Molecular Diagnostics in Tomato: Chip Digital PCR Assays Targeted to Identify and Quantify Clavibacter michiganensis subsp. michiganensis and Ralstonia solanacearum in planta
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Samples
2.2. Plant Samples
2.3. Naturally Infected Plant Samples
2.4. Design of Primers and Probes
2.5. Real-Time qPCR
2.6. Chip Digital PCR
3. Results
3.1. Primers/Probe Sets
3.2. Assays Efficiency, Specificity and Repeatability Evaluation in Real-Time PCR
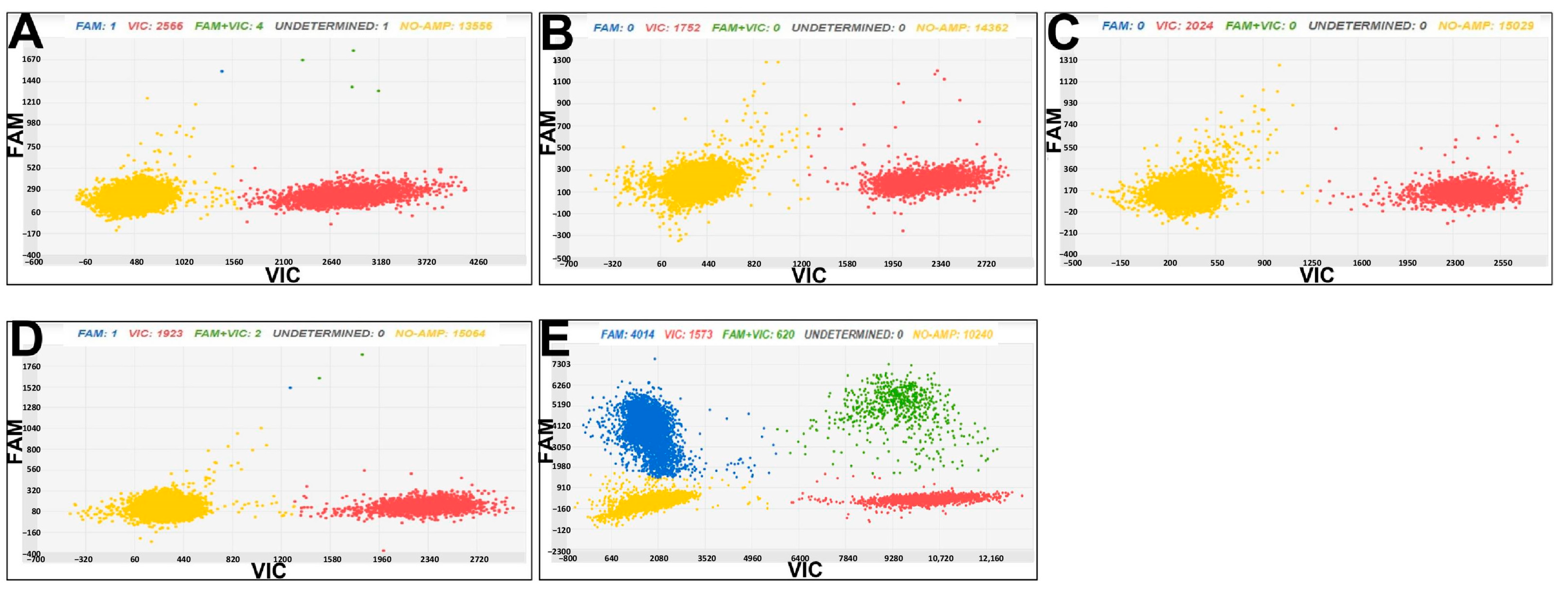
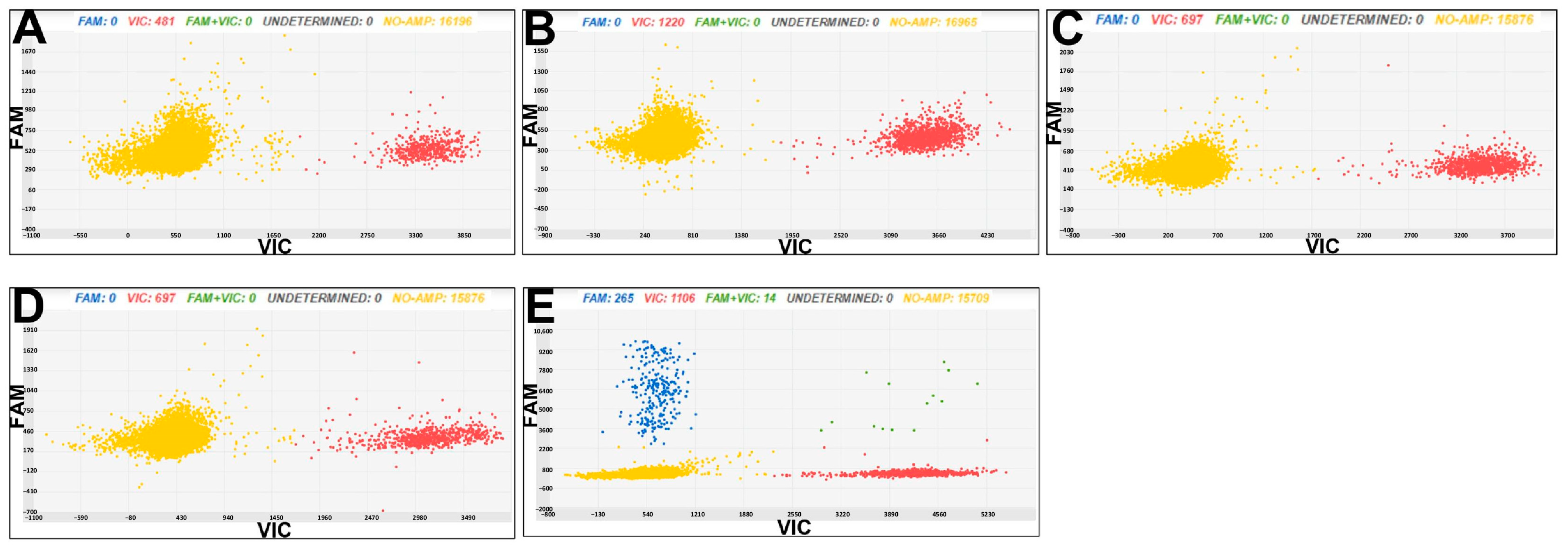
3.3. Chip Digital PCR for Cmm and Rs Diagnostics
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tripathi, R.; Vishunavat, K.; Tewari, R.; Kumar, S.; Minkina, T.; De Corato, U.; Keswani, C. Defense Inducers Mediated Mitigation of Bacterial Canker in Tomato through Alteration in Oxidative Stress Markers. Microorganisms 2022, 10, 2160. