Abstract
Basic helix-loop-helix (bHLH) transcription factors (TFs) play irreplaceable roles in plant growth and development, especially in plant secondary metabolism. However, the functions of most bHLH TFs in Lilium ‘Siberia’ are still unknown, especially their roles in regulating floral fragrance. In this study, two bHLH TFs in lily, i.e., LibHLH22 and LibHLH63, were identified and functionally characterized. A bioinformatics analysis revealed that LibHLH22 and LibHLH63 were unstable proteins. Subcellular localization demonstrated that LibHLH22 and LibHLH63 proteins were in the cell nucleus. Quantitative real-time PCR showed that the highest expression level of LibHLH22 was at the initial flowering stage and in the stigma, and the highest expression level of LibHLH63 was at the budding stage and in the filaments. The results of transient overexpression and virus-induced gene silencing (VIGS) of LibHLH22 and LibHLH63 in lily petals showed that these two transcription factors significantly promoted the expression of LiDXR and LiTPS2, and thus, markedly enhanced the release of floral fragrance. Our results indicated that LibHLH22 and LibHLH63 could effectively regulate the fragrance of Lilium ‘Siberia’, laying the foundation for fragrance breeding and improving the terpenoid transcriptional regulatory pathway.
1. Introduction
The floral fragrance is an important characteristic of ornamental plants, which has various physiological functions, such as attracting pollinators and avoiding herbivores/pathogens. Moreover, floral fragrance has a commercial value associated with the production of perfumes, relieving stress, assisting in calm people’s minds, and influencing consumer choice, etc. [1,2]. To make better use of floral fragrance in order to meet people’s daily life needs, the compounds, release rules, and molecular regulatory mechanisms of various fragrant flowers have been studied deeply, such as rose, peony, orchid, wintersweet, freesia, dendrobium, osmanthus, oncidium, plum blossom, and Hedychium coronarium [3,4,5,6,7,8,9,10,11,12,13,14]. Lilies are also known as famous fragrant flowers, and therefore, the composition and biosynthesis of their floral fragrance have been extensively studied. Previous studies have found that there were clear differences in the floral volatile compounds between Lilium ‘Siberia’ (oriental lily) with a strong fragrance and Lilium ‘Novano’ (asiatic lily) with a very faint fragrance. Moreover, they have found that the class of compounds known as terpenes was the main reason for the differences in their floral fragrance [15]. Among the terpene compounds, (E)-β-ocimene and linalool have been identified as the key components of the fragrance in most of the lily cultivars [16]. The key genes located in the skeleton of terpenoid biosynthesis in the MEP pathway, such as LiCMK, LiHDS, LiDXS, LiDXR, LiTPS2, LoTPS2, LoTPS4, LoTPS3, and LoTPS1 have been cloned and functionally characterized; however, the transcriptional regulation of these key genes participating in monoterpene biosynthesis in Lilium remains to be largely unclear [17,18,19,20,21,22].
Members of the bHLH superfamily of transcription factors are widespread in plants. Usually, they have two functionally conserved domains containing 50 to 60 amino acids: The basic region and the helix-loop-helix (HLH) domain. The HLH domain is located at the C-terminus and contains 40~50 amino acids. The basic region is located at the N-terminus and contains 13–17 amino acids that can recognize and specifically bind with cis-elements of target genes, such as E-box (5′-CANNTG-3′) and G-box (5′-CACGTG-3′) [23]. Recent studies have shown that bHLH transcription factors are involved in plant growth and development. For example, bHLH TFs regulate cell cycle progression, stomatal development, photosynthesis, carpel development, and seedling germination [24,25,26]. Furthermore, bHLH transcription factors help plants in resisting diverse environmental stresses, such as cold, salt, and drought [27,28,29,30,31,32]. The biosynthesis of secondary metabolites has also been shown to be regulated by bHLH TFs, such as flavonoids and anthocyanins [33,34,35,36,37]. Terpenoids are essential members of secondary metabolites, and extensive research has shown that the biosynthesis of terpenoids is closely correlated with bHLH transcription factors in many species. In tomatoes, SlMYC1 positively regulates monoterpene biosynthesis in tomato leaves, but negatively regulates sesquiterpene biosynthesis in stem trichomes [38]. The CpMYC2 and CpbHLH13 transcription factors from wintersweet have been proven to have roles in the positive regulation and biosynthesis of monoterpenoids (linalool) and sesquiterpenoids (β-caryophyllene) in transgenic plants [6]. LaMYC4 isolated from lavender has been shown to be responsive to UV, MeJA, drought, low temperature, and Pseudomonas syringae infection, and overexpression of LaMYC4 has been shown to increase the level of sesquiterpenoids in model plants [39]. However, few bHLH transcription factors have been reported to be relevant to the biosynthesis of terpenoids in lily.
Lilium ‘Siberia’ is one of the most popular lily cultivars in the world. Our previous study demonstrated that LiDXS, LiDXR, and LiTPS2 could positively regulate terpene biosynthesis in lily. In other research, LiMCT, LiMCS, LiCMK, and LiHDS of the MEP pathway have been cloned and functionally characterized, which indicated that the skeleton of terpene biosynthesis had been almost built [17,18,19,20,21,22]. Nevertheless, the transcriptional regulatory network of terpenoid biosynthesis is incomplete, especially the roles of bHLH transcription factors. To fill the gaps, in this study, a correlation analysis was conducted to screen out the bHLH transcription factors, LibHLH22 and LibHLH63, from the transcriptome data, and their functions were preliminarily verified by transient overexpression and virus-induced gene silencing (VIGS) techniques. Considered together, this research provides insight into fragrance breeding or the development of high-value terpenoids, and aims to improve the regulatory network of terpenoid metabolism and transcription.
2. Materials and Methods
2.1. Plant Material
In the present study, the cultivar Lilium ‘Siberia’ was used as the experimental plant material and purchased from the Dongfeng International Flower Market in Beijing. The flowering stages were defined as budding, initial flowering, full-blooming, and wilting stages. Figure 1 shows the different tissues of lily petals and the four different flowering stages that were prepared for expression analysis. Inner petals of the lily at each different stage and different organ were harvested and randomly divided into groups containing the same amounts of samples to ensure that each treatment included three biological replicates. After being frozen in liquid nitrogen, the sample was stored at −80 °C for further experiments.

Figure 1.
Plant material in the experiment: (A) Budding stage; (B) initial flowering stage; (C) full-blooming stage; (D) wilting stage; (E) petals; (F) filaments; (G) anthers; (H) style; (I) stigma.
2.2. RNA Extraction and Gene Cloning
The total RNA of lily petals at different stages (budding stage, initial flowering stage, full-blooming stage, wilting stage) and in different organs (petals, filaments, anther, style, and stigma) was extracted using a total RNA extraction kit (Omega, GA, USA), and then RNA quality and concentration were analyzed using a NanoDrop micro nucleic acid assay and 1% gel electrophoresis. The first strand of cDNA was synthesized using a PrimeScriptTM RT reagent kit (Perfect Real Time) (TaKaRa, Shiga, Japan) and then stored at −20 °C.
