Systematic Analysis of Two Tandem GGDEF/EAL Domain Genes Regulating Antifungal Activities in Pseudomonas glycinae MS82
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Media, and Growth Conditions
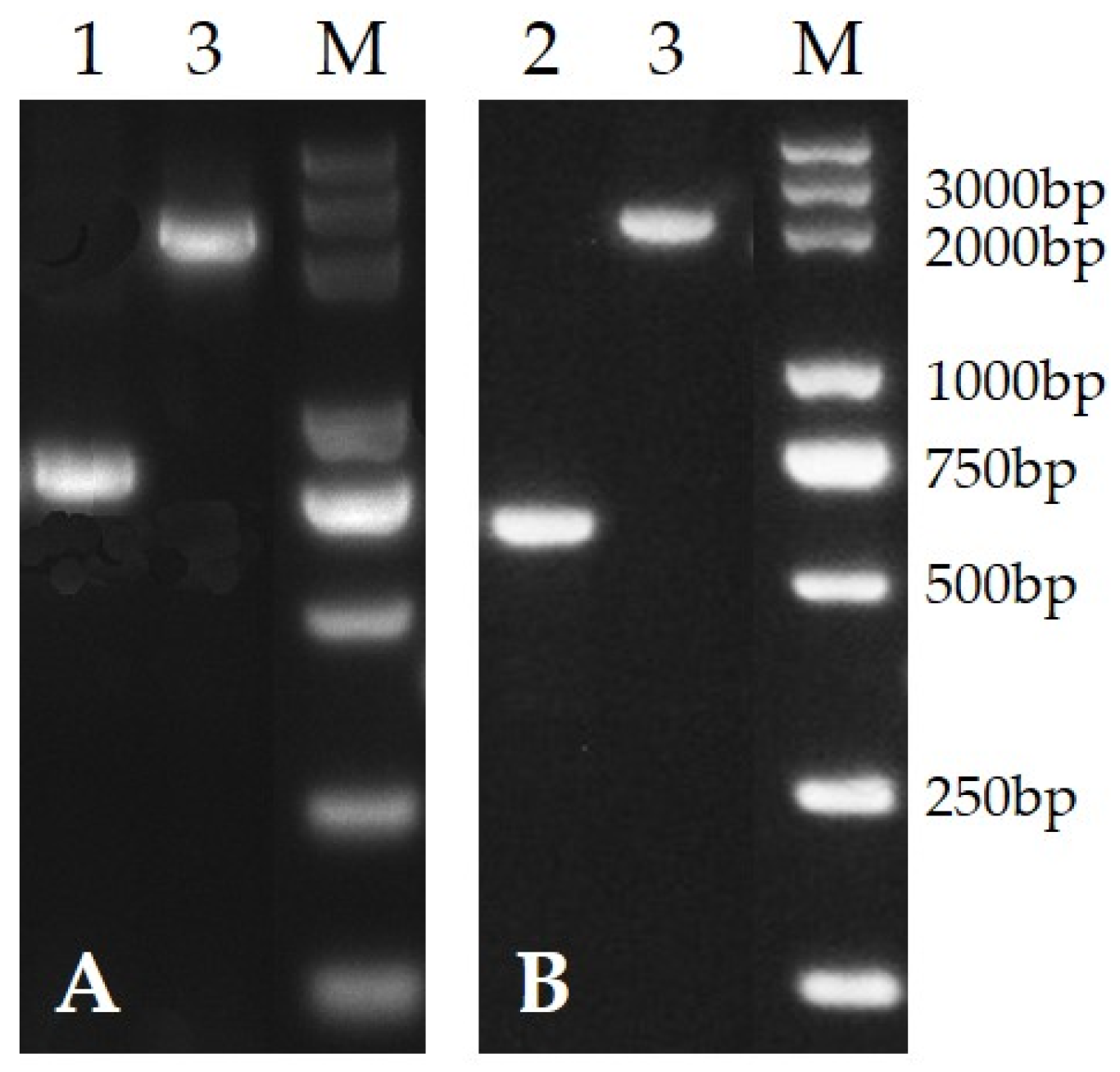
2.2. Construction of PafQ and PafR Deletion Mutants
2.3. Bioassay of Fungistatic Activity
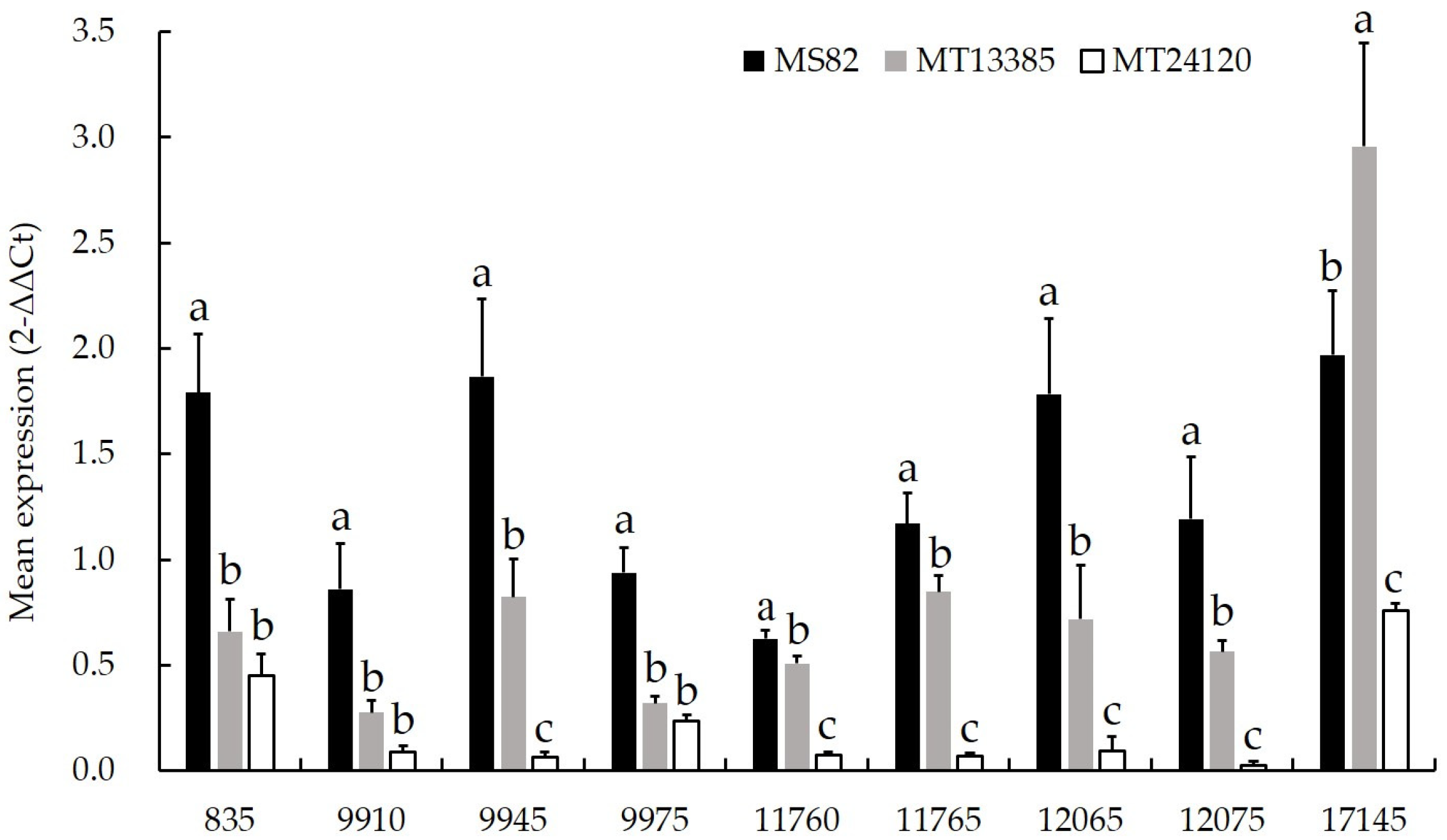
2.4. qRT-PCR Analysis
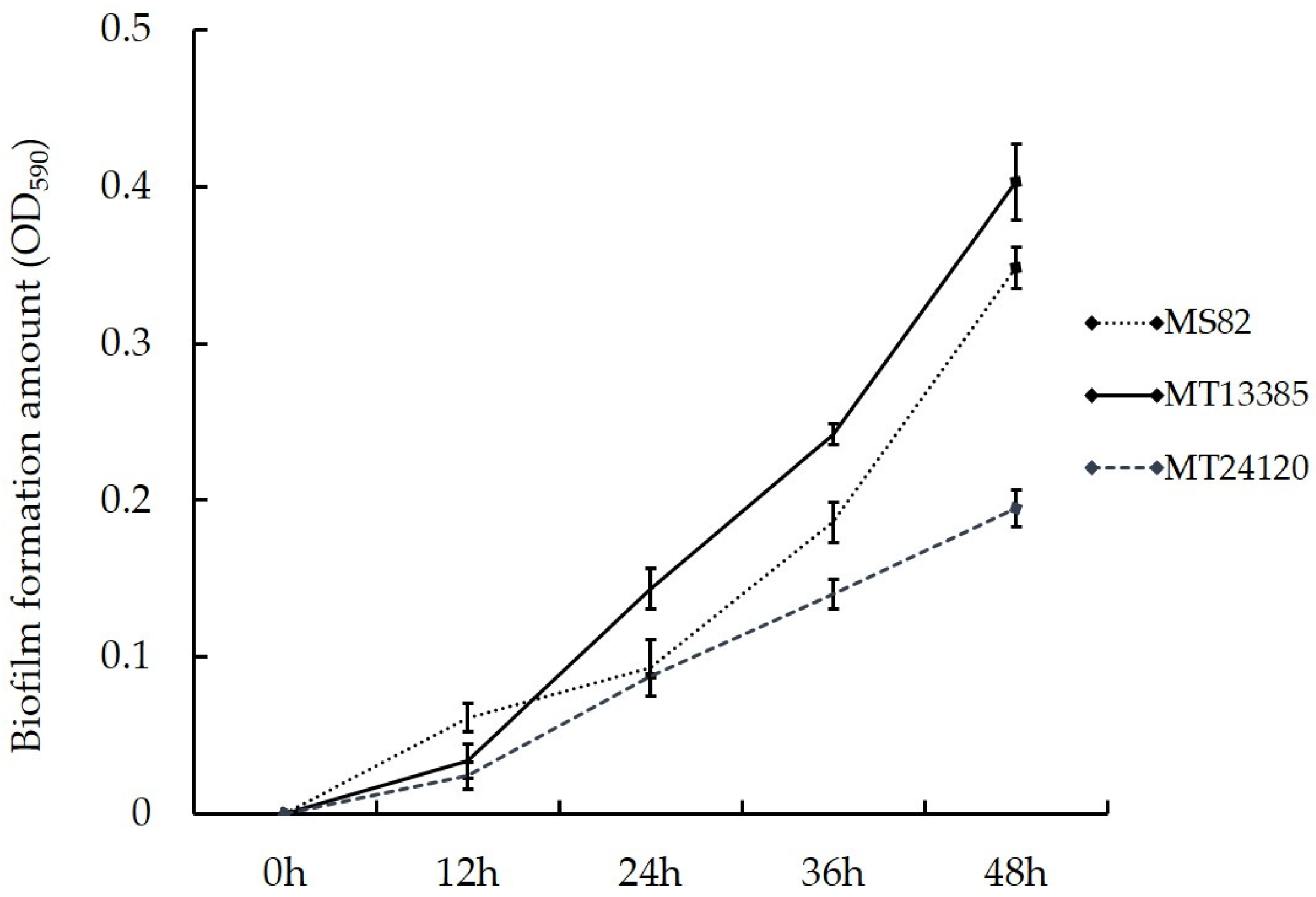
2.5. Analysis of Biofilm Formation
2.6. Analysis of Motility
3. Results
3.1. Deletion of PafQ and PafR
3.2. Bioassay of Fungistatic Activity
3.3. qRT-PCR Analysis
3.4. Analysis of Biofilm Formation
3.5. Analysis of Motility
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, K.; Newman, M.; McInroy, J.A.; Hu, C.H.; Kloepper, J.W. Selection and assessment of plant growth-promoting rhizobacteria for biological control of multiple plant diseases. Phytopathology 2017, 107, 928–936. [Google Scholar] [CrossRef] [PubMed]
- Montelongo-Martínez, L.F.; Hernández-Méndez, C.; Muriel-Millan, L.F.; Hernández-Estrada, R.; Fabian-Del Olmo, M.J.; González-Valdez, A.; Soberón-Chávez, G.; Cocotl-Yañez, M. Unraveling the regulation of pyocyanin synthesis by RsmA through MvaU and RpoS in Pseudomonas aeruginosa ID4365. J. Basic Microbiol. 2023, 63, 51–63. [Google Scholar] [CrossRef]
- Zhang, J.; Mavrodi, D.V.; Yang, M.; Thomashow, L.S.; Weller, D.M. Pseudomonas synxantha 2-79 transformed with pyrrolnitrin biosynthesis genes has improved biocontrol activity against soilborne pathogens of wheat and canola. Phytopathology 2020, 110, 1010–1017. [Google Scholar] [CrossRef]
- Gutiérrez-García, K.; Neira-González, A.; Pérez-Gutiérrez, R.M.; Granados-Ramírez, G.; Zarraga, R.; Wrobel, K.; Barona-Gómez, F.; Flores-Cotera, B.L. Phylogenomics of 2,4-diacetylphloroglucinol-producing Pseudomonas and novel antiglycation endophytes from Piper auritum. J. Nat. Prod. 2017, 80, 1955–1963. [Google Scholar] [CrossRef]
- Zhang, Q.X.; Kong, X.W.; Li, S.Y.; Chen, X.J.; Chen, X.J. Antibiotics of Pseudomonas protegens FD6 are essential for biocontrol activity. Australas. Plant Pathol. 2020, 49, 307–317. [Google Scholar] [CrossRef]
