Abstract
Pomegranate (Punica granatum L.) is regarded as one of the functional fruits because of its large amounts of secondary metabolites. The glycosylation processes mediated by UDP-glycosyltransferases (UGTs) play a decisive role in regulating secondary metabolite availability. In this study, a genome-wide search identified 145 UGT genes in pomegranate, and further phylogenetic analysis defined 17 distinct groups: A to P and R. PgUGTs were dispersed unevenly across all eight chromosomes. Duplication events analysis revealed that both segmental and tandem duplications were the main mechanisms leading to gene family expansions. The comparison of exon–intron patterns identified 53 intron-less genes. A total of 24 types of cis-acting elements related to hormone, stress, and developmental responses were predicted in the promoter regions. Expression analysis of PgUGT genes using RNA-seq data and quantitative real-time PCR (qRT-PCR) verification suggested that PgUGT genes were expressed at specific stages of fruit development, and different PgUGT members likely played different roles in specific fruit developmental stages. In an attempt to identify the UGTs involved in the glycosylation of flavonoids, 44 PgUGTs were putatively determined, and 5 well-defined orthologous groups (OGs) were characterized by the regioselectivity of these enzymes. These results provide significant insight into the UGT multi-gene family in pomegranate, and will be helpful to further elucidate their roles involved in secondary and specialized metabolism in pomegranate.
1. Introduction
Pomegranate (Punica granatum L.), native to Central Asia and the surrounding areas, is one of the members of the Lythraceae family [1]. Recent studies have proposed that pomegranate-derived secondary metabolites, especially polyphenolic compounds, are capable of exerting powerful antioxidant, anti-cancer, anti-inflammatory, antiparasitic, antihypertensive, and vascular-protective properties [2,3,4]. Consequently, pomegranate is well recognized as a ‘super fruit’ owing to its repository of bioactive substances, and thus, the plant acreages and fruit production of pomegranate have increased substantially over recent decades [5]. Given the potential applications of these bioactive compounds, it would be meaningful to comprehensively analyze the genes involved in bioactive compound biosynthesis.
Pomegranate accumulates a myriad of phenylpropanoid secondary metabolites, many of which are glycosylated in planta. Glycosylation, largely catalyzed by the glycosyltransferases (GTs), is a prominent modification reaction occurring in various biological processes. In higher plants, glycosylation is usually regarded as the last step in the biosynthesis of secondary metabolites which affects the stability, solubility, and subsequent bioavailability of these metabolites [6]. GTs are a ubiquitous group of enzymes that can transfer sugar moieties from donor molecules to a wide range of acceptor substrates [7], thus generating a broad range of structurally diverse compounds. GTs account for approximately 1–2% of the gene products in the whole genome of an organism [8]. Additionally, GTs are one of the highly divergent multigene families due to their high degree of specificity and selectivity towards substrates. Until now, 115 GT families have been identified in the CAZy database (Carbohydrate Active enzymes Database, available online http://www.cazy.org), of which the GT1 family is the largest in plants and is commonly referred to as UDP-glycosyltransferases (UGTs) [9]. The majority of plant UGTs use UDP-glucose in the transfer reaction, while UDP-rhamnose, UDP-xylose, and UDP-galactose also exist [6]. Plant UGT sequences are characterized by a unique and conserved Plant Secondary Product Glycosyltransferase (PSPG) motif, a 44-amino-acid fragment at the C-terminal end of the protein [7,10], which is considered to be involved in the glycosylation of plant secondary metabolites or other natural products [11].
By taking advantage of the current wealth of omics-based resources, the comprehensive analyses of UGT families have been carried out, covering more than 40 plant species, and a large number of UGT family members have been found. In pomegranate, despite the fact that a wide range of secondary metabolites have been reported, including those glycosylated forms [12,13,14,15], only a few UGT genes have been identified and functionally characterized to be involved in secondary metabolite regulation. For instance, UGT95B2 preferentially glycosylates flavones/flavonols at more than one position in the molecule [16]. UGT84A23 and UGT84A24 exhibited β-glucogallin-forming activities [17]. UGT72BD1 used gallic acid as a substrate and produced a regiospecific product: gallic acid 4-O-glucoside [18]. Overall, the number of functionally characterized UGTs is still relatively low given the large abundance of UGTs in the genome of P. granatum L. Hence, in order to understand the biosynthesis pathway of secondary metabolites, it is necessary to systematically identify the UGT multigene family at the whole-genome level. The availability of the chromosomal-level genome of pomegranate presents an opportunity to explore the properties of UGT family genes in this versatile horticultural crop. In this study, we present a genome-wide analysis of UGT genes in pomegranate (PgUGT) based on the ‘Tunisia’ pomegranate genome data. All candidate PgUGT genes in the pomegranate genome were fully screened and a phylogenetic tree was constructed. Next, gene structural characteristics, including chromosome location, exon–intron structures, conserved motifs and cis-acting elements were analyzed. RNA-seq was carried out to investigate the expression profiles of PgUGT genes during fruit development phases. Finally, the phylogenetic trees were reconstructed and combined with those function-known UGTs from other plant species to explore UGT members acting towards flavonoid substrates. These results will contribute to future research elucidating the functions of UGTs in pomegranate.
2. Materials and Methods
2.1. Genome-Wide Identification of UGT Genes and Basic Physicochemical Properties of Proteins
The Hidden Markov Model (HMM) profile of the UDPGT domain (PF00201) was downloaded from Pfam website (http://pfam.xfam.org/, accessed on 6 July 2022). Then, the HMM model was built using the HMMER v3.0 software package (https://www.ebi.ac.uk/Tools/hmmer/, accessed on 7 July 2022) and was also searched against the pomegranate protein database (ASM765513V2) with E-values less than 1e−5. The candidate protein sequences of each UGT were further verified through the PFAM (http://pfam.xfam.org/, accessed on 8 July 2022), SMART (http://smart.embl-heidelberg.de/, accessed on 8 July 2022), and InterProScan (http://www.ebi.ac.uk/interpro/search/sequence/, accessed on 9 July 2022) databases to confirm the presence of the UDP-glycosyltransferase domain in order to remove the redundant sequences and isoforms.
Various physical and chemical parameters, including amino acid length (aa), molecular weight (MW), isoelectric point (pI), and instability index of all UGT proteins, were obtained using the online ExPASy program (http://web.expasy.org/protgaram, accessed on 11 July 2022) [19]. Transcript count for each PgUGT was obtained from the genome dataset. The subcellular localization of each PgUGT protein was predicted using the online CELLO V2.5 (http://cello.life.nctu.edu.tw, accessed on 12 July 2022).
2.2. Sequence Alignment and Phylogenetic Analysis
In order to classify PgUGTs based on a phylogenic tree, the selected amino acid sequences of PgUGTs were aligned with that from Arabidopsis thaliana, Zea mays, Oryza sativa, and Camellia sinensis (Table S1) by MUSCLE in MEGA 11 (The Pennsylvania State University, University Park, PA, USA, 1993) with default settings [20]. A neighbor-joining (NJ) tree was constructed based on the JTT model and partial deletion using a 1000 bootstrap value. The phylogenetic tree was visualized and optimized by the online software Interactive Tree of Life program iTOL (https://itol.embl.de/, accessed on 15 July 2022) [21]. Grouping of PgUGTs was inferred by their phylogenetic position to the reference UGTs. To compare the phylogenetic groups of the UGTs in pomegranate and other species, we collected and summarized published records on the UGT families from other plant species.
2.3. Chromosomal Distribution of PgUGTs and Gene Duplications
The physical location of each PgUGT on the chromosome was retrieved based on pomegranate gene annotation files (GFF3 format) and visualized by MapChart 2.20 (Wageningen University & Research, Wageningen, The Netherlands, 2002) [22]. Segmental duplication events of the PgUGTs in the pomegranate genome were analyzed using TBtools software (South China Agricultural University, Guangzhou, Guangdong, China, 2020) [23]. The tandem duplications were defined according to the criteria published by Holub [24] and Gu et al. [25].
2.4. Exon–Intron Structures and Conserved Motifs
According to the general feature format file of P. granatum, the exon–intron structures of the PgUGTs were obtained and graphed with Tbtools software. The conserved motifs of the putative PgUGT proteins were predicted by using the on-line MEME procedure (http://meme-suite.org/, accessed on 2 August 2022) with maximum 15 motifs per sequence, and annotated with InterPro database (https://www.ebi.ac.uk/interpro/, accessed on 4 August 2022). An image combined NJ phylogenetic tree, conserved motifs and gene exon–intron structure was drawn.
2.5. Promoter Cis-Acting Element Analysis
The 2000-bp sequences upstream of the start codons of PgUGT genes were extracted from pomegranate genome sequence by TBtools software. The presence of cis-acting elements was predicted by PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 8 August 2022), and the results were visualized using TBtools.
2.6. Gene Expression Analysis Using RNA-seq
The fruits of four different developmental stages (10 June, 10 July, 10 August, and 10 September, designated as S1~S4, respectively) of the pomegranate ‘Hongbaoshi’ were used for transcriptomic analysis. For this, total RNA was firstly extracted according to the methods described by Yuan et al. [1], and the quality was assessed by electrophoresis and A260/A280. RNA-seq libraries were conducted and then sequenced on the Illumina HiSeq 2000 platform (5200 Illumina Way, San Diego, CA, USA, 2010). Three biological replicates for fruit development stages were prepared. The values of fragments per kilobase of per million mapped reads (FPKM) were used to calculate and evaluate transcript abundance. Genes with FPKM values <1.0 were defined to be minimally expressed and were removed from the data set. The FPKM values of PgUGTs were normalized with Log2, and heatmaps visualized by TBTools were presented to display the expression level of each PgUGT.