Based on the transcriptome data of the cultivar Lilium ‘Siberia’, the open reading frames (ORFs) of LibHLH22 and LibHLH63 transcription factors were obtained using an online website, the ORF finder in the NCBI (Table 1), which was amplified using PrimeSTAR® Max DNA Polymerase (TaKaRa, Shiga, Japan) by the general PCR cloning method; two pairs of specific primers, LibHLH22-F/R and LibHLH63-F/R, were shown in Table 2 [20]. The full-length fragments of LibHLH22 and LibHLH63 would be linked to the TOPO vector and transformed into Escherichia coli DH5α. After PCR identification, the positive bacteria would be sent for sequencing.

Table 1.
The names and websites of the bioinformatics analysis.

Table 2.
All primers in this article.
2.3. Bioinformatics Analysis and Phylogenetic Analysis
The cis-acting elements of the LiTPS2 promoter and LiDXR promoter were analyzed and visualized using the PlantCARE database (Table 1) and the Gene Structure Display Server (GSDS 2.0) (Table 1), respectively. The conserved domains of LibHLH22 and LibHLH63 were analyzed using the CD-Search tool in the NCBI database (Table 1). The physicochemical properties of the LibHLH22 and LibHLH63 proteins were analyzed using the ExPASy-ProtParam tool. The secondary and tertiary structures of LibHLH22 and LibHLH63 were predicted with the help of SOPMA (Table 1) and SWISS-MODEL, respectively (Table 1). The alignments of the protein sequences of these two bHLH transcription factors from the lily and other plants were analyzed using the DNAMAN 9.0 software (Lynnon BioSoft, San Ramon, CA, USA) and the Clustalx 2.0 software (The Conway Institute UCD Dublin, Dublin, Ireland). The phylogenetic tree was constructed using the neighbor-joining method with 1000 bootstrap replicates based on the aligned protein sequences by running the MEGA11 software (Mega Limited, Auckland, New Zealand).
2.4. Vector Construction
The plant expression vector pSuper1300-GFP was digested with KpnI and XbaI enzymes at 37 °C for 3~4 h. The ORFs of LibHLH22 and LibHLH63 were amplified with the specific primers, pSuper1300-LibHLH22-F/R and pSuper1300-LibHLH63-F/R (Table 2), that had 15 bp homologous sequences around restriction sites in the plant expression vector pSuper1300-GFP. The recombinant plasmids, pSuper1300-LibHLH22-GFP and pSuper1300-LibHLH63-GFP, were constructed using In-Fusion® Snap Assembly Master Mix enzyme (TaKara, Shiga, Japan), which ligated the linearized vector and the targeted gene fragments. The correct recombinants were transformed into Agrobacterium tumefaciens GV3101 for further infection work.
Two vectors, TRV1 and TRV2, were used in the virus-induced gene silencing (VIGS) system. The TRV2 vector was digested with EcoRI and BamHI for 4 h. LibHLH22-VIGS-F and LibHLH22-VIGS-R (Table 2), LibHLH63-VIGS-F and LibHLH63-VIGS-R (Table 2), were used to amplify 360 bp long fragments of LibHLH22 and LibHLH63, respectively. Then, they were connected to the TRV2′s linearized vector using In-Fusion® Snap Assembly Master Mix enzyme (TaKara, Shiga, Japan). TRV2-LibHLH22 and TRV2-LibHLH63 were imported into Agrobacterium GV3101 for further work.
2.5. Subcellular Localization
The recombinant plasmids, pSuper1300-LibHLH22-GFP and pSuper1300-LibHLH63-GFP, were transferred into leaves of Nicotiana benthamiana using the Agrobacterium-mediated method. The competent cell of Agrobacterium tumefaciens GV3101 stored at −80 °C was slowly melted at room temperature. When mixed with ice water, 0.01–1 μg of plasmid DNA was added into 100 μL competent cell, and then the mixture would stand in ice for 5 min, liquid nitrogen for 5 min, water bath at 37 °C for 5 min, and ice bath for 5 min. Thereafter, 500 μL LB of liquid medium without antibiotics was added and shaken at 28 °C for 2–3 h. After shaking, it was centrifuged at 6000 rpm for 1 min to collect the bacteria and about 100 μL of supernatant remained to gently blow a bacterial block and apply the bacterial solution evenly to an LB plate containing 50 μg/mL Kana and 50 μg/mL rifampicin antibiotics. Then, it was placed upside down in an incubator at 28 °C for 2 days. After injection, Nicotiana benthamiana was placed in a light incubator with 16 h light/8 h dark for 2~3 days, and the locations of the LibHLH22 and LibHLH63 proteins were observed through a laser confocal microscope (TCS SP8, Leica, Mannheim, Germany). The subcellular localizations of LibHLH22 and LibHLH63 were also predicted using the online software Plant-mPLoc (Table 1) and WoLF POST (Table 1).
2.6. Gene Expression Analysis
To determine the expression levels of the LibHLH22 and LibHLH63 after infection, and their expression levels during different flowering stages and in different floral organs, RNA was extracted from these materials and the cDNA was synthesized using PrimeScriptTM RT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, Shiga, Japan), then quantitative real-time PCR (qRT-PCR) was conducted using the PikoReal real-time PCR system (Thermo Fisher Scientific, MA, USA). TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) (TaKaRa, Shiga, Japan) was necessary for this experiment. The sequences of gene-specific primers, qLibHLH22-F/R and qLibHLH63-F/R, are listed in Table 2. Each sample was pooled in three replicates, and for each replicate, qRT-PCR was performed three times. The procedure for RT-qPCR was a three-step method. The qRT-PCR profile comprised 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 55 °C for 30 s, and 72 °C for 30 s. The relative transcript levels were calculated using the 2−ΔΔCt method. The lily Actin gene (Accession number: JX826390) was used as the internal reference.
2.7. Transformation of Lilium ‘Siberia’
After PCR identification, the bacterial liquid was rapidly propagated in an LB liquid medium containing Rif (50 μg/mL) + Kan (50 μg/mL), and shaken at 28 °C and 200 rpm overnight. The bacterial solution was centrifuged at 5000 rpm for 10 min in a refrigerated centrifuge when its OD600 value was between 0.6 and 1. After centrifugation, the bacteria was resuspended with an infection solution (0.2 M MES + 150 mM AS + 2 M MgCl2 + ddH2O) with an adjusted OD600 value of 0.6–0.8. Then, the mixed infection solution was left to sit for 2 h. The lily petals were at the best stage for injection on the first day of blooming. A mixture of Agrobacterium cultures was injected into petals with 1 mL of bacterial suspension per petal using a 2 mL syringe. After injection, the lily petals were left in the dark for 12~24 h, and then transferred to a light incubator for 3 days, and the gene expression levels of LibHLH22 and LibHLH63 and floral fragrance content of lily were determined later. There were three biological replicates for the infections of each gene.
The bacterial liquid was rapidly propagated in an LB liquid medium containing Rif (50 μg/mL) + Kan (50 μg/mL), and TRV1 was mixed with TRV2-LibHLH22 and TRV2-LibHLH63 bacterial solution at a ratio of 1:1 before standing for 2 h, with a liquid mixture consisting of TRV2 and TRV2 as the control. After being left in the dark for 12~24 h, the injected lily petals were transferred to the light incubator. Then, 2–3 days later, the injected samples were collected at 14:00, and the expression of the genes involved and the fragrance of the flowers were measured later.