- Pandey, S.; Gupta, S. Evaluation of Pseudomonas sp. for its multifarious plant growth promoting potential and its ability to alleviate biotic and abiotic stress in tomato (Solanum lycopersicum) plants. Sci. Rep. 2020, 10, 20951. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z. Analysis of the complete genome sequence of a rhizosphere-derived Pseudomonas sp. HN3-2 leads to the characterization of a cyclic lipopeptide-type antibiotic bananamide C. 3 Biotech 2022, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Soler-Rivas, C.; Arpin, N.; Olivier, J.M.; Wichers, H.J. WLIP, a lipodepsipeptide of Pseudomonas ‘reactans’, as inhibitor of the symptoms of the brown blotch disease of Agaricus bisporus. J. Appl. Microbiol. 1999, 86, 635–641. [Google Scholar] [CrossRef]
- Tajalipour, S.; Hassanzadeh, N.; Jolfaee, H.K.; Heydarib, A.; Ghasemib, A. Biological control of mushroom brown blotch disease using antagonistic bacteria. Biocontrol Sci. Technol. 2014, 24, 473–484. [Google Scholar] [CrossRef]
- Ma, L.; Wang, X.; Deng, P.; Baird, S.M.; Liu, Y.; Qu, S.; Lu, S. The PafR gene is required for antifungal activity of strain MS82 against Mycogone perniciosa. Adv. Microbiol. 2017, 7, 217–230. [Google Scholar] [CrossRef]
- Ma, L.; Qu, S.; Lin, J.; Jia, J.; Baird, S.M.; Jiang, N.; Li, H.; Hou, L.; Lu, S. The complete genome of the antifungal bacterium Pseudomonas sp. strain MS82. J. Plant Dis. Protect. 2019, 126, 153–160. [Google Scholar] [CrossRef]
- Tagua, V.G.; Molina-Henares, M.A.; Travieso, M.L.; Nisa-Martínez, R.; Quesada, J.M.; Espinosa-Urgel, M.; Ramos-González, M.I. C-di-GMP and biofilm are regulated in Pseudomonas putida by the CfcA/CfcR two-component system in response to salts. Environ. Microbiol. 2022, 24, 158–178. [Google Scholar] [CrossRef] [PubMed]
- Cutruzzolà, F.; Paiardini, A.; Rossi, C.S.; Spizzichino, S.; Paone, A.; Giardina, G.; Rinaldo, S. A conserved scaffold with heterogeneous metal ion binding site: The multifaceted example of HD-GYP proteins. Coordin. Chem. Rev. 2022, 450, 214228. [Google Scholar] [CrossRef]
- Verma, R.K.; Biswas, A.; Kakkar, A.; Lomada, S.K.; Pradhan, B.B.; Chatterjee, S. A bacteriophytochrome mediates interplay between light sensing and the second messenger cyclic di-GMP to control social behavior and virulence. Cell Rep. 2020, 32, 108202. [Google Scholar] [CrossRef]
- Ribbe, J.; Baker, A.E.; Euler, S.; O’Toole, G.A.; Maier, B. Role of cyclic di-GMP and exopolysaccharide in type IV pilus dynamics. J. Bacteriol. 2017, 199, e00859-16. [Google Scholar] [CrossRef]