2.7. RNA Isolation, Reverse Transcription and Quantitative Real-Time PCR (qRT-PCR) Analysis
To validate the expression pattern of the selected genes, total RNA was extracted from fruit peels at the S1~S4 stages, respectively, by using RNA isolation system (Tiangen, Beijing, China), according to the manufacturer’s instructions. The first-strand cDNAs were synthesized from the total RNA by using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA). qRT-PCR was performed with SuperReal PreMix Plus kit (Tiangen, Beijing, China) using the SYBR Green to detect gene expression. The primers used for qRT-PCR were listed in Table S2. The conditions for PCR conditions were performed following the previous reports [26]. The pomegranate PgActin (GenBank accession No. GU376750) was selected as an internal reference gene. Data from the individual runs were collated using the 2−ΔΔCT method [27]. All the reactions were performed using at least three replicates.
2.8. Functional Prediction of PgUGT Involved in Flavonoid Biosynthesis
In order to identify PgUGTs which participated in flavonoid biosynthetic pathway, 58 UGT proteins with known functions from other plant species were retrieved from Uniprot or NCBI database (Table S3). We constructed a NJ phylogenetic tree based on a collection of screened PgUGTs and 58 flavonoid UGT proteins to pre-screen flavonoid PgUGTs with parameters as described above in MEGA 11. The clustered PgUGTs were further filtered by removing those that do not use flavonoids as substrates when their functions were compared with annotation of P. granatum and A. thaliana. Subsequently, a phylogenetic tree based on the candidate flavonoid PgUGTs was reconstructed, and the filtered flavonoid UGTs were classified according to the method proposed by Yonekura-Sakakibara et al. [28].
3. Results
3.1. Identification of UGTs in Pomegranate
The comprehensive sequencing of the pomegranate genome greatly facilitated the identification of multi-gene families. In total, 180 candidate PgUGT genes were initially identified by HMM search. By subsequent verification of UDP-glycosyltransferase domain and removal of redundant sequences, a total of 145 putative UGT genes were screened and used for further analysis (Table S4). Most of the genes encoded proteins with the length in range of 400 to 500 amino acids, while only 24 were above 500 and 8 below 300 amino acids in size. Transcript counts for each gene were in a range of 1 to 4. The MW and pI ranged from 17.3 kDa to 60.9 kDa (average MW = 51.11 kDa) and from 4.62 to 9.89 (average pI = 5.82), respectively (Table S4). There were only 28 proteins with an instability index lower than 40, indicating that these UGT proteins were unstable. The predicted subcellular localization showed that 51, 28, 9, 2, and 2 of the encoded proteins were preferentially localized into cytoplasm, chloroplast, plasma membrane, mitochondria, and nucleus, respectively, while the rest were localized into any of these compartments (Table S4), revealing the various sub-cellular location of UGTs.
3.2. Phylogenetic Analysis of UGTs in Pomegranate
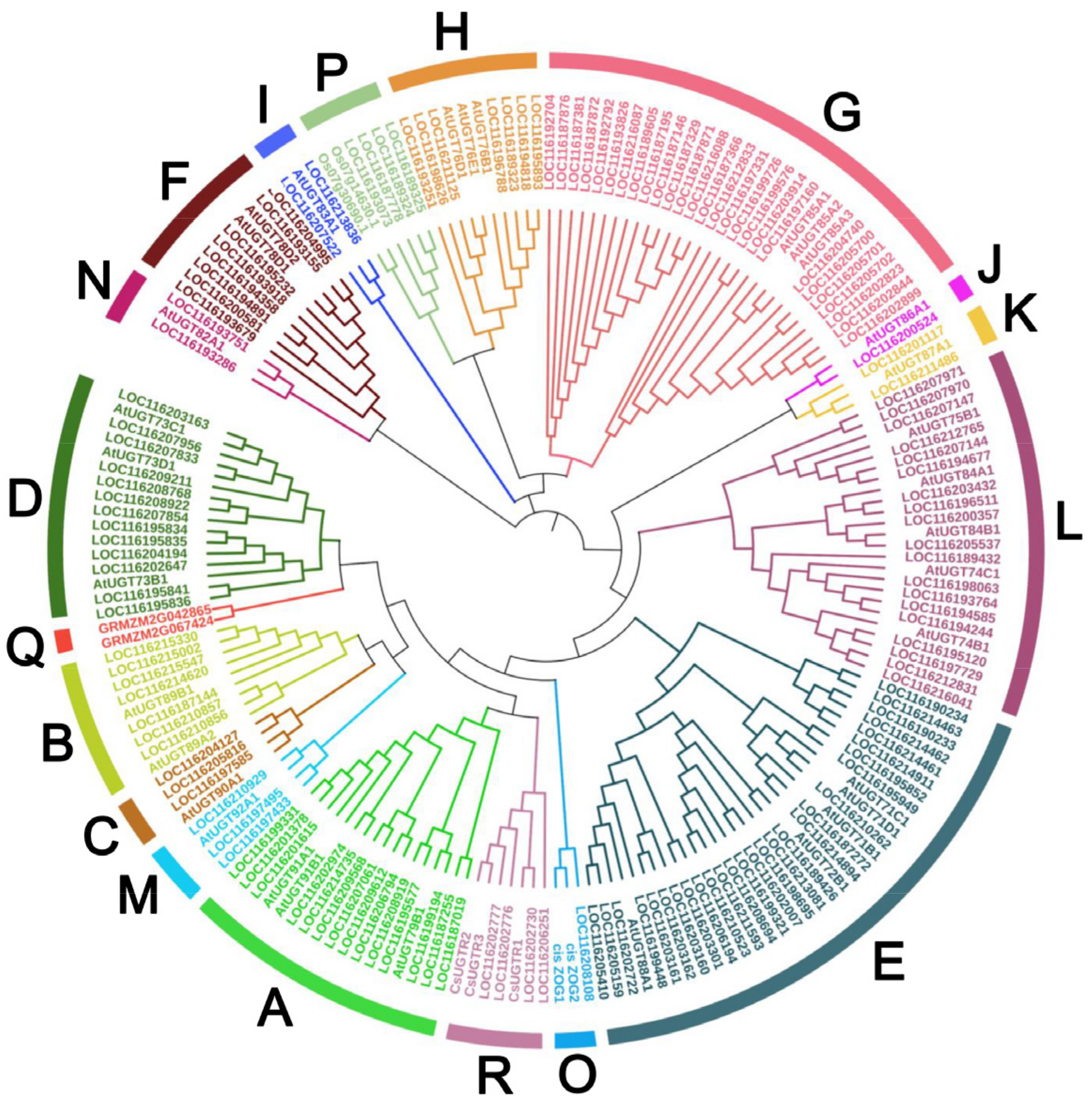
Accumulating phylogenetic analyses have demonstrated that plants UGTs could form 14 to 18 distinct groups depending on different plant species. In this study, we aligned all candidate 145 PgUGTs with 41 reference UGTs, including 32 from Arabidopsis thaliana, 4 from Zea mays, 3 from Camellia sinensis, and 2 from Oryza sativa to generate a phylogenetic tree for classification of PgUGTs. From Figure 1, we can see that 145 PgUGTs were clearly divided into 17 major phylogenetic groups, including 14 conserved that were defined as A-N according to the incorporated Arabidopsis UGTs in each group, and 3 additional new groups (O, P, R). Group Q did not contain any members of pomegranate.
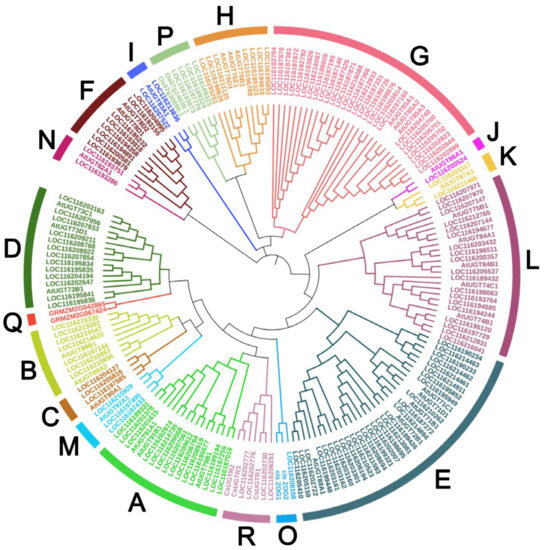
Figure 1.
Phylogenetic analysis of UGT family genes in P. granatum. The phylogenetic tree was constructed by using the full-length sequences of 145 PgUGTs and 41 UGTs from Arabidopsis thaliana, Zea mays, Camellia sinensis, and Oryza sativa.
Next, we compared the number of PgUGTs in each phylogenetic group with that of other species available (Table 1). The results showed that the total number of PgUGTs was higher than that in white pear (Pyrus bretschneideri, 139); lower than apple (Malus × domestica, 237/241), Chinese bayberry (Morella rubra, 152), grape (Vitis vinifera, 228/181), and peach (Prunus persica, 168); and equal to pomelo (Citrus grandis). The members of the PgUGT family were unevenly distributed in groups A-P and R (Figure 1, Table 1). The most PgUGTs was observed in group E (28), followed by G (26), L (19), A (14), and D (13). There were 1, 4, and 4 members in the three new groups O, P, and R, respectively.

Table 1.
Statistics of the number of UGTs in each phylogenetic groups from different plant species.