2.8. Volatile Compound Analysis
The method of headspace solid-phase microextraction gas chromatography-mass spectrometry (HS-SPME-GC-MS) was used to detect the differences in volatiles between the control group and the experimental group. The control group refers to lily petals injected with an empty expression vector, and the experimental group refers to petals injected with an expression vector containing the target gene. Weighed frozen samples of lily petals were placed in sampling glasses. After adding the internal standard of 5 μL ethyl decanoate methanol solution (1:10,000), the glasses equipped with lily petal samples were sealed immediately, and heated at 40 °C in a water bath for 30 min. Meanwhile, an SPME extraction needle was inserted in the glass sample and the extraction head was pushed out. At the end of sampling, the fiber was inserted into the injection port of the gas chromatograph and analyzed at 250 °C for 5 min, and the volatile compound analysis was performed later. The chromatographic column was a DB-5 column (30 m × 0.25 mm × 0.25 mm). The chromatographic conditions were as follows: The initial temperature was 50 °C for 4 min, and then the temperature was increased to 270 °C (at a rate of 10 °C/min) for 5 min. The temperature of the GC injection port was 250 °C. The mass spectrum condition was an ion source of 70 eV and the mass charge ratios recorded were from 30 to 500 m/z.
The formula for quantitative calculation of floral volatile scent content is as follows: mi(μg/gh) = Fi × (Ai/As) × ms/mt, where mi is the mass of volatile matter, Fi is the correction factor of volatiles to be measured (default of 1 in this test), Ai is the peak area of volatiles to be measured, As is the peak area of internal standard, ms is the quality of internal standard, m is the quality of measured flowers, and t is the solid phase microextraction time.
2.9. Statistical Analysis
The statistical significance was determined with SPSS23.0 (SPSS, Chicago, IL, United States) using the t-test. The correlation coefficient diagram was drawn using the Lianchuan Biologic Cloud platform (Table 1). The bar charts representing the data were drawn with Origin (OriginLab, Northampton, MA, USA).
3. Results
3.1. Analysis of LiDXR and LiTPS2 Promoters and Screening of LibHLH22 and LibHLH63 from Transcriptome Data
Based on the transcription data we obtained, the differentially expressed bHLH transcription factors were filtrated out by drawing a correlation analysis heat map with the reported terpenoid biosynthesis related genes LiDXR and LiTPS2. As shown in Figure 2C, we found that the genes, LibHLH22 and LibHLH63, had a high correlation with the terpenoid biosynthesis related gene LiDXR; LibHLH63 also had a high correlation with LiTPS2, while LibHLH22 did not. In addition, the promoter analysis of LiDXR and LiTPS2 showed that their promoters contained the binding site G-box of bHLH transcription factors (Figure 2A,B), suggesting that these screened transcription factors might regulate the expression of LiDXR and LiTPS2.
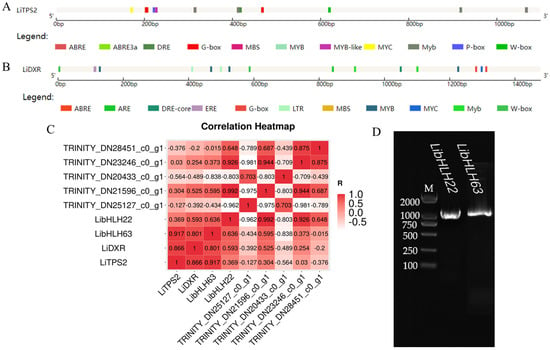
Figure 2.
(A) Visual analysis of cisacting elements of LiTPS2 promoter; (B) visual analysis of cis-acting elements of LiDXR promoter; (C) heat map of correlation analysis among the bHLH transcription factors, LiDXR and LiTPS2; (D) agarose gel electrophoresis diagram of LibHLH22 and LibHLH63. M, 2000 bp marker.
3.2. Cloning of LibHLH22 and LibHLH63
The CDS sequence of transcription factors was determined using the NCBI ORF Finder tool (Table 1), and the cDNA in different flowering stages was used as the template for PCR amplification. As shown in Figure 2D, 1% agarose gel electrophoresis showed that the product bands were close to 1000 bp without hetero bands. The sequencing results showed that LibHLH22 (left) was 1089 bp in length and coded for 362 amino acids (Figure 3A), and that LibHLH63 (right) was 1092 bp in length and coded for 363 amino acids (Figure 3B).

Figure 3.
(A) Nucleotide sequence and amino acid sequence of LibHLH22; (B) nucleotide sequence and amino acid sequence of LibHLH63.
3.3. Bioinformatics Analysis of LibHLH22 and LibHLH63
The ExPASy ProtParam tool was used to analyze the primary structures of LibHLH22 and LibHLH63. The results show that the molecular formula of LibHLH22 is C1705H2697N505O534S13, and the molecular formula of LibHLH63 is C1717H2736N504O553S26. The theoretical isoelectric points of LibHLH22 and LibHLH63 are 6.03 and 5.96, respectively, and the instability index values of the two proteins are 58.74 and 53.47, which indicates that they are unstable proteins. SOPMA was used to analyze the secondary structures of LibHLH22 and LibHLH63. Regarding the LibHLH22 and LibHLH63 proteins, the data show that the alpha helix accounts for 30.39% and 32.23%, respectively; the beta turn accounts for 5.25% and 4.13%, respectively; random coil accounts for 56.08% and 58.95%, respectively; and extended strand accounts for 8.29% and 4.68%, respectively (Figure 4A and Figure 5A). SWISS-MODEL was used to predict the tertiary structures of LibHLH22 and LibHLH63, and showed that the tertiary structures were consistent with the secondary structures (Figure 4B and Figure 5B). The conserved domain analysis in NCBI showed that LibHLH22 and LibHLH63 proteins contained basic regions and HLH conserved domains, showing that these two genes belong to the bHLH superfamily (bHLH-SF) (Figure 4C and Figure 5C).
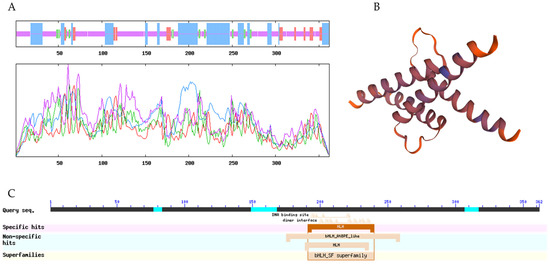
Figure 4.
Bioinformatics analysis of the LibHLH22 protein: (A) Secondary structure of the LibHLH22 protein; (B) tertiary structure of the LibHLH22 protein; (C) conserved domain analysis of the LibHLH22 protein.
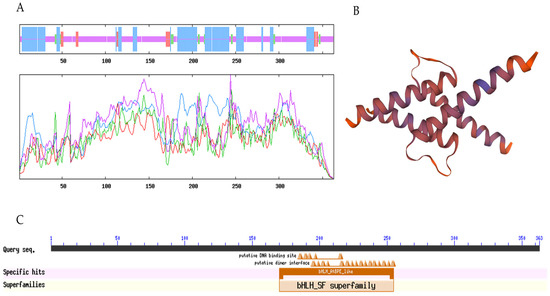
Figure 5.