- Gdaniec, B.G.; Allard, P.-M.; Queiroz, E.F.; Wolfender, J.-L.; van Delden, C.; Köhler, T. Surface sensing triggers a broad-spectrum antimicrobial response in Pseudomonas aeruginosa. Environ. Microbiol. 2020, 22, 3572–3587. [Google Scholar] [CrossRef]
- Blanco-Romero, E.; Garrido-Sanz, D.; Durán, D.; Rivilla, R.; Redondo-Nieto, M.; Martín, M. Regulation of extracellular matrix components by AmrZ is mediated by c-di-GMP in Pseudomonas ogarae F113. Sci. Rep. 2022, 12, 11914. [Google Scholar] [CrossRef] [PubMed]
- Christen, M.; Christen, B.; Folcher, M.; Schauerte, A.; Jenal, U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 2005, 280, 30829–30837. [Google Scholar] [CrossRef]
- Römling, U.; Galperin, M.Y.; Gomelsky, M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013, 77, 1–52. [Google Scholar] [CrossRef]
- Muriel, C.; Blanco-Romero, E.; Trampari, E.; Arrebola, E.; Durán, D.; Redondo-Nieto, M.; Malone, J.G.; Martín, M.; Rivilla, R. The diguanylate cyclase AdrA regulates flagellar biosynthesis in Pseudomonas fluorescens F113 through SadB. Sci. Rep. 2019, 9, 8096. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xia, J.; Su, Z.; Xu, G.; Gomelsky, M.; Qian, G.; Liu, F. Lysobacter PilR, the regulator of type IV pilus synthesis, controls antifungal antibiotic production via a cyclic di-GMP pathway. Appl. Environ. Microbiol. 2017, 83, e03397-16. [Google Scholar] [CrossRef] [PubMed]
- Lennox, E.S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1955, 1, 190–206. [Google Scholar] [CrossRef]
- Jia, J.; Wang, X.; Deng, P.; Ma, L.; Baird, M.S.; Li, X.; Lu, S.E. Pseudomonas glycinae sp. nov. isolated from the soybean rhizosphere. MicrobiologyOpen 2020, 9, e1101. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Qu, S.; Luo, X.; Lu, S.-E.; Liu, Y.; Li, H.; Hou, L.; Lin, J.; Jiang, N.; Ma, L. PafS containing GGDEF-domain regulates life activities of Pseudomonas glycinae MS82. Microorganisms 2022, 10, 2342. [Google Scholar] [CrossRef]
- Wang, X.Q.; Liu, A.X.; Guerrero, A.; Liu, J.; Yu, X.Q.; Deng, P.; Ma, L.; Baird, S.M.; Smith, L.; Li, X.D.; et al. Occidiofungin is an important component responsible for the antifungal activity of Burkholderia pyrrocinia strain Lyc2. J. Appl. Microbiol. 2015, 120, 607–618. [Google Scholar] [CrossRef]