3.3. Chromosome Distribution and Gene Duplication
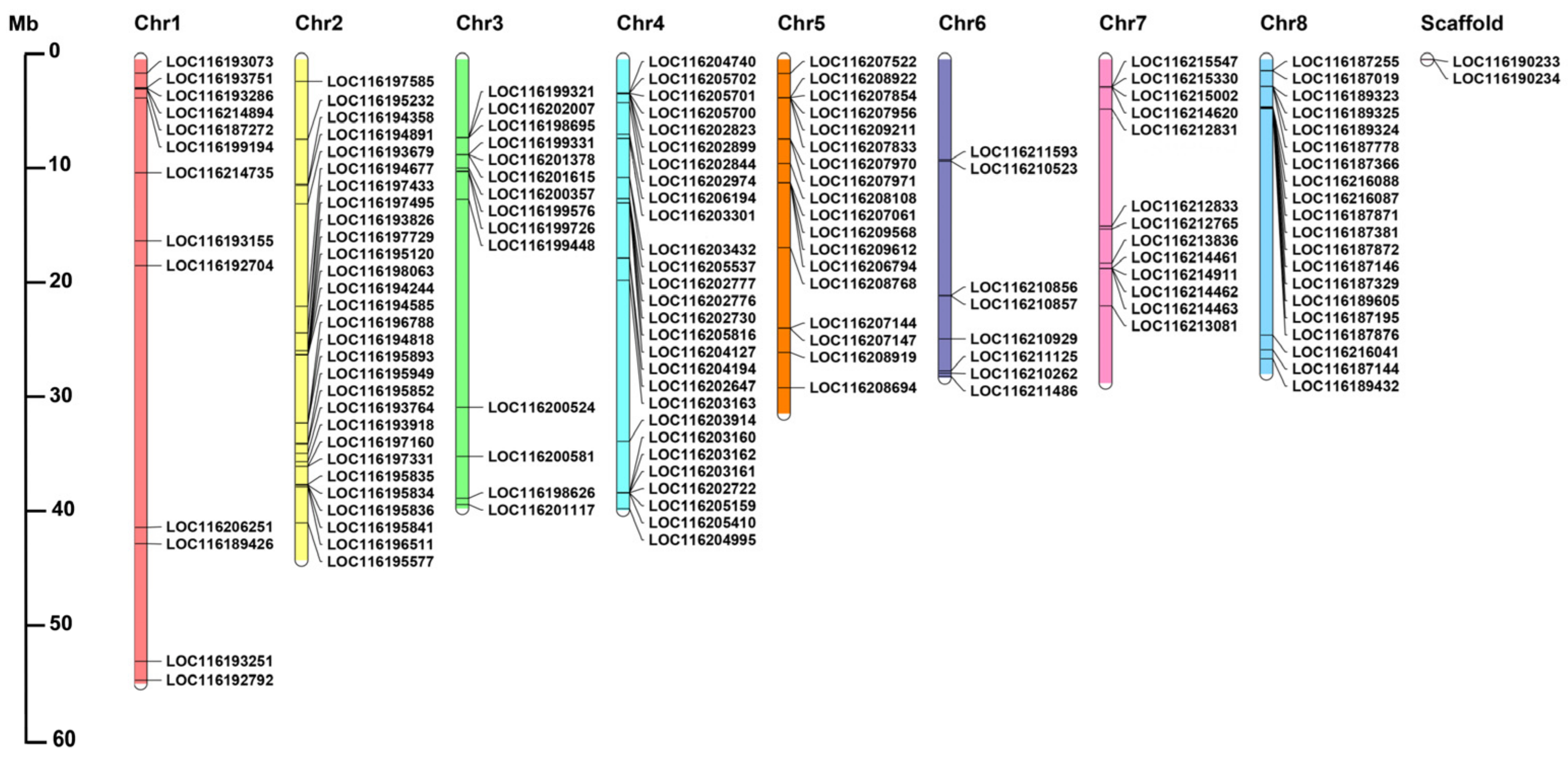
To summarize the genomic distribution of the PgUGT genes, the genetic mapping of UGTs on chromosomes was further investigated (Table S4, Figure 2). According to the genome annotation information retrieved from pomegranate genomic database, 143 PgUGTs were mapped on 8 specific chromosomes, and the remaining 2 PgUGTs, including LOC116190233 and LOC116190234, could not be found on any specific pomegranate chromosome and were set on scaffolds. Chromosome 2 had the maximum number of 29 PgUGTs, whereas chromosome 6 contained the minimum 8 PgUGTs.
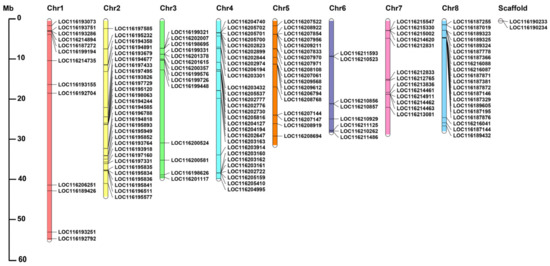
Figure 2.
Chromosome distribution of PgUGT genes. The chromosome numbers are shown at the top of each chromosome.
The chromosome distribution of genes belonging to different groups is presented in Figure S1. We found that PgUGTs are distributed on chromosomes in clusters with sizes ranging from 2 to 6 genes per cluster, with a maximum 11 genes of the G group on chromosome 8.
To further understand how PgUGT genes were evolved, gene duplication events were further investigated in the pomegranate genome. Eventually, 23 segmental duplication events involving 32 genes were identified (Figure S2). In addition, 30 gene pairs were considered to originate from tandem duplication events (Table S5), and they were unequally distributed on all chromosomes except chromosome 1. These results suggested that both segmental and tandem duplication might play important roles for the generation of clusters of duplicated genes and for the expansion of the PgUGT family.
3.4. Conserved Motifs and Exon–Intron Organization of PgUGTs
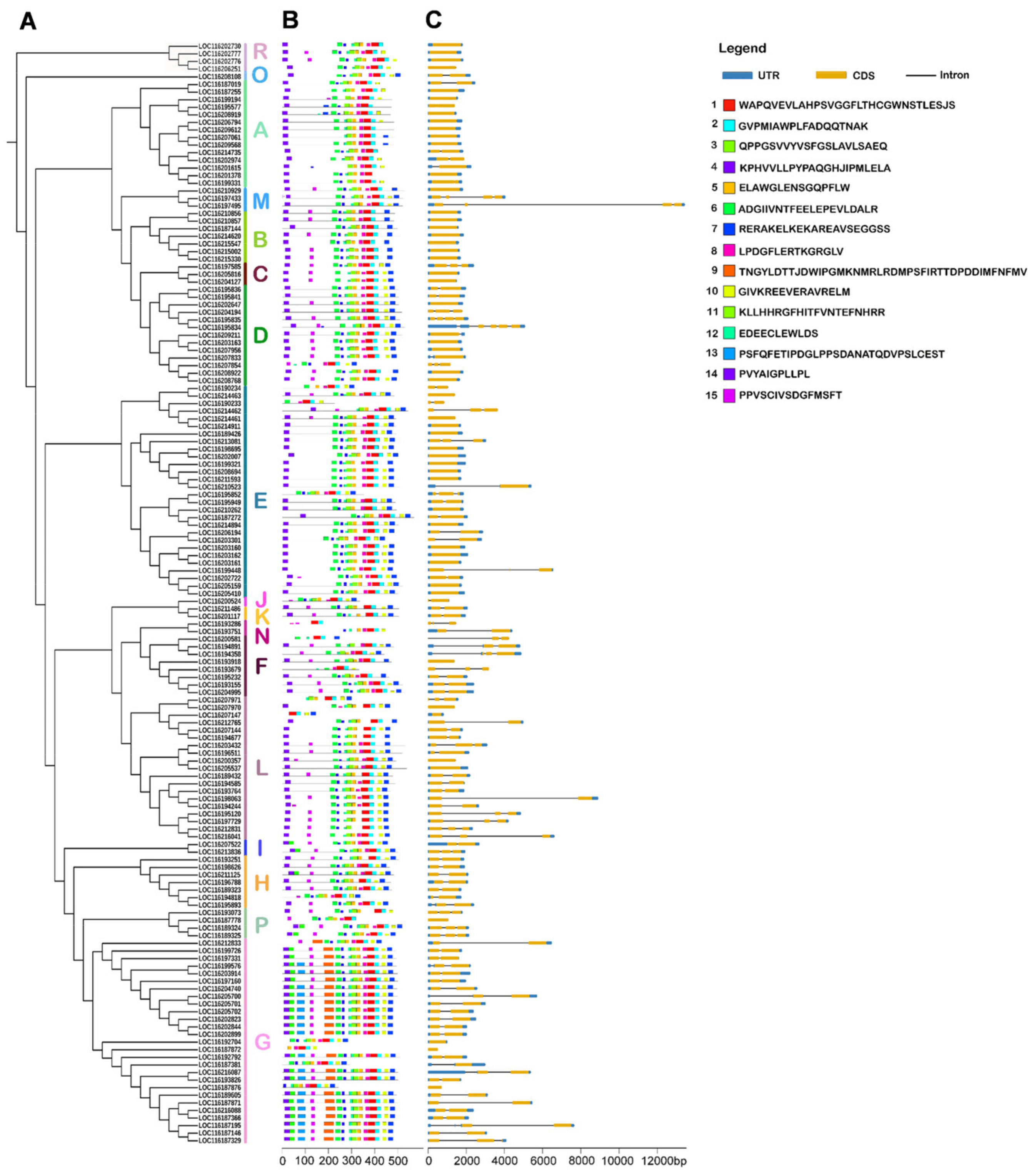
To elucidate the structural features of PgUGT genes, the gene exon/intron structures and the protein motif structures were analyzed (Figure 3). In total, 15 MEME-predicted motifs were identified and subsequently annotated with the InterPro database. Motifs 1 and 2 were referred to the UDP-glucoronosyl and UDP-glucosyltransferase (UDPGT) domain, which are typical conserved domains found in all plant UGT proteins. The results showed that all PgUGTs contained motifs 1 and 2, indicating that the identification of PgUGT family members was reliable.
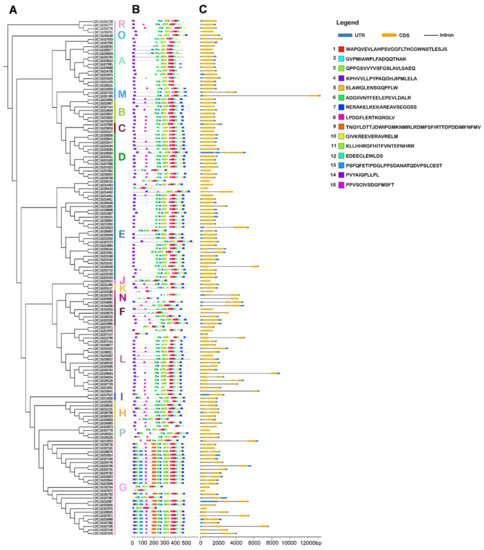
Figure 3.