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhong, L.; Huang, Z.; Li, C.; Lian, J.; Zheng, X.; Liang, Y. Real-time monitoring of Ralstonia solanacearum infection progress in tomato and Arabidopsis using bioluminescence imaging technology. Plant Methods 2022, 18, 7. [Google Scholar] [CrossRef]
- FAOSTAT. Available online: https://data.un.org/Data.aspx?d=FAO&f=itemCode%3A388 (accessed on 14 March 2023).
- Panno, S.; Davino, S.; Caruso, A.G.; Bertacca, S.; Crnogorac, A.; Mandić, A.; Noris, E.; Matić, S. A Review of the Most Common and Economically Important Diseases That Undermine the Cultivation of Tomato Crop in the Mediterranean Basin. Agronomy 2021, 11, 2188. [Google Scholar] [CrossRef]
- Yadeta, K.A.; Thomma, B.P. The xylem as battleground for plant hosts and vascular wilt pathogens. Front. Plant Sci. 2013, 4, 97. [Google Scholar] [CrossRef][Green Version]
- Huet, G. Breeding for resistances to Ralstonia solanacearum. Front. Plant Sci. 2014, 5, 715. [Google Scholar] [CrossRef][Green Version]
- EPPO (European and Mediterranean Plant Protection Organization). Available online: https://www.eppo.int/ (accessed on 10 March 2023).