- Huertas-Rosales, Ó.; Romero, M.; Heeb, S.; Espinosa-Urgel, M.; Camara, M.; Ramos-Gonzalez, M.I. The Pseudomonas putida CsrA/RsmA homologues negatively affect c-di-GMP pools and biofilm formation through the GGDEF/EAL response regulator CfcR. Envion. Microbiol. 2017, 19, 3551–3566. [Google Scholar] [CrossRef] [PubMed]
- Smet, J.D.; Wagemans, J.; Hendrix, H.; Staes, I.; Visnapuu, A.; Horemans, B.; Aertsen, A.; Lavigne, R. Bacteriophage mediated interference of the c-di-GMP signaling pathway in Pseudomonas aeruginosa. Microb. Biotechnol. 2020, 14, 967–978. [Google Scholar] [CrossRef]
- Cai, Y.; Hutchin, A.; Craddock, J.; Walsh, M.A.; Webb, J.S.; Tews, I. Differential impact on motility and biofilm dispersal of closely related phosphodiesterases in Pseudomonas aeruginosa. Sci. Rep. 2020, 10, 6232. [Google Scholar] [CrossRef]
- Meissner, A.; Wild, V.; Simm, R.; Rohde, M.; Erck, C.; Bredenbruch, F.; Morr, M.; Römling, U.; Hussler, S. Pseudomonas aeruginosa cupA-encoded fimbriae expression is regulated by a GGDEF and EAL domain-dependent modulation of the intracellular level of cyclic diguanylate. Envion. Microbiol. 2007, 9, 2475–2485. [Google Scholar] [CrossRef]
- Valentini, M.; Filloux, A. Biofilms and cyclic di-GMP (c-di-GMP) signaling: Lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 2016, 291, 12547–12555. [Google Scholar] [CrossRef]
- Liang, F.; Zhang, B.; Yang, Q.; Zhang, Y.; Zheng, D.; Zhang, L.Q.; Yan, Q.; Wu, X. Cyclic di-GMP regulates the quorum-sensing system and biocontrol activity of Pseudomonas fluorescens 2P24 through the RsmA and RsmE proteins. Appl. Environ. Microbiol. 2020, 86, e02016-20. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y. Functional Analysis of the GGDEF/EAL Domain Protein PigX in Serratia plymuthica G3. Ph.D. Thesis, Jiangsu University, Zhenjiang, China, 2016. [Google Scholar]
- Fineran, P.C.; Williamson, N.R.; Lilley, K.S.; Salmond, G.P.C. Virulence and prodigiosin antibiotic biosynthesis in Serratia are regulated pleiotropically by the GGDEF/EAL domain protein, PigX. J. Bacteriol. 2007, 189, 7653–7662. [Google Scholar] [CrossRef] [PubMed]