Conserved motifs and gene structure among PgUGTs. (A) The phylogenetic tree of 145 PgUGT proteins was constructed using the NJ method; (B) Conserved motifs in PgUGT proteins; (C) Exon–intron organization of PgUGT genes.
The number of motifs ranged from 4 to 15 in PgUGT sequences, and 19 PgUGT members contained all 15 motifs. In most PgUGT sequences, motif 4 was located at the N-terminal of the UGT sequence, and motif 7 was positioned in the C-terminal of the sequences. The results also demonstrated that the distribution of some motifs displayed group-specificity. For example, motif 13 was mainly present at the N-terminal of members in group G, while members of other groups did not contain motif 13. Similarly, motif 10 existed in group members at the C-terminal exclusive to B group. Motif 15 was not found in A, E, and O group members, but existed in most members of the D, F, H, G, and L groups. These differences in the distribution of these motifs might be related to the function differentiation of each group member. In general, the evolutionary relationship among PgUGT members was consistent with the types and locations of conserved motifs.
The characteristics of gene structure are an important basis for the analysis of the evolution and phylogeny of gene families. Exon–intron structure analysis indicated that the number of introns of 92 PgUTGs genes varied from 1 to 5, with 53 members lacking introns (Figure 3, Table S6). Out of the intron-containing PgUGTs, 58, 27, and 6 members had 1, 2, and 3 introns, respectively. Additionally, only one gene, LOC116195834, contained 5 introns. The characterization by fewer introns revealed a high conservation of gene structure in PgUGT gene family members.
In phylogenetic groups, the largest number of genes lacking introns was observed for group E, with 16 members, followed by 8 in A and 7 in B group. A total of 16 PgUGTs in group G contained 1 intron, followed by 9 in group L (Table S6). All members of the B and R groups had no introns. Generally, members within each group exhibited similar exon–intron organization style, which was consistent with the results obtained in the conserved motif structure. These results suggested that the PgUGT family members within groups were relatively conserved and diverged greatly among different groups.
3.5. Characterization of Cis-Acting Elements in the PgUGT Promoters
To understand the transcriptional regulation of PgUGT genes, the upstream promoter regions (2.0 kb in size) were used to predict potential cis-acting elements using the PlantCARE database. A total of 24 types of cis-acting elements were observed in this study (Table 2, Figure S3). These elements were randomly distributed in the promoter regions of PgUGTs and were predicted to participate in hormone responses, stress responses, and developmental responses.

Table 2.
Characteristics of cis-acting regulatory elements presented in the promoter regions of PgUGT genes.
Among the cis-acting elements belonging to the hormone responses, abscisic acid-, auxin-, gibberellin-, MeJA-, and salicylic acid-responsive elements were observed in the promoters of PgUGTs, respectively. Stress-related response elements contained ARE, GC-motif, LTR, MBS, TC-rich repeats, and WUN-motif, which were involved in anaerobic induction, anoxic specific inducibility, low-temperature responsiveness, drought inducibility, defense and stress responsiveness, and wound responsiveness, respectively. This suggest that the expression of those genes containing the elements in promoters might be regulated by ambient pressure. Plant developmental elements contain cis-acting regulatory elements related to meristem expression (CAT-box), circadian control (circadian), endosperm expression (GCN4-motif), palisade mesophyll cells differentiation (HD-Zip 1), zein metabolism regulation (O2-site), flavonoid biosynthetic regulation (MYSI), seed-specific regulation (RY-element), and cell-cycle regulation (MSA), indicating that the PgUGT genes play vital roles in regulating physiological processes.
The number of cis-acting elements in genes was uneven (Table 2, Table S7). In particular, the most common motif was the ABRE elements associated with abscisic acid responsiveness, accounting for 30% of the hormone-responsive motifs, followed by the CGTCA motif, which was related to MeJA and accounted for 24% of the hormone-responsive motifs. Furthermore, there were 11 MYSI and 81 MBS belonging to MYB recognition and binding elements which were found in 10 and 63 PgUGT genes, revealing that the expression of these genes may be regulated by MYB transcription factors. Additionally, there were 92 and 13 PgUGTs containing LTR and WUN cis-acting elements, indicating that their transcription may be activated by low temperature and wound, respectively. Thus, the various cis-acting elements in the gene promoter region suggested that PgUGT genes play crucial roles in the complex hormone regulatory network and participate in diverse stress responses and secondary metabolite biosynthesis.
3.6. Temporal Expression Profiles of PgUGTs during Fruit Development
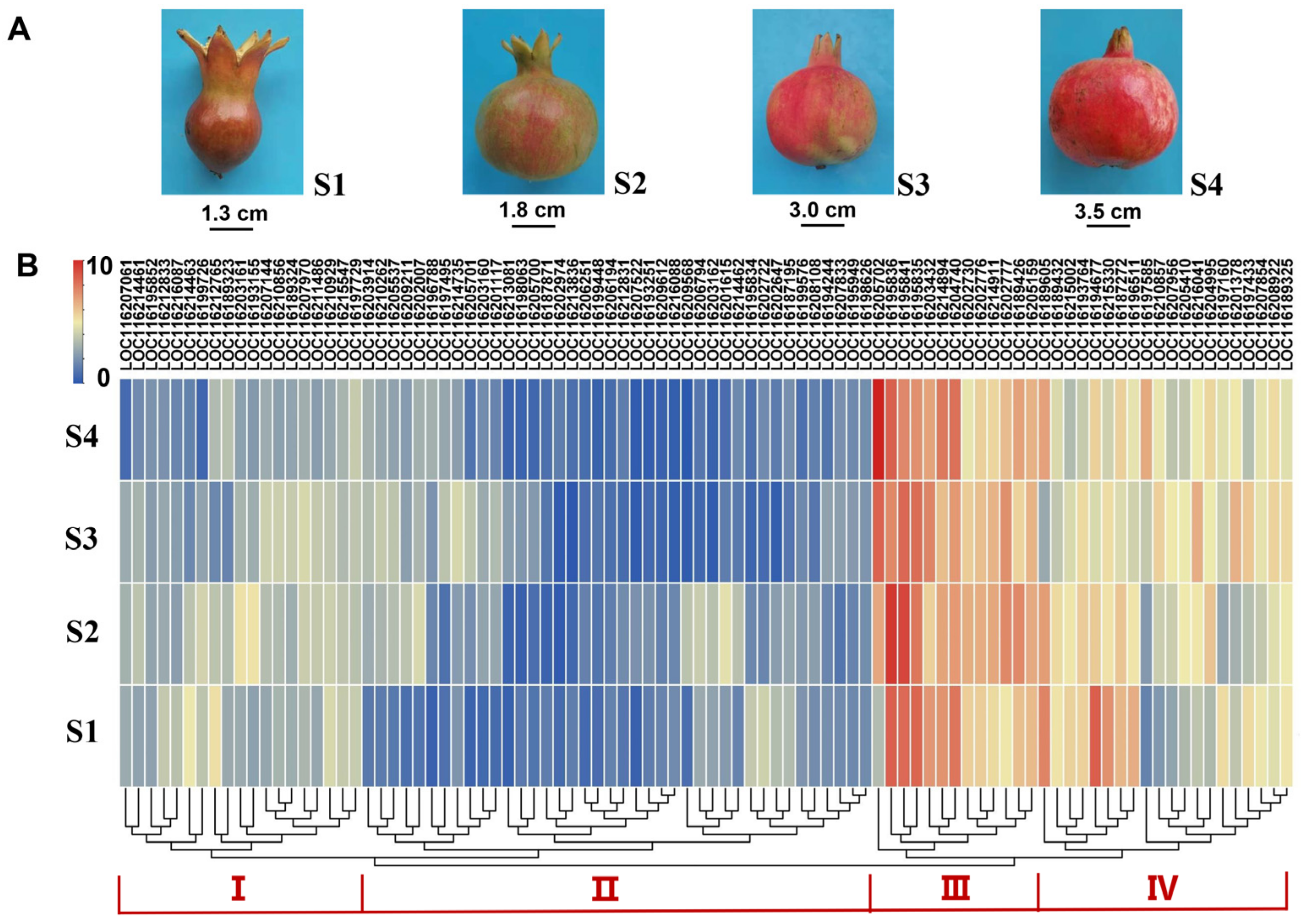
After filtering the low RNA-seq data, a total of 92 PgUGTs were further used to characterize the expression profiles in four fruit developmental stages. Next, the FPKM values were row-scaled, yielding normalized expression values for analysis. A hierarchical clustering analysis of their transcript levels indicated that these PgUGT genes could be divided into four expression patterns (Figure 4). For instance, the gene expression of cluster I and II exhibited low to medium levels, while cluster III and IV displayed a much higher level across fruit development. The 13 genes in cluster III maintained the highest level in four stages. Genes in Cluster IV were detected to display a fluctuated transcriptional abundance in S1~S4. These results suggested that the PgUGT genes in cluster III and IV may perform more vital glycosylation functions during pomegranate fruit development. Additionally, we found that the highly expressed UGTs were centered in groups D, E, G, L, and R (Figure S4).
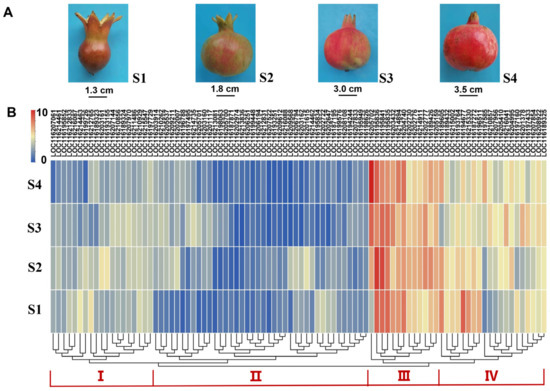
Figure 4.
Expression profiles of PgUGTs across fruit developmental stages. (A) The fruits in four developmental stages. (B) The expression levels of PgUGTs. The scale represents the signal intensity of FPKM values. Red indicates a higher expression level, while blue indicates a lower expression level.