Bioinformatics analysis of the LibHLH63 protein: (A) Secondary structure of LibHLH63 protein; (B) tertiary structure of the LibHLH63 protein; (C) conserved domain analysis of the LibHLH63 protein.
To further explore the potential functions of LibHLH22 and LibHLH63 in Lilium ‘Siberia’, after performing the amino acid sequence alignment between the two potential proteins and other bHLH transcription factors from different species (Figure 6A), a neighbor-joining phylogenetic analysis was constructed based on predicted protein sequences of selected bHLH transcription factors. The results showed high amino acid homology between the two potential genes and BpbHLH8 (Figure 6B), and they were in the same clade.
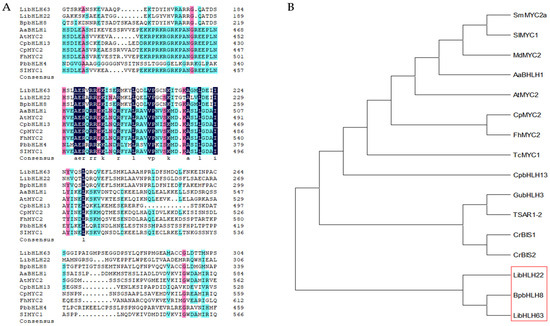
Figure 6.
(A) Multiple alignment of LibHLH22 and LibHLH63 with bHLH transcription factors related to terpenoid biosynthesis from other species, and different color areas in the picture show different consistency levels: dark blue is 100%; red is ≥75%; light blue is ≥50%. BpbHLH8 Betula platyphylla MH113336.1, AabHLH1 Artemisia annua AGH2505, AtMYC2 Arabidopsis thaliana At1g32640, FhMYC2 Freesia hybrida QNC43971, PbbHLH4 Phalaenopsis bellina KY979199, Solanum SlMYC1 lycopersicum KF430611; (B)analysis of the phylogenetic relationship of LibHLH22 and LibHLH63 in Lilium ‘Siberia’ using the neighbor-joining method, the role of the red highlight box in the picture is to emphasize the high homology of the transcription factors LibHLH22 and LibHLH63 with BpbHLH8. Other plant species are as follows: Fh, Freesia hybrida; Md, Malus domestica; Sl, Solanum lycopersicum; Aa, Artemisia annua; Cr, Catharanthus roseus; At, Arabidopsis thaliana; Tc, Taxus cuspidate; Bp, Betula platyphylla; Sm, Salvia miltiorrhiza; Cp, Chimonanthus praecox; Gu, Glycyrrhiza uralensis; Pb, Phalaenopsis bellina.
3.4. Subcellular Localization of LibHLH22 and LibHLH63
To further study the function of LibHLH22 and LibHLH63 in monoterpene biosynthesis, the localization of LibHLH22 and LibHLH63 in the cell was determined, and pSuper1300-LibHLH22-GFP and pSuper1300-LibHLH63-GFP were transiently transferred into the leaf of N. benthamiana by A. tumefaciens inoculation. The results showed that the green fluorescent protein signal (GFP) was localized in the nuclei, consistent with results obtained from Plant-mPLoc and WoLF POST (Figure 7).
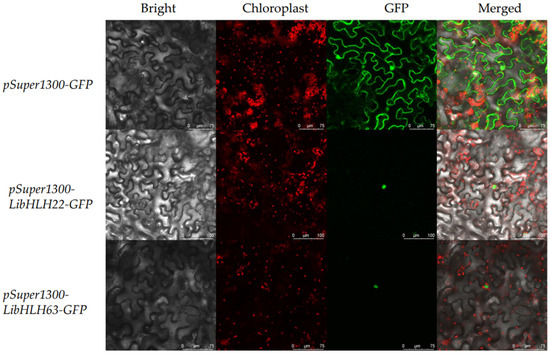
Figure 7.
Subcellular localization of LibHLH22 and LibHLH63. The second panel is the chloroplast autofluorescence channel. The third panel is constructed in the green fluorescence channel. The merged image is generated from the second and the third panels.
3.5. Expression Patterns of LibHLH22 and LibHLH63 in Lilium ‘Siberia’
qRT-PCR was used to identify the expression levels of LibHLH22 and LibHLH63 in lily petals at different flowering stages and in different floral organs. The results showed that the expression level of LibHLH22 was the highest at the initial flowering stage and the lowest at the wilting stage (Figure 8A), and the expression level of LibHLH63 was the highest at the budding stage (Figure 8C). The expression level of LibHLH22 peaked at the initial flowering stage and declined rapidly at the wilting stage, which was consistent with the transcriptome data and the pattern of floral fragrance release. LibHLH63 had high expression in different stages, which indicated that this transcription factor might have other functions in different periods.
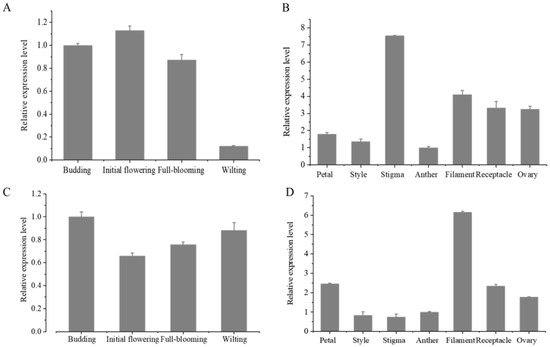
Figure 8.
The expression levels of LibHLH22 and LibHLH63 at different flowering stages and in different floral tissues: (A) The expression levels of LibHLH22 at different flowering stages; (B) the expression levels of LibHLH22 in different floral tissues; (C) the expression levels of LibHLH63 at different flowering stages; (D) the expression levels of LibHLH63 in different floral tissues.
The expression levels of LibHLH22 and LibHLH63 in different tissues were also investigated. The results showed that the expression levels of LibHLH22 and LibHLH63 were the highest in stigma and filaments, respectively, and they also had higher expression levels in lily petals than in anthers (Figure 8B,D). Furthermore, both LibHLH22 and LibHLH63 had high expression levels in other tissues, suggesting that LibHLH22 and LibHLH63 were involved in regulating floral fragrance, and also may have played important roles in the development and growth of the whole flower organs.
3.6. LibHLH22 and LibHLH63 Promote Volatile Terpenoids Accumulation in Lilium ‘Siberia’
To investigate whether LibHLH22 and LibHLH63 regulated terpenoid biosynthesis in the native species, pSuper1300-LibHLH22-GFP and pSuper1300-LibHLH63-GFP were transferred into lily following the Agrobacterium-mediated method. The results showed that LibHLH22 was significantly overexpressed, and its expression level was about 6 times higher than the control group. LiTPS2 and LiDXR were significantly higher than those of the control group (Figure 9A). Accordingly, linalool and ocimene, the most important components of floral fragrance in lily, showed a marked increase in the experimental group (Figure 9B and Figure 10A). In Figure 11A, the expression level of LibHLH63 in the experimental group was about 2 times higher than the control group, and the expression levels of LiTPS2, LiDXR, and LiDXS also improved several times (Figure 11B). In addition, there were significant improvements in linalool and ocimene components in the experimental group compared with those in the control group (Figure 12A).