- Blake, C.; Christensen, M.N.; Kovács, T. Molecular aspects of plant growth promotion and protection by Bacillus subtilis. Mol. Plant Microbe Interact. 2021, 34, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Allard-Massicotte, R.; Tessier, L.; Lecuyer, F.; Lakshmanan, V.; Lucier, J.-F.; Garneau, D.; Caudwell, L.; Vlamakis, H.; Bais, H.P.; Beauregard, P.B. Bacillus subtilis early colonization of Arabidopsis thaliana roots involves multiple chemotaxis receptors. mBio 2016, 7, e01664-16. [Google Scholar] [CrossRef]
- Baker, A.E.; Diepold, A.; Kuchma, S.L.; Scott, J.E.; Ha, D.G.; Orazi, G.; Armitage, J.P.; O’Toole, G.A. PilZ domain protein FlgZ mediates cyclic di-GMP-dependent swarming motility control in Pseudomonas aeruginosa. J. Bacteriol. 2016, 198, 1837–1846. [Google Scholar] [CrossRef]
- Xu, L.; Xin, L.; Zeng, Y.; Yam, J.K.H.; Ding, Y.; Venkataramani, P.; Cheang, Q.W.; Yang, X.; Tang, X.; Zhang, L.-H.; et al. A cyclic di-GMP-binding adaptor protein interacts with a chemotaxis methyltransferase to control flagellar motor switching. Sci. Signal. 2016, 9, ra102/1–ra102/12. [Google Scholar] [CrossRef]
- Yan, X.-F.; Xin, L.; Yen, J.T.; Zeng, Y.; Jin, S.; Cheang, Q.W.; Fong, R.A.C.Y.; Chiam, K.-H.; Liang, Z.-X.; Gao, Y.-G. Structural analyses unravel the molecular mechanism of cyclic di-GMP regulation of bacterial chemotaxis via a PilZ adaptor protein. J. Biol. Chem. 2018, 293, 100–111. [Google Scholar] [CrossRef]
- Deepa, N.; Chauhan, S.; Kumari, P.; Rai, A.K.; Tandon, S.; Singh, A. Linalool reduces the virulence of Pseudomonas syringae pv. tomato DC 3000 by modulating the PsyI/PsyR quorum-sensing system. Microb. Pathog. 2022, 173, 105884. [Google Scholar] [CrossRef]
- Bense, S.; Witte, J.; Preuße, M.; Koska, M.; Pezoldt, L.; Dröge, A.; Hartmann, O.; Müsken, M.; Schulze, J.; Fiebig, T.; et al. Pseudomonas aeruginosa post-translational responses to elevated c-di-GMP levels. Mol. Microbiol. 2022, 117, 1213–1226. [Google Scholar] [CrossRef]
- Jain, R.; Sliusarenko, O.; Kazmierczak, B.I. Interaction of the cyclic-di-GMP binding protein FimX and the Type 4 pilus assembly ATPase promotes pilus assembly. PLoS Pathog. 2017, 13, e1006594. [Google Scholar] [CrossRef]
- Whitfield, G.B.; Marmont, L.S.; Ostaszewski, A.; Rich, J.D.; Whitney, J.C.; Parsek, M.R.; Harrison, J.J.; Howell, P.L. Pel polysaccharide biosynthesis requires an inner membrane complex comprised of PelD, PelE, PelF, and PelG. J. Bacteriol. 2020, 202, e00684-19. [Google Scholar] [CrossRef]