3.7. The Validation of PgUGT Expression with qRT-PCR
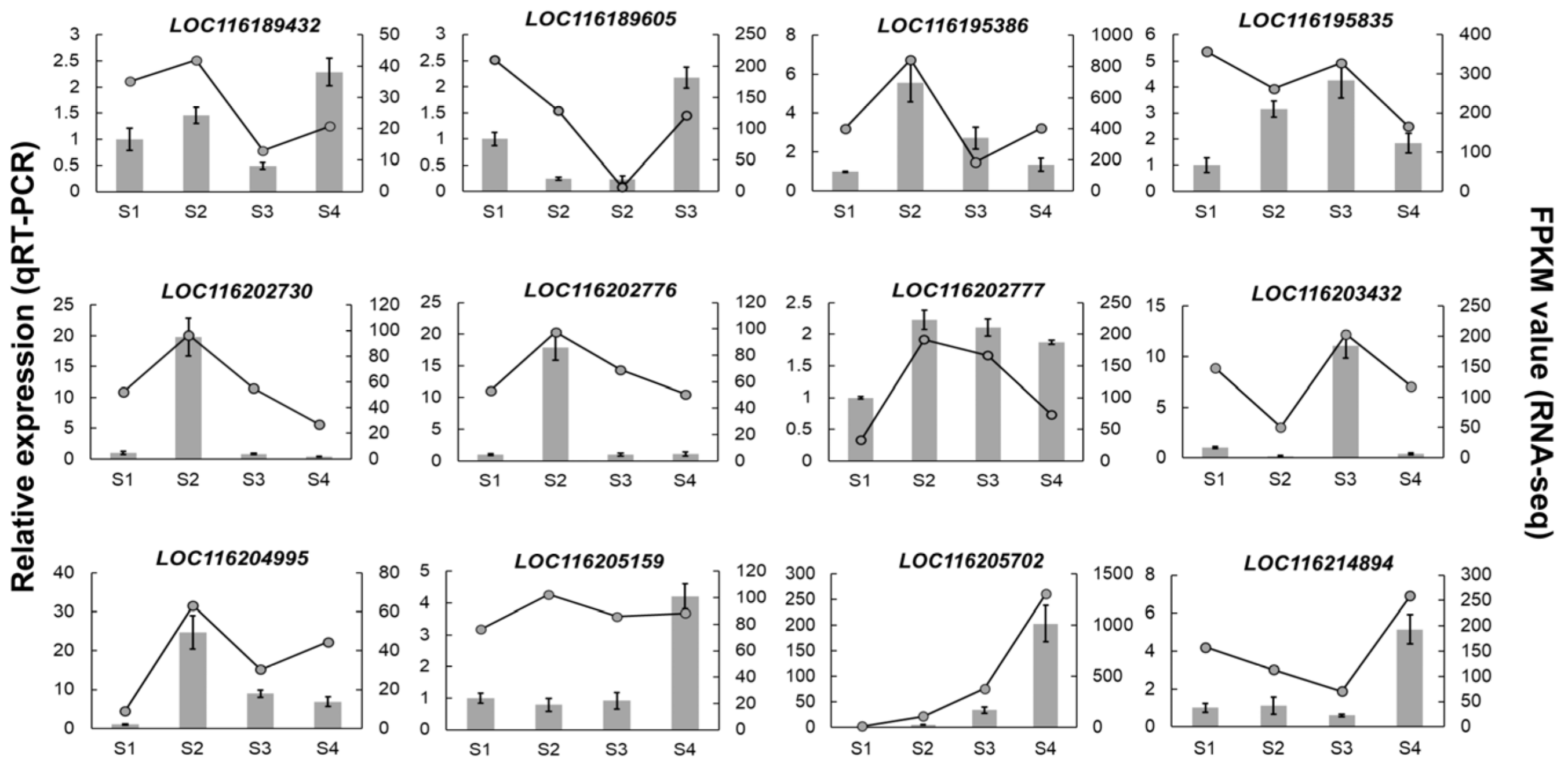
To confirm the reliability of the RNA-seq results, 12 representative PgUGT genes were selected and assayed using qRT-PCR (Figure 5). Most genes showed similar expression patterns with the FPKM values obtained by RNA-seq. The results of qRT-PCR in samples from S1 to S4 supported the reliability of the transcriptomic analysis described above.
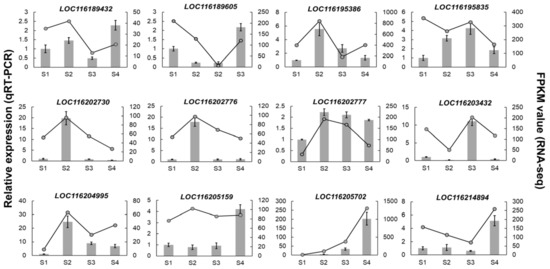
Figure 5.
Validation of the expression of PgUGTs by qRT-PCR. Gray columns represent the expression levels tested by qRT-PCR; error bars indicate the standard deviation of qRT-PCR data (n = 3). For RNA-seq, each point is the mean of three biological replicates.
It can be seen that the selected PgUGTs exhibited different expression patterns during fruit development. Seven genes, including LOC116189432, LOC116195386, LOC116202730, LOC116202776, LOC116202777, LOC116204995, and LOC116205159, were highly expressed in the S2 stage, while there were two (LOC116189605, LOC116195835), one (LOC116203432), and two (LOC116205702, LOC116214894) genes which reached transcription summit in S1, S3, and S4, respectively. These temporal gene expression patterns demonstrated that PgUGTs were more active in the S2 stage during fruit development.
3.8. Functional Prediction of PgUGT Involved in Flavonoid Biosynthesis
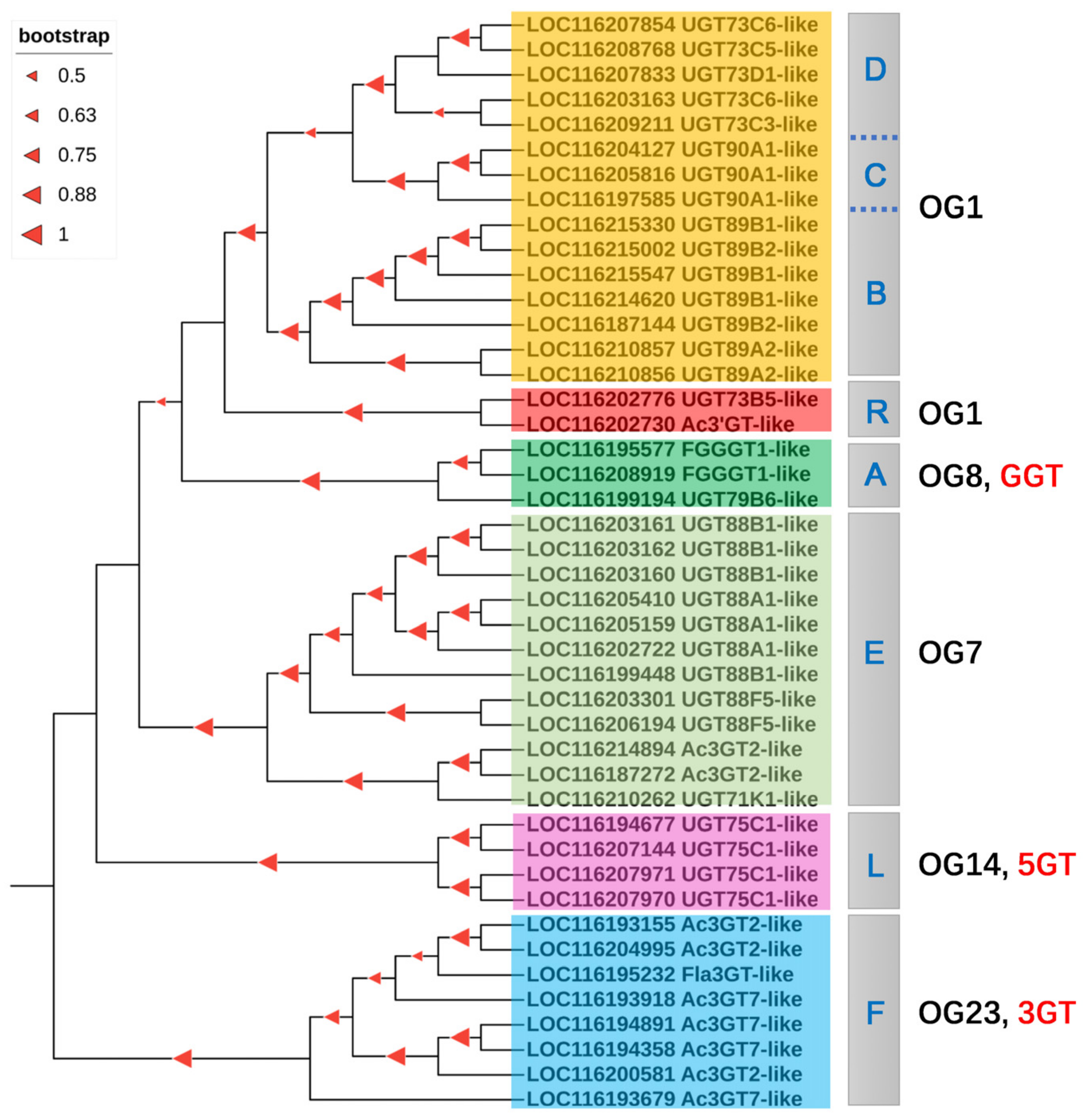
To determine putative UGTs for glycosylation reaction of flavonoids in pomegranate, we generated an unrooted NJ phylogenetic tree based on the full-length proteins of PgUGTs and 58 UGTs which use mainly flavonoids as substrate acceptors. The preliminary results showed that 80 PgUGTs appeared to be involved in glycosylation of flavonoids by inferring their close phylogenetic relationships with known flavonoid UGT proteins in plants (Figure S5). The PgUGTs with flavonoid specificity fell into nearly all clustering groups, except several minor groups, such as the J, K, M, N, and O groups. By subsequent functional screening, a total of putative 44 flavonoid PgUGTs were further used to reconstruct a NJ tree (Figure 6). The candidate UGTs for flavonoid biosynthesis were those distributed in A, B, C, D, E, F, L, and R groups. When these flavonoid UGTs were classified according to their regiospecificity, it can be seen that orthologous group 1 (OG1) occupied four PgUGT groups: B, C, D, and R. The other OGs each contained a single UGT group. Additionally, the high bootstrap values (>50%) on most nodes demonstrated the good reliability of the phylogenetic tree.