- Tancos, M.A.; Chalupowicz, L.; Barash, I.; Manulis-Sasson, S.; Smart, C.D. Tomato fruit and seed colonization by Clavibacter michiganensis subsp. Michiganensis through external and internal routes. Appl. Environ. Microbiol. 2013, 79, 6948–6957. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nandi, M.; Macdonald, J.; Liu, P.; Weselowski, B.; Yuan, Z.-C. Clavibacter michiganensis ssp. Michiganensis: Bacterial canker of tomato, molecular interactions and disease management. Mol. Plant Pathol. 2018, 19, 2036–2050. [Google Scholar] [CrossRef][Green Version]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef][Green Version]
- Álvarez, B.; Biosca, E.G.; López, M.M. On the life of Ralstonia solanacearum, a destructive bacterial plant pathogen. In Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology; Méndez-Vilas, A., Ed.; Formatex: Badajoz, Spain, 2010; pp. 267–279. [Google Scholar]
- Peňázová, E.; Dvořák, M.; Ragasová, L.; Kiss, T.; Pečenka, J.; Čechová, J.; Eichmeier, A. Multiplex real-time PCR for the detection of Clavibacter michiganensis subsp. Michiganensis, Pseudomonas syringae pv. Tomato and pathogenic Xanthomonas species on tomato plants. PLoS ONE 2020, 15, e0227559. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, D.; Liu, E.; Liu, H.; Jin, X.; Niu, C.; Gao, Y.; Su, X. A droplet digital PCR assay for detection and quantification of Verticillium nonalfalfae and V. albo-atrum. Front. Cell. Infect. Microbiol. 2023, 12, 1110684. [Google Scholar] [CrossRef]
- Morcia, C.; Ghizzoni, R.; Delogu, C.; Andreani, L.; Carnevali, P.; Terzi, V. Digital PCR: What Relevance to Plant Studies? Biology 2020, 9, 433. [Google Scholar] [CrossRef]
- PM 7/21 (3) Ralstonia solanacearum, R. pseudosolanacearum and R. syzygii (Ralstonia solanacearum species complex). EPPO Bull. 2022, 52, 225–261. [CrossRef]
- PM 7/42 (3) Clavibacter michiganensis subsp. michiganensis. Eppo Bull. 2016, 46, 202–225. [CrossRef]
- PM 7/110 (1) Xanthomonas spp. (Xanthomonas euvesicatoria, Xanthomonas gardneri, Xanthomonas perforans, Xanthomonas vesicatoria) causing bacterial spot of tomato and sweet pepper. EPPO Bull. 2013, 43, 7–20. [CrossRef]
- ISTA 7-021 (2022), International Rules for Seed Testing. 7-021: Ver 3.2 Detection of Xanthomonas axonopodis pv. phaseoli and Xanthomonas axonopodis pv. phaseoli var. fuscans on Phaseolus vulgaris. In Annexe to Chapter 7: Seed Health Testing Methods; International Seed Testing Association: Bassersdorf, Switzerland, 2022.
- Mohan, S.K.; Schaad, N.W. An improved agar plating assay for detecting Pseudomonas syringae pv. syringae and P. s. pv. phaseolicola in contaminated bean seed. Phytopathology 1987, 77, 1390–1395. [Google Scholar] [CrossRef]
- Weller, S.A.; Elphinstone, J.G.; Smith, N.C.; Boonham, N.; Stead, D.E. Detection of Ralstonia solanacearum strains with a quantitative, multiplex, real-time, fluorogenic PCR (TaqMan) assay. Appl. Environ. Microbiol. 2000, 66, 2853–2858. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Collier, R.; Dasgupta, K.; Xing, Y.P.; Hernandez, B.T.; Shao, M.; Rohozinski, D.; Kovak, E.; Lin, J.; de Oliveira, M.L.P.; Stover, E.; et al. Accurate measurement of transgene copy number in crop plants using droplet digital PCR. Plant J. 2017, 90, 1014–1025. [Google Scholar] [CrossRef][Green Version]
- Dreo, T.; Pirc, M.; Ramšak, Ž.; Pavšič, J.; Milavec, M.; Zel, J.; Gruden, K. Optimising droplet digital PCR analysis approaches for detection and quantification of bacteria: A case study of fire blight and potato brown rot. Anal. Bioanal. Chem. 2014, 406, 6513–6528. [Google Scholar] [CrossRef]
- Hougs, L.; Gatto, F.; Goerlich, O.; Grohmann, L.; Lieske, K.; Mazzara, M.; Narendja, F.; Ovesna, J.; Papazova, N.; Scholtens, I.; et al. Verification of analytical methods for GMO testing when implementing interlaboratory validated methods. In Testing and Analysis of GMO-Containing Foods and Feed; EUR 29015 EN; Publication Office of the European Union: Luxembourg, 2017; ISBN 978-92-79-77310-5. [Google Scholar] [CrossRef]
- Longchar, B.; Phukan, T.; Yadav, S.; Senthil-Kumar, M. An efficient low-cost xylem sap isolation method for bacterial wilt assays in tomato. Appl. Plant Sci. 2020, 8, e11335. [Google Scholar] [CrossRef]
- Lowe-Power, T.M.; Hendrich, C.G.; von Roepenack-Lahaye, E.; Li, B.; Wu, D.; Mitra, R.; Dalsing, B.L.; Ricca, P.; Naidoo, J.; Cook, D.; et al. Metabolomics of tomato xylem sap during bacterial wilt reveals Ralstonia solanacearum produces abundant putrescine, a metabolite that accelerates wilt disease. Environ. Microbiol. 2018, 20, 1330–1349. [Google Scholar] [CrossRef]
- NPPO. Available online: https://www.fao.org/documents/card/en/c/CA6375EN (accessed on 14 March 2023).