- Roelofs, K.G.; Jones, C.J.; Helman, S.R.; Shang, X.; Orr, M.W.; Goodson, J.R.; Galperin, M.Y.; Yildiz, F.H.; Lee, V.T. Systematic identification of cyclic-di-GMP binding proteins in Vibrio cholerae reveals a novel class of cyclic-di-GMP-binding ATPases associated with type II secretion systems. PLoS Pathog. 2015, 27, e1005232. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Ahmad, I.; Blanka, A.; Schottkowski, M.; Cimdins, A.; Galperin, M.Y.; Römling, U.; Gomelsky, M. GIL, a new c-di-GMP-binding protein domain involved in regulation of cellulose synthesis in enterobacteria. Mol. Microbiol. 2014, 93, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Zhou, E.; Seminara, A.B.; Kim, S.; Hall, C.L.; Wang, Y.; Lee, V.T. Thiol-benzotriazolo-quinazolinone inhibits Alg44 binding to c-di-GMP and reduces alginate production by Pseudomonas aeruginosa. ACS Chem. Biol. 2017, 12, 3076–3085. [Google Scholar] [CrossRef] [PubMed]
- Molina-Henares, M.A.; Ramos-González, M.I.; Daddaoua, A.; Fernández-Escamilla, A.M.; Espinosa-Urgel, M. FleQ of Pseudomonas putida KT2440 is a multimeric cyclic diguanylate binding protein that differentially regulates expression of biofilm matrix components. Res. Microbiol. 2017, 168, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Chuah, M.L.C.; Dong, X.; Xie, K.; Luo, Z.; Tang, K.; Liang, Z.X. Binding of cyclic diguanylate in the non-catalytic EAL domain of FimX induces a long-range conformational change. J. Biol. Chem. 2011, 286, 2910–2917. [Google Scholar] [CrossRef]
- Nie, H.; Xiao, Y.; He, J.; Liu, H.; Nie, L.; Chen, W.; Huang, Q. Phenotypic–genotypic analysis of GGDEF/EAL/HD-GYP domain-encoding genes in Pseudomonas putida. Environ. Microbiol. Rep. 2020, 12, 38–48. [Google Scholar] [CrossRef]
- Tal, R.; Wong, H.C.; Calhoon, R.; Gelfand, D.; Benziman, M. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: Genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 1998, 180, 4416–4425. [Google Scholar] [CrossRef]
- Wang, F.; Wang, Y.; Cen, C.; Fu, L.; Wang, Y. A tandem GGDEF-EAL domain protein-regulated c-di-GMP signal contributes to spoilage-related activities of Shewanella baltica OS155. Appl. Microbiol. Biotechnol. 2020, 104, 2205–2216. [Google Scholar] [CrossRef]
- Wang, Y.; Hay, I.D.; Rehman, Z.U.; Rehm, B.H.A. Membrane-anchored MucR mediates nitrate-dependent regulation of alginate production in Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2015, 99, 7253–7265. [Google Scholar] [CrossRef]