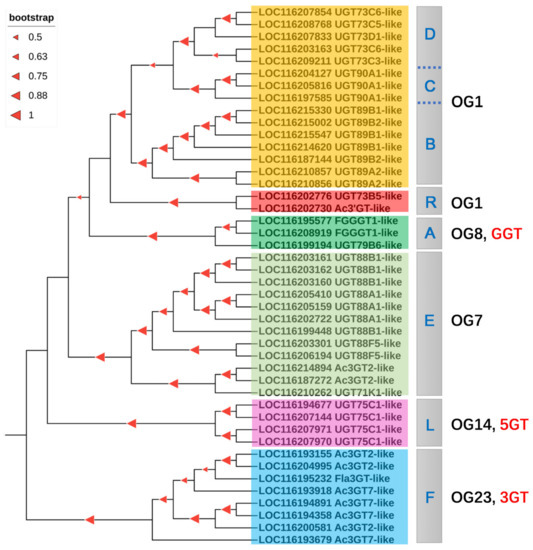
Figure 6.
Phylogenetic tree of the flavonoid UGTs. A, B, C, D, E, F, L, and R are PgUGT groups as described in 3.2; OG, orthologous group; 3GT, 3-O-glycosyltransferase; 5GT, 5-O-glycosyltransferase; GGT, glycoside glycosyltransferase.
4. Discussion
Plant UGT is a large and functionally diverse family which plays an important role in the diversification of plant secondary metabolites. PgUGTs in ‘Dabenzi’ pomegranate have been recently reported [50]; however, the characterization of the PgUGT family in ‘Tunisia’, a noted soft-seeded cultivar, remains undefined. To deepen our understanding about the UGT family in pomegranate, we performed a genome-wide analysis on PgUGTs. Furthermore, the UGTs involved in flavonoid biosynthesis were identified and classified based on phylogenetic analysis, providing valuable information for further investigations into the catalytic functions of pomegranate UGTs.
4.1. The Classification of PgUGTs Based on Phylogenetic Tree
It has been reported that the ratio of UGT genes to total genes in vascular plants was in the range of 0.18% to 0.72%. In the present study, a total of 145 UGT genes were identified in the pomegranate ‘Tunisia’, accounting for approximately 0.4% of the total number of genes in the whole genome [55], lower than 0.5% in pomelo [34] and 0.6% in peach [49], but higher than maize (0.23%) [54] and soybean (0.26%) [30]. Recently, 120 UGT genes were screened in the ‘Damenzi’ pomegranate [50]. A possible reason for this discrepancy derived from the genome data of the two different cultivars ‘Tunisia’ and ‘Dabenzi’.
Previous studies divided UGTs into A-N [56], O and P [7], Q [54], and R [33] groups successively. Based on phylogenetic analysis, we identified a total of 17 groups, including 14 groups (A-N) highly conserved in plants and 3 newly discovered ones: O, P, and R. However, this result was not consistent with that reported by Li et al. [50], who classified PgUGTs into 15 groups, lacking members in the K, O, and Q groups (Table 1). It was previously thought that group Q was specific to monocots, such as maize [54] and wheat [52]; however, the occurrence of Q members in Malus × domestica [41], Epimedium pubescens [35], and Triticum aestivum [52] suggested that group Q was not exclusive to monocots. Only one UGT in group R was found in Camellia sinensis [33] and other four species (Lotus japonicus, Medicago truncatula, Phaseolus vulgaris, and Trifolium pratense) [38], while there were 4, 4, and 6 UGT members belonging to the R group in P. granatum, Malus × domestica [41], and Gossypium raimondii [37], respectively, suggesting that group R may make an important contribution to the glycosylation of specific metabolites. Quantitatively, PgUGTs were concentrated in groups A, D, E, G, and L, indicating that members of these five groups expanded more rapidly than any other groups during plant evolution, as described by Caputi et al. [7]. In addition, group E contained the highest number of UGT members, consistent with views that group E expanded more than any other phylogenetic groups [7].
4.2. Segmental and Tandem Duplication Contribute to the Expansion of PgUGT Family
Gene duplication plays an important role in the occurrence of new gene functions and gene amplification. In plants, genomic duplications mainly arise from whole-genome duplication (WGD), segmental duplication, or tandem duplication [57]. Yuan et al. [1] reported that pomegranate underwent a paleotetraploidy event, resulting in at least two whole-genome duplications in the P. granatum genome. In this study, we identified segmental and tandem duplications between 23 and 30 PgUGT gene pairs, respectively, revealing that both tandem and segmental duplication events were the main driving forces for PgUGT expansion, which corresponded with previous work on the ‘Dabenzi’ pomegranate [50]. This phenomenon was also consistent with previous findings in A. thaliana [58], Vitis vinifera [53], and Epimedium pubescens [35]. However, other reports showed opposing observations that tandem duplication, rather than segmental duplication, was the major cause of UGT gene expansion [31,35,46]. In soybean, tandem duplication was not observed, and a series of segmental duplications caused UGT evolution [30]. In pear, segmental duplication was the dominate gene duplication event [51]. Together, these results demonstrated that the expansion of the UGT family driven by duplication events was species-specific.
4.3. PgUGT Transcription Analysis
Transcriptomic sequencing data can provide powerful complementary information to genomic analysis, guiding subsequent screening novel candidate genes for glycosylated secondary metabolite biosynthesis. The analysis results highlighted differential expression patterns, such as 33 PgUGTs (35.9%) showing high expression levels while 59 genes (64.1%) showed low transcriptional levels. This result was inconsistent with the expression data of Cier arietinum, which showed 87.5% high-expression CaUGTs and 12.5% low-expression genes in developmental stages, indicating that the expression profiles were often species-, development-, and tissue-specific.
The highly expressed PgUGTs were centered in groups D, E, G, L, and R. It has been reported that members of these groups are involved in the glycosylation of a variety of polyphenols [7,56]. Therefore, the occurrence and high transcriptional levels of UGT members in D, E, G, and L groups may be associated with the diversity of phenolic compounds.
4.4. Identification of PgUGTs Involved in Flavonoid Biosynthesis
Flavonoids are abundant in vegetables, fruits, grains, and tea, and are known to have powerful antioxidant activity and provide a broad spectrum of health benefits [59]. In pomegranate, an increasing number of studies have described a large amount of flavonoids in fruits, flowers, leaves, seeds, and barks [60]. In plants, flavonoids are mostly glycosylated by UGTs with one or more sugar groups, leading to the diversity of flavonoids. Therefore, it is essential to identify UGT members with flavonoid substrate preferences.
In this study, a phylogenetic tree, constructed using stepwise-screened UGT sequences, identified 44 PgUGTs highly probably involved in flavonoid biosynthesis. Previously, the strategy of UGT specificity prediction has been used in Vitis vinifera [53], Cicer arietinum [61], Epimedium pubescens [35], and Citrus sinensis [62] by phylogenetic analysis with known UGT functions. According to the sugar acceptors and glycosidic linkages of characterized UGTs, Wilson et al. [63] placed UGTs with flavonoid acceptors into 11 groups, including A, B, C, D, E, F, G, H, L, Q, and R, while the A, B, D, E, F, and L groups were the most concentrated [7,33,35]. Our analysis confirmed the same distribution pattern of flavonoid PgUGTs among phylogenetic groups, suggesting the diversity and prosperity of glycosylation for flavonoid compounds in pomegranate.
It is generally accepted that sugar acceptor regiospecificity, rather than sugar donor specificity, is the basis for the clustering of flavonoid UGTs [64]. According to this criterion, flavonoid UGTs were categorized into unique clusters, including 3GT, 5GT, 7GT/3′GT, GGT, and CGT subfamilies [65,66]. Yonekura-Sakakibara et al. [28] classified the UGT families into 24 orthologous groups (OGs), while A. thaliana UGTs could be divided into ten OGs. In pomegranate, eight PgUGT members in the F group were categorized into OG23, which were regarded as flavonoid 3-O-glycosyltransferase belonging to the 3GT subfamily [28]. Four pomegranate UGT members in the L group were defined as UGT75C1-like proteins which may function as anthocyanin-5-O-glucosyltransferase as previously described [67]. Therefore, subfamily 5GT in group L clustered into OG14. There were three members belonging to A group, namely LOC116195577, LOC116208919, and LOC116199194; the former two were annotated as FGGT1-like (cyanidin 3-O-galactoside 2″-O-xylosyltransferase) and the last was UGT79B6-like. In Arabidopsis, UGT79B6 encodes a flavonoid GGT, also known as flavonoid 3-O-glucoside: 2″-O-glucosyltransferase [68]. Thus, flavonoid GGTs belonging to UGT79 group were divided into OG8.
The UGTs in both OG1 and OG7 showed functions as 7GT, 3′GT, GGT, and 3GT, demonstrating a broad plasticity in the position of glycosylation and formation of more than one glycoside product [28,65]. These UGT groups distributed into two OGs revealed that their function were established after divergence of OG1 and OG7 [69]. Until now, no function validation of UGT members in R group has been performed, though Cui et al. [33] inferred that UGTs belonging to R group in Camellia sinensis may be involved in the reaction of flavonoid glycosylation. Using phylogenetic analysis, members in R group were clustered closely with OG1, but distantly with OG7, indicating that the R group can be grouped into OG1 rather than OG7. Given that the genes in an OG diverged from a common ancestor, UGTs in R group may share a common origin with B, C, and D groups during plant lineage evolution.
