevost, A.; Wilkinson, M.J. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor. Appl. Genet. 1999, 98, 107–112. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Nei, M.; Roychoudhury, A.K. Sampling variances of heterozygosity and genetic distance. Genetics 1974, 76, 379–390. [Google Scholar] [CrossRef]
- Varshney, R.K.; Chabane, K.; Hendre, P.S.; Aggarwal, R.K.; Graner, A. Comparative assessment of EST-SSR, EST-SNP and AFLP markers for evaluation of genetic diversity and conservation of genetic resources using wild, cultivated and elite barleys. Plant Sci. 2007, 173, 638–649. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Gamborg, O.L.; Miller, R.A.; Ojima, K. Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 1968, 50, 151–158. [Google Scholar] [CrossRef]
- Gomez, K.A.; Gomez, A.A. Statistical Procedures for Agricultural Research; John Wiley & Sons: Hoboken, NJ, USA, 1984. [Google Scholar]
- Chen, S.-Y.; Dai, T.-X.; Chang, Y.-T.; Wang, S.-S.; Ou, S.-L.; Chuang, W.-L.; Chuang, C.-Y.; Lin, Y.-H.; Lin, Y.-Y.; Ku, H.-M. Genetic diversity among “Ocimum” species based on ISSR, RAPD and SRAP markers. Aust. J. Crop Sci. 2013, 7, 1463–1471. [Google Scholar]
- Rostami-Ahmadvandi, H.; Cheghamirza, K.; Kahrizi, D.; Bahraminejad, S. Comparison of morpho-agronomic traits versus RAPD and ISSR markers in order to evaluate genetic diversity among Cuminum cyminum L. accessions. Aust. J. Crop Sci. 2013, 7, 361–367. [Google Scholar]
- Mohammadizadeh, M.S.; Bahadori, F.; Hakimi, L.; Dehshiri, A. Evaluation of Molecular Diversity Analysis and Relation with Environmental Factors in Accessions of Cumin (Cuminum cyminum L.) in Iran, Revealed by Inter-Simple Sequence Repeat (ISSR) Markers and Start Codon Targeted (SCoT) Markers. 2020. Available online: https://www.researchsquare.com/article/rs-35133/v1 (accessed on 2 February 2023).
- Bahraminejad, A.; Mohammadinejad, G. Use of microsatellite markers for molecular characterization of cumin (Cuminum cyminum L.) ecotypes. Iran. J. Genet. Plant Breed. 2013, 2, 35–41. [Google Scholar]
- Mohamamadizadeh, M.S.; Bahadori, F.; Hakimi, L.; Khalighi, A.; Dehshiri, A. Genetic Diversity of Iranian Cumin (Cuminum cyminum L.) Accessions, using Inter-Simple Sequence Repeat (ISSR) and Start Codon Targeted (SCoT) Markers. J. Med. Plants By-Prod. 2021, 11, 25–35. [Google Scholar] [CrossRef]
- Nakasha, J.J.; Sinniah, U.R.; Kemat, N.; Mallappa, K.S. Induction, subculture cycle, and regeneration of callus in safed musli (Chlorophytum borivilianum) using different types of phytohormones. Pharmacogn. Mag. 2016, 12, S460–S464. [Google Scholar] [PubMed]
- Lo, K.; Nadali, B.J.; Chan, L.-K. Investigation on the effect of subculture frequency and inoculum size on the artemisinin content in a cell suspension culture of Artemisia annua L. Aust. J. Crop Sci. 2012, 6, 801–807. [Google Scholar]
- Soorni, J.; Kahrizi, D. Effect of Genotype, Explant Type and 2,4-D on Cell Dedifferentiation and Callus Induction in Cumin (Cuminum cyminum L.) Medicinal Plant. J. Appl. Biotechnol. Rep. 2015, 2, 265–270. [Google Scholar]
- Tawfik, A.A.; Noga, G.J. Differentiation of somatic embryos in suspension cultures and plant regeneration of cumin (Cuminum cyminum L.). J. Appl. Bot. 2002, 76, 144–149. [Google Scholar]
- Suthar, R.; Bhatt, P.N.; Bhatt, D.P. Selection of vascular wilt resistance cumin callus to culture filtrate of Fusarium equiseti and regeneration of plants. Vegetos 2021, 34, 318–324. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).