- Chalam, V.C.; Gupta, K.; Sharma, R.; Sharma, V.D.; Maurya, A.K. Pest Risk Analysis and Plant Quarantine Regulations. In Emerging Trends in Plant Pathology; Singh, K.P., Jahagirdar, S., Sarma, B.K., Eds.; Springer: Singapore, 2021. [Google Scholar] [CrossRef]
- Gautam, A.K.; Shashank, K. Techniques for the detection, identification, and diagnosis of agricultural pathogens and diseases. In Natural Remedies for Pest, Disease and Weed Control; Academic Press: Cambridge, MA, USA, 2020; pp. 135–142. [Google Scholar]
- Fang, Y.; Ramasamy, R. Current and prospective methods for plant disease detection. Biosensors 2015, 5, 537–561. [Google Scholar] [CrossRef][Green Version]
- Khater, M.; de la Escosura-Muñiz, A.; Merkoçi, A. Biosensors for plant pathogen detection. Biosens. Bioelectron. 2017, 93, 72–86. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Patel, R.; Mitra, B.; Vinchurkar, M.; Adami, A.; Patkar, R.; Giacomozzi, F.; Lorenzelli, L.; Baghini, M.S. A review of recent advances in plant-pathogen detection systems. Heliyon 2022, 8, e11855. [Google Scholar] [CrossRef]
- Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastHome (accessed on 18 April 2023).
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 18 April 2023).
- Morcia, C.; Tumino, G.; Gasparo, G.; Ceresoli, C.; Fattorini, C.; Ghizzoni, R.; Carnevali, P.; Terzi, V. Moving from qPCR to Chip Digital PCR Assays for Tracking of some Fusarium Species Causing Fusarium Head Blight in Cereals. Microorganisms 2020, 8, 1307. [Google Scholar] [CrossRef]
- Demeke, T.; Dobnik, D. Critical assessment of digital PCR for the detection and quantification of genetically modified organisms. Anal. Bioanal. Chem. 2018, 410, 4039–4050. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mehle, N.; Gregur, L.; Bogožalec Košir, A.; Dobnik, D. One-step reverse-transcription digital PCR for reliable quantification of different Pepino mosaic virus genotypes. Plants 2020, 9, 326. [Google Scholar] [CrossRef][Green Version]
- Dong, X.; Gao, D.; Dong, J.; Chen, W.; Li, Z.; Wang, J.; Liu, J. Mass ratio quantitative detection for kidney bean in lotus seed paste using duplex droplet digital PCR and chip digital PCR. Anal. Bioanal. Chem. 2020, 412, 1701–1707. [Google Scholar] [CrossRef] [PubMed]
- Pavšič, J.; Žel, J.; Milavec, M. Assessment of the real-time PCR and different digital PCR platforms for DNA quantification. Anal. Bioanal. Chem. 2016, 408, 107–121. [Google Scholar] [CrossRef][Green Version]
- Debski, P.; Gewartowski, K.; Bajer, S.; Garsteki, P. Calibration-free assays on standard real-time PCR devices. Sci. Rep. 2017, 7, 44854. [Google Scholar] [CrossRef][Green Version]



| Strain | Source | Isolation Procedure | Morphology | Ref. | |
|---|---|---|---|---|---|
| Isolation Media | Incubation Temperature/Time | ||||
| Cmm UCSCC | Symptomatic tomato stem | Isolation on semi-selective SCM agar; purification on YDC medium | 28 °C/7–10 days | On SCM: translucent grey, mucoid, often irregularly shaped with a variable grey to almost black center. On YDC: yellow, mucoid, confluent and convex, becoming deeper yellow with longer incubation. | [16] |
| Rs UCSCR | Symptomatic tomato stem | Isolation on semi-selective SMSA agar; purification on semi-selective SMSA agar | 28 °C/2–6 days | On SMSA: fluidal, confluent, irregular and creamy-white with pinkish center. | [15] |