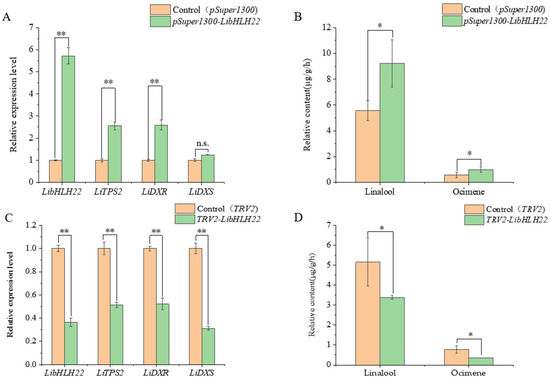
Figure 9.
Overexpression and instantaneous silence of LibHLH22: (A) Overexpression of LibHLH22; (B) determination of floral volatile content in the case of overexpression of LibHLH22; (C) instantaneous silence of LibHLH22; (D) determination of floral volatile content in the case of instantaneous silence of LibHLH22. Values shown are mean ± SD of at least three replicates, and standard errors are indicated as vertical lines on the top of each bar. Statistical significance was determined by Student’s t-test (* p < 0.05, ** p < 0.01, the letters n.s. stand for no significance).
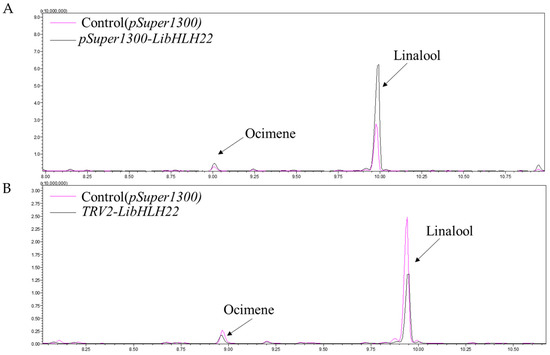
Figure 10.
GC-MS analysis for the fragrance in samples of LibHLH22: (A) Total ion chromatogram for the fragrance of LibHLH22 under overexpression conditions; (B) total ion chromatogram for the fragrance of LibHLH22 under instantaneous silence conditions. The red lines represent the control group and the black lines represent the experimental group.
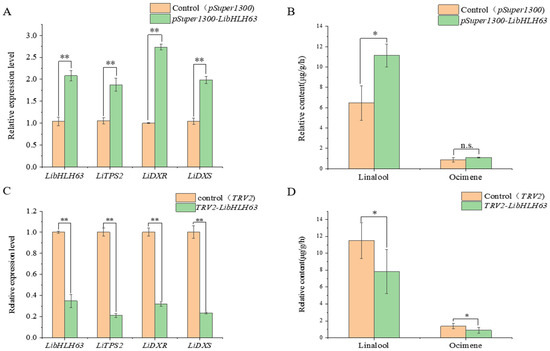
Figure 11.
Overexpression and instantaneous silence of LibHLH63: (A) Overexpression of LibHLH63; (B) determination of floral volatile content in the case of overexpression of LibHLH63; (C) instantaneous silence of LibHLH63; (D) determination of floral volatile content in the case of instantaneous silence of LibHLH63. Values shown are mean ± SD of at least three replicates, and standard errors are indicated as vertical lines on the top of each bar. Statistical significance was determined by Student’s t-test (* p < 0.05, ** p < 0.01, the letters n.s. stand for no significance).
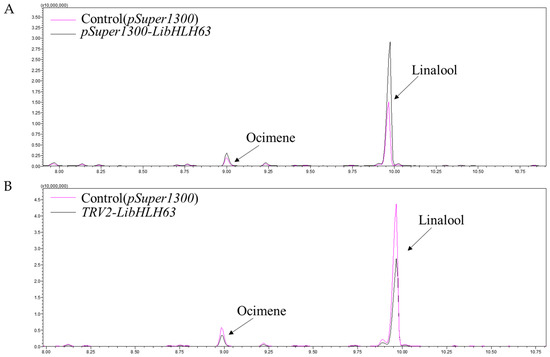
Figure 12.
GC-MS analysis for the fragrance in samples of figureLibHLH63: (A) Total ion chromatogram for the fragrance of LibHLH63 under overexpression conditions; (B) total ion chromatogram for the fragrance of LibHLH63 under instantaneous silence conditions. The red lines represent the control group and the black lines represent the experimental group.
The virus-induced gene silencing (VIGS) vectors, pTRV2-LibHLH22 and pTRV2-LibHLH63, were also constructed to be transferred into lily. The results indicated that the expression level of LibHLH22 was significantly lower than the control group. In the same way, the expression levels of LiTPS2, LiDXR, and LiDXS (Figure 9C), and the content of fragrance declined significantly (Figure 9D and Figure 10B). In Figure 12C, the expression levels of LibHLH63, LiTPS2, LiDXR, and LiDXS dropped dramatically in the experimental group; the fragrance of flowers also dropped observably compared to the control group (Figure 12B). In summary, the results indicated that LibHLH22 and LibHLH63 had a certain effect on fragrance biosynthesis.
4. Discussion
As important components of floral fragrance, many key enzyme genes and transcription factors regulate the biosynthesis and release of terpenoids. The bHLH superfamily has a wide range of functions and regulates terpenoids in many species, such as Betula platyphylla and Medicago truncatula [40,41]. Terpenoids are an important component of lily flower fragrance and previous studies have fully studied the key genes LoTPS1, LoTPS2, LoTPS3, LoTPS4, LiTPS2, LiDXR, LiDXS, and other genes in the terpenoid biosynthesis skeleton of lily. Moreover, we found that the transcription factors of the MYB family, LiMYB105 were directly bound and activated the LiOcS promoter to increase the synthesis of monoterpenes; however, the researches on how bHLH transcription factors regulate terpenoids are not sufficient [17,18,19,20,21,22,42]. Therefore, this research aims to explore the molecular mechanism of bHLH transcription factors on the regulation of terpenoids biosynthesis, which could improve the regulation network of fragrance metabolism in lily. Moreover, it has an important guiding significance for the directional breeding of aromatic lily.
As one of the most popular varieties in the market, Lilium ‘Siberia’ is also a good material for studying the floral aroma among the wide variety of lily species. Based on the transcriptome database of lily and correlation analysis, two transcription factors possibly involved in regulating flower aroma, LibHLH22 and LibHLH63, were screened out. The open reading frames of these two transcription factors were further cloned. The amino acid sequence alignment results showed that LibHLH22 and LibHLH63 had conserved domains of the bHLH superfamily with other species. We found that LibHLH22 and LibHLH63 have the highest homology with BpbHLH8, indicating that they have similar biological functions. The conserved domain of the bHLH superfamily contains nearly 60 amino acid residues, and the basic amino acid regions of LibHLH22 and LibHLH63 contain typical His5-Glu9-Arg13 sequences, which play an important role in the specific binding of target genes to bHLH proteins [43]. LibHLH22 and LibHLH63 not only have bHLH conserved domains, but also have similar conservative N-terminal and C-terminal regions with BpbHLH8 in birch.