This study tentatively identifies 44 PgUGTs involved in flavonoid biosynthesis on the basis of phylogenetic relationships. However, it is important to take into consideration that sometimes there were incongruences between the phylogenetic position and substrate specificities [7,69], which is to say that the same group may not use the same flavonoid as substrates, or distantly related UGTs glycosylate the same substrate, as reported in Medicago truncatula [70]. Thus, the selectivity for acceptors cannot only be inferred by their primary sequence information. Our results on flavonoid PgUGTs prediction should only be regarded as suggestive. It is essential to characterize the functions of glycosyltransferase enzyme combining with in vitro or in vivo assays.
5. Conclusions
In this study, a total of 145 UGT genes were identified from the genome of Punica granatum ‘Tunisia’, and the phylogenetic relationships, gene structure, gene duplications, and gene expression profiles of PgUGT were analyzed. In addition, 44 PgUGT members involved in flavonoid biosynthesis were tentatively identified. Their distribution in distinct OGs revealed the regiospecificity towards flavonoid substrates. Taken together, these data provide a useful basis for more precise annotation of each PgUGT and evaluation of those involved in glycosylation of a variety of secondary metabolites, especially flavonoid compounds. Further function confirmation of PgUGTs is required to be established by experimental evidence.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae9050540/s1, Figure S1: The chromosome distribution of genes belonging to different groups; Figure S2: Segmental gene duplication events exhibited by PgUGT genes across eight pomegranate chromosomes; Figure S3: Cis-acting element analysis of the promoter regions of PgUGT genes; Figure S4: The expression profile of PgUGT members in different groups. Figure S5: Function preliminary prediction of PgUGTs involved in flavonoid biosynthesis; Table S1: Referenced UGTs used to construct phylogenetic tree; Table S2: The primers used in qRT-PCR; Table S3: Sequence information of 58 UGTs dataset; Table S4: Basic information of UGT genes identified in pomegranate; Table S5: The identified tandem duplication pairs of PgUGT genes; Table S6: Number of PgUGTs in each group according to intron amount; Table S7: The cis-acting elements of PgUGTs.
Author Contributions
Conceptualization, X.Z.; methodology, Y.F.; software, X.Z. and D.K.; validation, X.Z.; formal analysis, X.Z. and Y.T.; investigation, X.Z.; resources, Y.C. and R.L.; data curation, X.Z. and D.K.; writing—original draft preparation, X.Z.; writing—review and editing, X.Z., Y.F., D.K., Y.T.; Y.C. and R.L.; visualization, Y.F. and Y.T.; project administration, X.Z.; funding acquisition, X.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China (31901341) and the Priority Academic Program Development of Jiangsu High Education Institutions (PAPD).
Data Availability Statement
The RNA-seq data in this study were deposited in the NCBI Sequence Read Archive under the BioProject with the accession number PRJNA952822.
Acknowledgments
The authors are grateful to Zhaoxiang Hao of Zaozhuang Pomegranate Research Center for his help with the preparation of pomegranate samples.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yuan, Z.; Fang, Y.; Zhang, T.; Fei, Z.; Han, F.; Liu, C.; Liu, M.; Xiao, W.; Zhang, W.; Wu, S.; et al. The pomegranate (Punica granatum L.) genome provides insights into fruit quality and ovule developmental biology. Plant Biotechnol. J. 2018, 16, 1363–1374. [Google Scholar] [PubMed]
- Ge, S.S.; Duo, L.; Wang, J.; Yang, J.F.; Li, Z.Y.; Tu, Y. A unique understanding of traditional medicine of pomegranate, Punica granatum L. and its current research status. J. Ethnopharmacol. 2021, 271, 113877. [Google Scholar] [PubMed]
- Ranjha, M.M.A.N.; Shafique, B.; Wang, L.; Irfan, S.; Safdar, M.N.; Murtaza, M.A.; Nadeem, M.; Mahmood, S.; Mueen-ud-Din, G.; Nadeem, H.R. A comprehensive review on phytochemistry, bioactivity and medicinal value of bioactive compounds of pomegranate (Punica granatum). Adv. Tradit. Med. 2023, 23, 37–57. [Google Scholar] [CrossRef]
- Viuda-Martos, M.; Fernandez-Lopez, J.; Perez-Alvarez, J.A. Pomegranate and its many functional components as related to human health: A review. Compr. Rev. Food Sci. Food Saf. 2010, 9, 635–654. [Google Scholar]
- Zhao, X.; Yuan, Z. Anthocyanins from pomegranate (Punica granatum L.) and their role in antioxidant capacities in vitro. Chem. Biodivers. 2021, 18, e2100399. [Google Scholar] [PubMed]
- Paquette, S.; Moller, B.L.; Bak, S. On the origin of family 1 plant glycosyltransferases. Phytochemistry 2003, 62, 399–413. [Google Scholar] [CrossRef]
- Caputi, L.; Malnoy, M.; Goremykin, V.; Nikiforova, S.; Martens, S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012, 69, 1030–1042. [Google Scholar] [CrossRef]
- Albesa-Jove, D.; Guerin, M.E. The conformational plasticity of glycosyltransferases. Curr. Opi. Struc. Biol. 2016, 40, 23–32. [Google Scholar] [CrossRef]
- Le Roy, J.; Huss, B.; Creach, A.; Hawkins, S.; Neutelings, G. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front. Plant Sci. 2016, 7, 735. [Google Scholar] [CrossRef]
- Gachon, C.M.M.; Langlois-Meurinne, M.; Saindrenan, P. Plant secondary metabolism glycosyltransferases: The emerging functional analysis. Trends Plant Sci. 2005, 10, 542–549. [Google Scholar] [CrossRef]
- Vogt, T.; Jones, P. Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 2000, 5, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Fischer, U.A.; Carle, R.; Kammerer, D.R. Identification and quantification of phenolic compounds from pomegranate (Punica granatum L.) peel, mesocarp, aril and differently produced juices by HPLC-DAD-ESI/MSn. Food Chem. 2011, 127, 807–821. [Google Scholar]
- Fourati, M.; Smaoui, S.; Ben Hlima, H.; Elhadef, K.; Ben Braiek, O.; Ennouri, K.; Mtibaa, A.C.; Mellouli, L. Bioactive compounds and pharmacological potential of pomegranate (Punica granatum) seeds—A review. Plant Food Hum. Nutr. 2020, 75, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Man, G.; Xu, L.; Wang, Y.; Liao, X.; Xu, Z. Profiling phenolic composition in pomegranate peel from nine selected cultivars using UHPLC-QTOF-MS and UPLC-QQQ-MS. Front Nutr. 2022, 8, 807447. [Google Scholar] [CrossRef] [PubMed]
- Zeghad, N.; Abassi, E.A.; Belkhiri, A.; Demeyer, K.; Heyden, Y.V. Phenolic compounds profile from Algerian pomegranate fruit extract (Punica granatum L.) by UPLC-DAD-ESI-MS. Chem. Afr. 2022, 5, 1295–1303. [Google Scholar]
- Wilson, A.E.; Wu, S.; Tian, L. PgUGT95B2 preferentially metabolizes flavones/flavonols and has evolved independently from flavone/flavonol UGTs identified in Arabidopsis thaliana. Phytochemistry 2019, 157, 184–193. [Google Scholar] [CrossRef]
- Ono, N.N.; Qin, X.; Wilson, A.E.; Li, G.; Tian, L. Two UGT84 family glycosyltransferases catalyze a critical reaction of hydrolyzable tannin biosynthesis in pomegranate (Punica granatum). PLoS ONE 2016, 11, e0156319. [Google Scholar] [CrossRef]
- Chang, L.; Wu, S.; Tian, L. Effective genome editing and identification of a regiospecific gallic acid 4-O-glycosyltransferase in pomegranate (Punica granatum L.). Hortic. Res. 2019, 6, 123. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Cavalcanti, A.; Chen, F.; Bouman, P.; Li, W. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yuan, Z.; Feng, L.; Fang, Y. Cloning and expression of anthocyanin biosynthetic genes in red and white pomegranate. J. Plant Res. 2015, 128, 687–696. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR. Methods 2002, 25, 402–408. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Hanada, K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011, 66, 182–193. [Google Scholar] [CrossRef]
- Akere, A.; Chen, S.H.; Liu, X.; Chen, Y.; Dantu, S.C.; Pandini, A.; Bhowmik, D. Structure-based enzyme engineering improves donor-substrate regognition of Arabidopsis thaliana glycosyltransferases. Bichem. J. 2020, 477, 2791–2805. [Google Scholar] [CrossRef]
- Rehman, H.M.; Nawaz, M.A.; Bao, L.; Shah, Z.H.; Lee, J.; Ahmad, M.Q.; Chung, G.; Yang, S.H. Genome-wide analysis of family-1 UDP-glycosyltransferases in soybean confirms their abundance and varied expression during seed development. J. Plant Physiol. 2016, 206, 87–97. [Google Scholar] [CrossRef]
- Wang, F.; Su, Y.; Chen, N.; Shen, S. Genome-wide analysis of the UGT gene family and identification of flavonids in Broussonetia papyrifera. Molecules 2021, 26, 3449. [Google Scholar] [CrossRef]
- Song, Z.; Niu, L.; Yang, Q.; Dong, B.; Wang, L.; Dong, M.; Fan, X.; Jian, Y.; Meng, D.; Fu, Y. Genome-wide identification and characterization of UGT family in pigeonpea (Cajanus cajan) and expression analysis in abiotic stress. Trees 2019, 33, 987–1002. [Google Scholar] [CrossRef]