- Hay, I.D.; Remminghorst, U.; Rehm, B.H.A. MucR, a novel membrane-associated regulator of alginate biosynthesis in Pseudomonas aeruginosa. Appl. Environ. Microb. 2009, 75, 1110–1120. [Google Scholar] [CrossRef]
- Li, Y.; Heine, S.; Entian, M.; Sauer, K.; Frankenberg-Dinkel, N. NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by an MHYT domain-coupled phosphodiesterase. J. Bacteriol. 2013, 195, 3531–3542. [Google Scholar] [CrossRef] [PubMed]
- Phippen, C.W.; Mikolajek, H.; Schlaefli, H.G.; Keevil, C.W.; Webb, J.S.; Tews, I. Formation and dimerization of the phosphodiesterase active site of the Pseudomonas aeruginosa MorA, a bi-functional c-di-GMP regulator. FEBS Lett. 2014, 588, 4631–4636. [Google Scholar] [CrossRef]
- Choy, W.-K.; Zhou, L.; Syn, C.K.-C.; Zhang, L.-H.; Swarup, S. MorA defines a new class of regulators affecting flagellar development and biofilm formation in diverse Pseudomonas species. J. Bacteriol. 2004, 186, 7221–7228. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, A.; Ramachandran, M.; Suriyanarayanan, T.; Wong, C.C.; Swarup, S. Global regulator MorA affects virulence-associated protease secretion in Pseudomonas aeruginosa PAO1. PLoS ONE 2015, 10, e0123805. [Google Scholar] [CrossRef] [PubMed]
- Katharios-Lanwermeyer, S.; Whitfield, G.B.; Howell, P.L.; O’Toole, G.A. Pseudomonas aeruginosa uses c-di-GMP phosphodiesterases RmcA and MorA to regulate biofilm maintenance. mBio 2021, 12, e03384-20. [Google Scholar] [CrossRef]


| Strains and Plasmids | Genotype or Phenotype 1 | Source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | supE44 ΔlacU169 (Φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | TSINGKE, Beijing, China |
| S17-1λ | RP4-2(Km::Tn7,Tc::Mu-1), pro-82, LAMpir, recA1, endA1, thiE1, hsdR17, creC510 | WEIDI, Shanghai, China |
| P. gylcinae | ||
| MS82 | Ampr 1, Wild type | [23] |
| MT13385 | Ampr, PafQ deletion mutant derived from MS82 | This work |
| MT24120 | Ampr, PafR deletion mutant derived from MS82 | This work |
| T. virens | ||
| NJ1 | A fungal pathogen collected from Pleurotus ostreatus substrate | Lab stock |
| Plasmids and vectors | ||
| pEX18GM | Gmr, Suicide vector, SacB | FENGHUI, Changsha, China |
| pEX18-13385 | Gmr, up- and down-stream region of PafQ in PEX18GM | This work |