Previous studies have shown that the release pattern of volatiles in lily was consistent with the expression pattern of key enzyme genes LiTPS2, LiDXR, and LiDXS, which all peaked at the flowering stage and petals [19,20,44]. Our study found that LibHLH63 and LibHLH22 were located in the nucleus, while the key enzyme genes LiTPS2, LiDXR, and LiDXS were located in chloroplasts [19,20]. The expression levels of LibHLH63 and LibHLH22 were the highest in the bud and initial flowering stages of the lily, respectively, while the expression levels of LiTPS2, LiDXR, and LiDXS were the highest in the full-blooming stage [19,20]. The results showed that there might be spatial and temporal disparities in terms of expression peaks between two transcription factors and key enzyme genes. The transcription factors may first promote the transcription of related key enzyme genes in the nucleus, and then proteins of these key enzyme genes were transferred to the plastids to promote the biosynthesis of floral fragrance. We also found that LibHLH63 and LibHLH22 had different expression levels in different organs of flowers, and the bHLH transcription factors have various functions; therefore, LibHLH63 and LibHLH22 may not only participate in the release of terpenoids from petals, but also be involved in the development of different organs in different flowering stages.
Although the genetic transformation system of lily has been established, it still has some areas for improvement, such as low transformation efficiency and long transformation time; therefore, many key terpenoid synthase genes have been transformed into model plants to verify their functions [45]. Compared with a stable genetic transformation system, the transient expression has the advantage of a short cycle and high efficiency, which could be used for gene function analysis and subcellular localization analysis [46]. In addition, mature systems of transient expression have been established in Lilium, Catharanthus roseus, Nictiana tabacum, Vitis vinifera, Arabidopsis, Rosa hybrida, and Gerbera hybrida [47,48,49,50,51,52], and the transient overexpression method used in this study improved the transient expression system of lily, which was used to verify the functions of LibHLH63 and LibHLH22 in lily petals. Moreover, we found that both of them can regulate the release of lily flower fragrance. Through correlation analysis and visual analysis of promoters, it is speculated that these two transcription factors may regulate the expression of key genes LiDXR and LiTPS2 in terpenoids biosynthesis by combining their promoters. To further verify this conjecture, it is necessary to carry out subsequent experiments, such as yeast one-hybrid, EMSA, and dual-luciferase reporter assays.
5. Conclusions
In conclusion, this study used correlation analysis to screen and clone LibHLH22 and LibHLH63 from lily petals. A series of online software was used to perform bioinformatics analysis and to explore the expression patterns of LibHLH22 and LibHLH63 during different flowering periods and in different tissues. Finally, transient overexpression and virus-induced gene silencing (VIGS) techniques were used to verify the functions of LibHLH22 and LibHLH63. The results show that these two genes regulate the main components of lily’s fragrance, i.e., ocimene and linalool. In addition, their regulation model is illustrated intuitively in Figure 13.
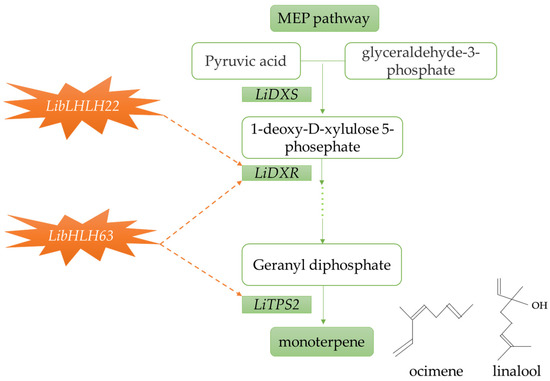
Figure 13.
A model diagram of LibHLH22 and LibHLH63 actively regulating the synthesis of linalool and ocimene in lily. The yellow dotted arrows indicate the predicted regulatory pathway, the green dotted line represents the path that has been omitted.
Author Contributions
Conceptualization, Y.F., Z.G. and M.S.; methodology, Y.F., M.S., J.Z. and Z.G.; software, Y.F.; validation, Y.F.; resources, Y.F., P.Z. and Y.L.; data curation, Y.F., Z.G. and J.Z.; writing—original draft preparation, Y.F. and Z.G.; writing—review and editing, Y.F., M.S. and J.Z.; visualization, Y.F.; supervision, M.S. and J.Z.; project administration, M.S.; funding acquisition, M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 31971708, 32271947), Science, Technology and Innovation Project of Xiongan New Area (2022XAGG0100), and the Beijing Municipal Natural Science Foundation (No. 6202022).
Data Availability Statement
Data supporting the reporting results are available and will be provided upon request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Suchet, C.; Dormont, L.; Schatz, B.; Giurfa, M.; Simon, V.; Raynaud, C.; Chave, J. Floral scent variation in two Antirrhinum majus subspecies influences the choice of naïve bumblebees. Behav. Ecol. Sociobiol. 2010, 65, 1015–1027. [Google Scholar] [CrossRef]
- Das, A.; Lee, S.H.; Hyun, T.K.; Kim, S.W.; Kim, J.Y. Plant volatiles as method of communication. Plant Biotechnol. Rep. 2012, 7, 9–26. [Google Scholar] [CrossRef]
- Caser, M.; Scariot, V. The contribution of volatile organic compounds (VOCs) emitted by petals and pollen to the scent of garden roses. Horticulturae 2022, 8, 1049. [Google Scholar] [CrossRef]
- Li, S.S.; Zhang, L.; Sun, M.; Lv, M.W.; Yang, Y.; Xu, W.Z.; Wang, L.S. Biogenesis of flavor-related linalool is diverged and genetically conserved in tree peony (Paeonia × suffruticosa). Hortic. Res. 2023, 10, 253. [Google Scholar] [CrossRef]
- Ramya, M.; Jang, S.; An, H.-R.; Lee, S.-Y.; Park, P.-M.; Park, P.H. Volatile organic compounds from Orchids: From synthesis and function to gene regulation. Int. J. Mol. Sci. 2020, 21, 1160. [Google Scholar] [CrossRef]
- Aslam, M.Z.; Lin, X.; Li, X.; Yang, N.; Chen, L. Molecular cloning and functional characterization of CpMYC2 and CpBHLH13 transcription factors from Wintersweet (Chimonanthus praecox L.). Plants 2020, 9, 785. [Google Scholar] [CrossRef]