- Cui, L.; Yao, S.; Dai, X.; Yin, Q.; Liu, Y.; Jiang, X.; Wu, Y.; Qian, Y.; Pang, Y.; Gao, L.; et al. Identification of UDP-glycosyltransferases involved in the biosynthesis of astringent taste compounds in tea (Camellia sinensis). J. Exp. Bot. 2016, 67, 2285–2297. [Google Scholar] [CrossRef]
- Wu, B.; Liu, X.; Xu, K.; Zhang, B. Genome-wide characterization, evolution and expression profiling of UDP-glycosyltransferase family in pomelo (Citrus grandis) fruit. BMC Plant Biol. 2020, 20, 459. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Gu, J.; Luo, Y.; Wang, Y.; Pang, Y.; Shen, G.; Guo, B. Genome-wide analysis of UGT gene family identified key gene for the biosynthesis of bioactive flavonol glycosides in Epimedium pubescens Maxim. Syn. Syst. Biotechnol. 2022, 7, 1095–1107. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Pang, C.; Fan, S.; Song, M.; Yu, J.; Wei, H.; Ma, Q.; Li, L.; Zhang, C.; Yu, S. Genome-wide analysis of the family 1 glycosyltransferases in cotton. Mol. Genet. Genom. 2015, 290, 1805–1818. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhao, L.; Huang, H.; Zhang, Y.; Wang, J.; Lu, X.; Wang, S.; Wang, D.; Chen, X.; Chen, C.; et al. Genome-wide identifcation, evolution and function analysis of UGTs superfamily in cotton. Front. Mol. Biosci. 2022, 9, 965403. [Google Scholar] [CrossRef]
- Krishnamurthy, P.; Tsukamota, C.; Ishimoto, M. Reconstruction of the evolutionary histories of UGT gene sumperfamily in Legumes clarifies the functional divergence of duplicates in specialized metabolism. Int. J. Mol. Sci. 2020, 21, 1855. [Google Scholar] [CrossRef]
- Barvkar, V.T.; Pardeshi, V.C.; Kale, S.M.; Kadoo, N.Y.; Gupta, V.S. Phylogenomic analysis of UDP glycosyltransferase 1 multigene family in Linum usitatissimum identified genes with varied expression patterns. BMC Genom. 2012, 13, 175. [Google Scholar] [CrossRef]
- Zhou, K.; Hu, L.; Li, P.; Gong, X.; Ma, F. Genome-wide identification of glycosyltransferases converting phloretin to phloridzin in Malus species. Plant Sci. 2017, 265, 131–145. [Google Scholar] [CrossRef]
- Li, Y.; Li, P.; Zhang, L.; Shu, J.; Court, M.H.; Sun, Z.; Jiang, L.; Zheng, C.; Shu, H.; Ji, L.; et al. Genome-wide analysis of the apple family 1 glycosyltransferases identified a flavonoid-modifying UGT, MdUGT83L3, which is targeted by MdMYB88 and contributes to stress adaptation. Plant Sci. 2022, 321, 111314. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Dai, J.; Chen, Z.; Tie, W.; Yan, Y.; Yang, H.; Zeng, J.; Hu, W. Comprehensive analysis and expression profiles of cassava UDP-glycosyltransferases (UGT) family reveal their involvement in development and stress responses in cassava. Genomics 2021, 113, 3415–3429. [Google Scholar] [CrossRef]
- Ao, B.; Han, Y.; Wang, S.; Wu, F.; Zhang, J. Genome-Wide analysis and profile of UDP-glycosyltransferases family in alfalfa (Medicago sativa L.) under drought stress. Int. J. Mol. Sci. 2022, 23, 7243. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.; Yan, Q.; Wu, F.; Wang, Y.; Wang, S.; Zong, X.; Zhou, X.; Zhang, J. Genome-wide analysis of the UDP-glycosyltransferase family reveals its role in coumarin biosynthesis and abiotic stress in Melilotus albus. Int. J. Mol. Sci. 2021, 22, 10826. [Google Scholar] [CrossRef]
- Ren, C.; Guo, Y.; Xie, L.; Zhao, Z.; Xing, Z.; Cao, Y.; Liu, Y.; Lin, J.; Grierson, D.; Zhang, B.; et al. Identification of UDP-rhamnosyltransferases and UDP-galactosyltransferase involved in flavonol glycosylation in Morella rubra. Hortic. Res. 2022, 9, uhac138. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, X.; Lu, M.; Chen, J.; Shi, T. Gene expression and evolution of family-1 UDPglycosyltransferases—Insights from an aquatic flowering plant (sacred lotus). Aquat. Bot. 2020, 166, 103270. [Google Scholar] [CrossRef]
- Dong, L.; Tang, Z.; Yang, T.; Hao, F.; Deng, X. Genome-wide analysis of UGT genes in Petunia and identification of PhUGT51 involved in the regulation of salt resistance. Plants 2022, 11, 2434. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhuo, X.; Yan, X.; Zhang, Q. Comparative genomic and transcriptomic analyses of family-1 UDP glycosyltransferase in Prunus Mume. Int. J. Mol. Sci. 2018, 19, 3382. [Google Scholar] [CrossRef]
- Wu, B.; Gao, L.; Gao, J.; Xu, Y.; Liu, H.; Cao, X.; Zhang, B.; Chen, K. Genome-wide identification, expression patterns, and functional analysis of UDP glycosyltransferase family in peach (Prunus persica L. Batsch). Front. Plant Sci. 2017, 8, 389. [Google Scholar] [CrossRef]
- Li, G.; Li, J.; Qin, G.; Liu, C.; Liu, X.; Cao, Z.; Jia, B.; Zhang, H. Characterization and expression analysis of the UDP glycosyltransferase family in pomegranate (Punica granatum L.). Horticulturae 2023, 9, 119. [Google Scholar] [CrossRef]
- Cheng, X.; Muhammad, A.; Li, G.; Zhang, J.; Cheng, J.; Qiu, J.; Jiang, T.; Jin, Q.; Cai, Y.; Lin, Y. Family-1 UDP glycosyltransferases in pear (Pyrus bretschneideri): Molecular identification, phylogenomic characterization and expression profiling during stone cell formation. Mol. Biol. Rep. 2019, 46, 2152–2175. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Ahmad, D.; Zhang, X.; Zhang, Y.; Wu, L.; Jiang, P.; Ma, H. Genome-wide analysis of family-1 UDP glycosyltransferases (UGT) and identification of UGT genes for FHB resistance in wheat (Triticum aestivum L.). BMC Plant Biol. 2018, 18, 67. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Mu, H.; Xu, G.; Wang, Y.; Li, S.; Wang, L. Genome-wide analysis and functional characterization of the UDP-glycosyltransferase family in grapes. Horticulturae 2021, 7, 204. [Google Scholar] [CrossRef]
- Li, Y.; Li, P.; Wang, Y.; Dong, R.; Yu, H.; Hou, B. Genome-wide identification and phylogenetic analysis of family-1 UDP-glycosyltransferases in maize (Zea mays). Planta 2014, 239, 1256–1279. [Google Scholar] [CrossRef]
- Luo, X.; Li, H.; Wu, Z.; Yao, W.; Cao, S. The pomegranate (Punica granatum L.) draft genome dissects genetic divergence between soft- and hard-seeded cultivars. Plant Biotechnol. J. 2019, 18, 955–968. [Google Scholar]
- Ross, J.; Li, Y.; Lim, E.; Bowles, D.J. Higher plant glycosyltransferases. Genome Biol. 2001, 2, REVIEWS3004. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S. Evolution of gene dulication in plants. Plant Physiol. 2016, 171, 1194–2316. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef]
- Zhao, X.; Shen, Y.; Yan, M.; Yuan, Z. Flavonoid profiles in peels and arils of pomegranate cutlivars. J. Food Meas. Charact. 2022, 16, 880–890. [Google Scholar] [CrossRef]
- Sharma, R.; Rawat, V.; Suresh, C.G. Genome-wide identfication and tissue-specific expression analysis of UDP-glycosyltransferases genes confirm their abundance in Cicer arietinum (Chickea). PLoS ONE 2014, 9, e109715. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lin, C.; Ma, X.; Tan, Y.; Wang, J.; Zeng, M. Functional characterization of a flavonoid glycosyltransferase in sweet orange (Citrus sinensis). Front. Plant Sci. 2018, 9, 166. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.E.; Tian, L. Phylogenomic analysis of UDP-dependent glycosyltransferases provides insights into the evolutionary landscape of glycosylation in plant metabolism. Plant J. 2019, 100, 1273–1288. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, A.; Horikawa, M.; Fukui, Y.; Fukuchi-Mizutani, M.; Iuchi-Okada, A.; Ishiguro, M.; Kiso, Y.; Nakayama, T.; Ono, E. Local differentiation of sugar donor specificity of flavonoid glycosyltransferase in Lamiales. Plant Cell 2009, 21, 1556–1572. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Saito, K. Function, structure, and evolution of flavonoid glycosyltransferases in Plants. In Recent Advances in Polyphenol Research; Romani, A., Lattanzio, V., Quideau, S., Eds.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2014; pp. 61–82. [Google Scholar]
- Peng, M.; Shahzad, R.; Gul, A.; Subthain, H.; Shen, S.; Lei, L.; Zheng, Z.; Zhou, J.; Lu, D.; Wang, S.; et al. Differentially evolved glucosyltransferases determine natural variation of rice flavone accumulation and UV-tolerance. Nat. Commun. 2017, 8, 1875. [Google Scholar] [CrossRef]
- Tohge, T.; Nishiyama, Y.; Hirai, M.Y.; Yano, M.; Nakajima, J.; Awazuhara, M.; Inoue, E.; Takahashi, H.; Goodenowe, D.B.; Kitajima, M.; et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005, 42, 218–235. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Nakabayashi, R.; Sugawara, S.; Tohge, T.; Ito, T.; Koyanagi, M.; Kitajima, M.; Takayama, H.; Saito, K. A flavonoid 3-O-glucoside:2″-O-glucosyltransferase responsible for terminal modification of pollen-specific flavonols in Arabidopsis thaliana. Plant J. 2014, 79, 769–782. [Google Scholar] [CrossRef]
- Osmani, S.A.; Bak, S.; Møller, B.L. Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry 2009, 70, 325–347. [Google Scholar] [CrossRef]
- Modolo, L.V.; Blount, J.W.; Achnine, L.; Naoumkina, M.; Wang, X. A functional genomics approach to (iso)flavonoid glycosylation in the model legume Medicago truncatula. Plant Mol. Biol. 2007, 64, 499–518. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).