| Xv UCSCV | Symptomatic tomato berries | Isolation on mTMB agar; purification on YDC agar | 28 °C/3–6 days | On mTMB: yellow, slightly mucoid, raised and circular. On YDC: pale or bright yellow, circular, mucoid and slightly raised. | [17] |
| Xap UCSCX | Bean seeds | Isolation on MT agar medium; purification on YDC agar | 28 °C/3–5 days | On MT: yellow, with two zones of hydrolysis, i.e., a large clear zone of casein hydrolysis and a smaller milky zone of Tween TM 80 lysis. On YDC: yellow and mucoid. | [18] |
| Pst UCSCP | Symptomatic tomato leaves | Isolation on KB agar medium; purification on KB agar medium | 25 °C/2–4 days | On KB: production of a pale-blue pigment fluorescent under UV light. Flat, clear and cream-colored colonies. | [19] |
| Assay Code | Probe and Primers ID | Probe and Primers Sequences | Biological Target | Target Gene | Amplicon Size | Reference |
|---|---|---|---|---|---|---|
| Cmm-dig | Cmm-digF | tctgggtgtgtctggtttcttg | Clavibacter michiganensis subsp michiganensis | 16S-23S GenBank: HM18741.1 | 61 bp | This Work |
| Cmm-digR2 | ccccaccaccatccacaa | |||||
| Cmm-Pr | FAM-cggaccctttccgtcgt-MGB | |||||
| Rs-dig | RS-I-F | gcatgccttacacatgcaagtc | Ralstonia solanacearum | 16S GenBank: OP269681.1 | 93 bp | [20] |
| RS-II-R | ggcacgttccgatgtattactca | |||||
| RS-Pr | FAM-agcttgctacctgccggcgagtg-MGB | |||||
| Tom-dig | Tom-F | gcaatatcaagagccccgtc | Solanum lycopersicum | Prosystemin GenBank: M84800.1.1 | 91 bp | [21] |
| Tom-R | ggagcgcttagcacacat | |||||
| Tom-Pr | VIC-tgcaacatccttctttcttctcgtg-MGB |
| Bacteria DNA Amount | Mean Ct Cmm-dig Assay ± dev.stnd | Mean Ct Rs-dig Assay ± dev.stnd |
|---|---|---|
| 10 ng | 12.96 ± 0.21 | 13.26 ± 0.08 |
| 1 ng | 16.05 ± 0.014 | 16.68 ± 0.12 |
| 0.1 ng | 19.4 ± 0.14 | 20.01 ± 0.18 |
| 0.01 ng | 22.82 ± 0.23 | 23.19 ± 0.052 |
| 0.001 ng | 25.76 ± 0.035 | 27.34 ± 0.011 |
| 0.0001 ng | 29.36 ± 0.26 | 30.09 ± 0.19 |
| Slope | −3.27 | −3.4 |
| Y-intercept | 17.13 | 18.25 |
| Efficiency | 102% | 96% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morcia, C.; Piazza, I.; Ghizzoni, R.; Terzi, V.; Carrara, I.; Bolli, G.; Chiusa, G. Molecular Diagnostics in Tomato: Chip Digital PCR Assays Targeted to Identify and Quantify Clavibacter michiganensis subsp. michiganensis and Ralstonia solanacearum in planta. Horticulturae 2023, 9, 553. https://doi.org/10.3390/horticulturae9050553
Morcia C, Piazza I, Ghizzoni R, Terzi V, Carrara I, Bolli G, Chiusa G. Molecular Diagnostics in Tomato: Chip Digital PCR Assays Targeted to Identify and Quantify Clavibacter michiganensis subsp. michiganensis and Ralstonia solanacearum in planta. Horticulturae. 2023; 9(5):553. https://doi.org/10.3390/horticulturae9050553
Chicago/Turabian StyleMorcia, Caterina, Isabella Piazza, Roberta Ghizzoni, Valeria Terzi, Ilaria Carrara, Giovanni Bolli, and Giorgio Chiusa. 2023. "Molecular Diagnostics in Tomato: Chip Digital PCR Assays Targeted to Identify and Quantify Clavibacter michiganensis subsp. michiganensis and Ralstonia solanacearum in planta" Horticulturae 9, no. 5: 553. https://doi.org/10.3390/horticulturae9050553
APA StyleMorcia, C., Piazza, I., Ghizzoni, R., Terzi, V., Carrara, I., Bolli, G., & Chiusa, G. (2023). Molecular Diagnostics in Tomato: Chip Digital PCR Assays Targeted to Identify and Quantify Clavibacter michiganensis subsp. michiganensis and Ralstonia solanacearum in planta. Horticulturae, 9(5), 553. https://doi.org/10.3390/horticulturae9050553