- Weng, S.; Fu, X.; Gao, Y.; Liu, T.; Sun, Y.; Tang, D. Identification and evaluation of aromatic volatile compounds in 26 cultivars and 8 hybrids of Freesia hybrida. Molecules 2021, 26, 4482. [Google Scholar] [CrossRef]
- Wang, S.; Du, Z.; Yang, X.; Wang, L.; Xia, K.; Chen, Z. An integrated analysis of metabolomics and transcriptomics reveals significant differences in floral scents and related gene expression between two varieties of Dendrobium loddigesii. Appl. Sci. 2022, 12, 1262. [Google Scholar] [CrossRef]
- Liu, J.; Hu, H.; Shen, H.; Tian, Q.; Ding, W.; Yang, X.; Wang, L.; Yue, Y. Insights into the cytochrome P450 monooxygenase superfamily in Osmanthus fragrans and the role of OfCYP142 in linalool synthesis. Int. J. Mol. Sci. 2022, 23, 12150. [Google Scholar] [CrossRef]
- Yang, X.L.; Li, H.Y.; Yue, Y.Z.; Ding, W.J.; Xu, C.; Shi, T.T.; Chen, G.W.; Wang, L.G. Transcriptomic analysis of the candidate genes related to aroma formation in Osmanthus fragrans. Molecules 2018, 23, 1604. [Google Scholar] [CrossRef]
- Chiu, Y.T.; Chen, H.C.; Chang, C. The Variation of oncidium rosy sunset Flower volatiles with daily rhythm, flowering period, and flower parts. Molecules 2017, 22, 1468. [Google Scholar] [CrossRef]
- Wang, X.J.; Song, Z.Q.; Ti, Y.J.; Ma, K.F.; Li, Q.W. Comparative transcriptome analysis linked to key volatiles reveals molecular mechanisms of aroma compound biosynthesis in Prunus mume. BMC Plant Biol. 2022, 22, 395. [Google Scholar] [CrossRef]
- Li, T.; Zhao, X.; Cao, X. Volatile metabolome and aroma differences of six cultivars of Prunus mume blossoms. Plants 2023, 12, 308. [Google Scholar] [CrossRef]
- Abbas, F.; To, Y.G.; Zhou, Y.W.; Yu, R.C.; Imran, M.; Amanullah, S.; Rothenberg, D.O.; Wang, Q.; Wang, L.; Fan, Y.P. Functional characterization of Hedychium coronarium J. Koenig MYB132 confers the potential role in floral aroma synthesis. Plants 2021, 10, 2014. [Google Scholar] [CrossRef]
- Hu, Z.H.; Tang, B.; Wu, Q.; Zheng, J.; Leng, P.S.; Zhang, K.Z. Transcriptome sequencing analysis reveals a difference in monoterpene biosynthesis between scented Lilium ‘Siberia’ and unscented Lilium ‘Novano’. Front. Plant Sci. 2017, 8, 1351. [Google Scholar] [CrossRef]
- Kong, Y.; Bai, J.; Lang, L.X.; Bao, F.; Dou, X.Y.; Wang, H.; Shang, H.Z. Variation in floral scent compositions of different lily hybrid groups. J. Am. Soc. Hortic. Sci. 2017, 142, 175–183. [Google Scholar] [CrossRef]
- Liu, S.J.; Wang, H.N.; Fu, Y.C.; Yan, W.X.; Hu, Z.H.; Leng, P.S. Cloning and functional analysis of LiCMK gene in Lilium ‘Siberia’. Biotechnol. Bull. 2023, 39, 1–10. [Google Scholar] [CrossRef]
- Yan, W.X.; Zhang, Q.; Li, S.J.; Leng, P.S.; Hu, Z.H. Cloning and expression analysis of LiHDS gene from Lilium ‘Siberia’. Plant Physiol. J. 2023, 59, 1–11. [Google Scholar] [CrossRef]
- Zhang, T.X.; Sun, M.; Guo, Y.H.; Shi, X.J.; Yang, Y.J.; Chen, J.T.; Zheng, T.C.; Han, Y.; Bao, F.; Ahmad, S. Overexpression of LiDXS and LiDXR from Lily (Lilium ‘Siberia’) enhances the terpenoid content in tobacco flowers. Front. Plant Sci. 2018, 9, 909. [Google Scholar] [CrossRef]
- Zhang, T.X.; Guo, Y.H.; Shi, X.J.; Yang, Y.J.; Chen, J.T.; Zhang, Q.X.; Sun, M. Overexpression of LiTPS2 from a cultivar of lily (Lilium ‘Siberia’) enhances the monoterpenoids content in tobacco flowers. Plant Physiol. Biochem. 2020, 151, 391–399. [Google Scholar] [CrossRef]
- Abbas, F.; Ke, Y.G.; Zhou, Y.W.; Ashraf, U.; Li, X.Y.; Yu, Y.Y.; Yue, Y.C.; Ahmad, K.W.; Yu, R.C.; Fan, Y.P. Molecular cloning, characterization and expression analysis of LoTPS2 and LoTPS4 involved in floral scent formation in oriental hybrid Lilium variety ‘Siberia’. Phytochemistry 2020, 173, 112294. [Google Scholar] [CrossRef]
- Abbas, F.; Ke, Y.G.; Yu, R.C.; Fan, Y.P. Functional characterization and expression analysis of two terpene synthases involved in floral scent formation in Lilium ‘Siberia’. Planta 2019, 249, 71–93. [Google Scholar] [CrossRef]
- Li, J.X.; Fan, Y.Q.; Dai, S.T.; Qin, N.; Song, Y.H.; Zhu, C.C.; Wang, C.Y.; Weng, H.Y. Application progress of bHLH transcription factors in genetic engineering of plants against abiotic stress. Jiangsu Agric. Sci. 2022, 50, 1–9. [Google Scholar] [CrossRef]
- Li, T.T.; Yang, S.Y.; Kang, X.K.; Lei, W.; Qiao, K.; Zhang, D.W.; Lin, H.H. The bHLH transcription factor gene AtUPB1 regulates growth by mediating cell cycle progression in Arabidopsis. Biochem. Biophys. Res. Commun. 2019, 518, 565–572. [Google Scholar] [CrossRef]
- Dong, Y.; Wang, C.P.; Han, X.; Tang, S.; Liu, S.; Xia, X.; Yin, W.L. A novel bHLH transcription factor PebHLH35 from Populus euphratica confers drought tolerance through regulating stomatal development, photosynthesis and growth in Arabidopsis. Biochem. Biophys. Res. Commun. 2014, 450, 453–458. [Google Scholar] [CrossRef]
- Groszmann, M.; Bylstra, Y.; Lampugnani, E.R.; Smyth, D.R. Regulation of tissue-specific expression of SPATULA, a bHLH gene involved in carpel development, seedling germination, and lateral organ growth in Arabidopsis. J. Exp. Bot. 2010, 61, 1495–1508. [Google Scholar] [CrossRef]
- Yao, P.F.; Sun, Z.X.; Li, C.L.; Zhao, X.R.; Li, M.F.; Deng, R.Y.; Huang, Y.J.; Zhao, H.X.; Chen, H.; Wu, Q. Overexpression of fagopyrum tataricum FtbHLH2 enhances tolerance to cold stress in transgenic Arabidopsis. Plant Physiol. Biochem. 2018, 125, 85–94. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, S.; Tian, Y.; Wang, Q.; Chen, S.; Li, H.; Ma, C.; Li, H. Functional characterization of a sugar beet BvbHLH93 transcription factor in salt stress tolerance. Int. J. Mol. Sci. 2021, 22, 3669. [Google Scholar] [CrossRef]
- Xu, W.R.; Zhang, N.B.; Jiao, Y.T.; Li, R.M.; Xiao, D.M.; Wang, Z.P. The grapevine basic helix-loop-helix (bHLH) transcription factor positively modulates CBF-pathway and confers tolerance to cold-stress in Arabidopsis. Mol. Biol. Rep. 2014, 41, 5329–5342. [Google Scholar] [CrossRef]
- Waseem, M.; Rong, X.Y.; Li, Z.G. Dissecting the role of a basic Helix-Loop-Helix transcription factor, SlbHLH22, under salt and drought stresses in transgenic Solanum lycopersicum L. Front. Plant Sci. 2019, 10, 734. [Google Scholar] [CrossRef]
- Wang, X.R.; Wang, Y.H.; Jia, M.; Zhang, R.R.; Liu, H.; Xu, Z.S.; Xiong, A.S. The phytochrome-interacting factor DcPIF3 of carrot plays a positive role in drought stress by increasing endogenous ABA level in Arabidopsis. Plant Sci. 2022, 322, 111367. [Google Scholar] [CrossRef]
- Li, J.L.; Wang, T.; Han, J.; Ren, Z.H. Genome-wide identification and characterization of cucumber bHLH family genes and the functional characterization of CsbHLH041 in NaCl and ABA tolerance in Arabidopsis and cucumber. BMC Plant Biol. 2020, 20, 272. [Google Scholar] [CrossRef]
- Li, P.H.; Chen, B.B.; Zhang, G.Y.; Chen, L.X.; Dong, Q.; Wen, J.Q.; Mysore, K.S.; Zhao, J. Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytol. 2016, 210, 905–921. [Google Scholar] [CrossRef]
- Zhu, J.H.; Xia, D.N.; Xu, J.; Guo, D.; Li, H.L.; Wang, Y.; Mei, W.L.; Peng, S.Q. Identification of the bHLH gene family in Dracaena cambodiana reveals candidate genes involved in flavonoid biosynthesis. Ind. Crops Prod. 2020, 150, 112407. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Xu, Q.S.; Zhao, S.Q.; Xia, X.B.; Yan, X.M.; An, Y.L.; Mi, X.Z.; Guo, L.X.; Samarina, L.; Wei, C.L. Comprehensive co-expression analysis provides novel insights into temporal variation of flavonoids in fresh leaves of the tea plant (Camellia sinensis). Plant Sci. 2020, 290, 110306. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.Y.; Cheng, G.X.; Zhang, Z.; Sun, J.T.; Ali, M.; Jia, Q.L.; Luo, D.X.; Gong, Z.H.; Li, D.W. CaMYC, A Novel transcription factor, regulates anthocyanin biosynthesis in color-leaved pepper (Capsicum annuum L.). J. Plant Growth Regul. 2018, 38, 574–585. [Google Scholar] [CrossRef]
- Song, M.X.; Wang, H.M.; Wang, Z.; Huang, H.T.; Chen, S.; Ma, H.Q. Genome-wide characterization and analysis of bHLH transcription factors related to anthocyanin biosynthesis in Fig (Ficus carica L.). Front Plant Sci. 2021, 12, 730692. [Google Scholar] [CrossRef]
- Xu, J.S.; van Herwijnen, Z.O.; Drager, D.B.; Sui, C.; Haring, M.A.; Schuurink, R.C. SlMYC1 regulates type VI glandular trichome formation and terpene biosynthesis in tomato glandular Cells. Plant Cell 2018, 30, 2988–3005. [Google Scholar] [CrossRef]
- Dong, Y.M.; Zhang, W.Y.; Li, J.R.; Wang, D.; Bai, H.T.; Li, H.; Shi, L. The transcription factor LaMYC4 from lavender regulates volatile terpenoid biosynthesis. BMC Plant Biol. 2022, 22, 289. [Google Scholar] [CrossRef]
- Mertens, J.; Pollier, J.; Vanden Bossche, R.; Lopez-Vidriero, I.; Franco-Zorrilla, J.M.; Goossens, A. The bHLH transcription factors TSAR1 and TSAR2 regulate triterpene saponin biosynthesis in Medicago truncatula. Plant Physiol. 2016, 170, 194–210. [Google Scholar] [CrossRef]
- Yin, J.; Li, X.; Zhan, Y.G.; Li, Y.; Qu, Z.Y.; Sun, L.; Wang, S.; Yang, J.; Xiao, J.L. Cloning and expression of BpMYC4 and BpbHLH9 genes and the role of BpbHLH9 in triterpenoid synthesis in birch. BMC Plant Biol. 2017, 17, 214. [Google Scholar] [CrossRef]
- Yang, Y.Y.; Ma, B.; Li, Y.Y.; Han, M.Z.; Wu, J.; Zhou, X.F.; Tian, J.; Wang, W.H.; Leng, P.S.; Hu, Z.H. Transcriptome analysis identifies key gene LiMYB305 involved in monoterpene biosynthesis in Lilium ‘Siberia’. Front Plant Sci. 2022, 13, 1021576. [Google Scholar] [CrossRef]
- Sun, Y.Q.; Meng, Y.X.; Zhao, X.Y.; Wang, F.X.; Weng, Q.Y.; Zhao, Z.H.; Liu, Y.H.; Yuan, J.C. Identification and bioinformatics analysis of transcription factor family gene bHLH in Foxtail Millet. Seed 2021, 40, 45–55. [Google Scholar] [CrossRef]
- Zhang, H.X.; Leng, P.S.; Hu, Z.H.; Zhao, J.; Wang, W.H.; Xu, F. The floral scent emitted from Lilium ‘Siberia’ at different flowering stages and diurnal variation. Acta Hortic. Sin. 2013, 40, 693–702. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, M.F.; Xiao, W.; Cheng, X.Q.; Zhang, X.H.; Dong, R. Advances in research on genetic transformation and nanomagnetic beads of Lily. Mol. Plant Breed. 2020, 18, 4657–4664. [Google Scholar] [CrossRef]
- Qiu, N.; Tao, G.; Li, Q.K.; Qiu, Y.B.; Liu, Z.Y. Transient gene expression mediated by Agroinfiltration and its application. Mol. Plant Breed. 2009, 7, 1032–1039. [Google Scholar] [CrossRef]
- Tang, Y.; Li, L.F.; Wang, X.Q. Establishment of transient gene expression and virus-induced gene silencing (VIGS) system in Gerbera hybrida petals. Plant Physiol. J. 2017, 53, 505–512. [Google Scholar] [CrossRef]
- Xu, Y.; Han, Y.Q.; Yu, F.; Liu, Z.W.; Wang, Y.Y. Promoting the biosynthesis of catharanthine and vindoline in Catharanthus roseus by overexpressing JAR1. Biotechnol. Bull. 2017, 33, 62–68. [Google Scholar] [CrossRef]
- Huo, L.; Ji, X.Y.; Wang, Y.C. Transient expression conditions of tobacco transformation mediated by Agrobacterium. Mol. Plant Breed. 2016, 14, 80–85. [Google Scholar] [CrossRef]
- Shu, X.J.; Wen, T.J.; Xing, J.Y.; Lu, L.; Hu, Z.F. Isolation of protoplast and establishment of transient expression system in grapevine (Vitis vinifera L.). Acta Bot. Boreali-Occident. Sin. 2015, 35, 1262–1268. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, Y.; Wang, Z. Optimizing transient genetic transformation method on Arabidopsis plant mediated by Agrob acterium tumefaciens. J. Northeast. For. Univ. 2016, 44, 41–44+83. [Google Scholar] [CrossRef]
- Wang, L.; Chen, W.; Liu, Y.; Yang, R.; Yin, X.; Ma, N.; Gao, J. Optimization of Agrobacterium-mediated transient gene expression system and its utilization in RNAi based gene silencing of rose (Rosa hybrida) petals. Chin. J. Agric. Biotechnol. 2014, 22, 133–140. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).