| pEX18-24120 | Gmr, up- and down-stream region of PafR in PEX18GM | This work |
| Primer Name | Sequence 5′→3′ | Restriction Enzyme Site 1 | Length (bp) | Annealing Temperature (°C) |
|---|---|---|---|---|
| 13385-F1 13385-R1 | CGGAATTCGAATACCTCAACAGACACTC 1 GGGGTACCCCTTGGCAGTAGCAATGCGC | Eco RI Kpn I | 387 | 55 |
| 13385-F2 13385-R2 | GGGGTACCGGAAATCACCGAAACCACCG GCTCTAGAATGACCTGCGATTAGCGGCT | Kpn I Xba I | 425 | 55 |
| 24120-F1 24120-R1 | CGGAATTCCAGCCAGACCGCAGGATTAC GGGGTACCGCTGTCGCTGAGGATTTTTC | Eco RI Kpn I | 404 | 55 |
| 24120-F2 24120-R2 | GGGGTACCGCGACGAGGTTCAGGGTTAT GCTCTAGAATCGGGGGCAGAAAAGGGG | Kpn I Xba I | 302 | 55 |
| Gene Locus_tag 1 | Gene Function | Primer Name | Primer Sequence 5′→3′ |
|---|---|---|---|
| DBV33_00020 | DNA gyrase subunit B | gyrB-F | CGGCACCCAGATTCACTT |
| gyrB-R | GGAGTTGAGGAAGGACAGTT | ||
| DBV33_00835 | adenylyl-sulfate kinase | 835-F | AGTCGTGGTCTGCAAAGTGT |
| 835-R | CTGCACCAGACCTCGCAATA | ||
| DBV33_09910 | non-ribosomal peptide synthetase (NRPS) | 9910-F | CGTCAGACTGCTCAACACCT |
| 9910-R | TTGACCGATCGGCATTGTCA | ||
| DBV33_09945 | membrane dipeptidase | 9945-F | ATCGGGTTCAAGGACAACCC |
| 9945-R | TCCTTGTCGACTTCGTTCCC | ||
| DBV33_09975 | ornithine monooxygenase | 9975-F | CAACAATGCCACCGGTGAAG |
| 9975-R | AAGCCCTGCATGTACAGACC | ||
| DBV33_11760 | NRPS | 11760-F | CTGGCAGCGATCCATGTGTA |
| 11760-R | TGAATGACAACTCGCGACCA | ||
| DBV33_11765 | NRPS | 11765-F | AGCATCTGGACGAACCTGTG |
| 11765-R | CAGGTCGAGGCGGAAGTATC | ||
| DBV33_12065 | NRPS | 12065-F | GTTGAAGTGTGGCCGTTGTC |
| 12065-R | CGTTGCTGATCCGGACGATA | ||
| DBV33_12075 | NRPS | 12075-F | GGTATCGGGCCAATCCTGAG |
| 12075-R | ACCGCTGCCACTCAAATACA | ||
| DBV33_17145 | (2Fe-2S)-binding protein | 17145-F | CAATGCACTGCCTAGAAAGAACC |
| 17145-R | TGAGCACGGTTTCGCCAATAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, J.; Qu, S.; Chen, X.; Li, H.; Hou, L.; Lu, S.-E.; Xu, P.; Jiang, N.; Ma, L. Systematic Analysis of Two Tandem GGDEF/EAL Domain Genes Regulating Antifungal Activities in Pseudomonas glycinae MS82. Horticulturae 2023, 9, 446. https://doi.org/10.3390/horticulturae9040446
Lin J, Qu S, Chen X, Li H, Hou L, Lu S-E, Xu P, Jiang N, Ma L. Systematic Analysis of Two Tandem GGDEF/EAL Domain Genes Regulating Antifungal Activities in Pseudomonas glycinae MS82. Horticulturae. 2023; 9(4):446. https://doi.org/10.3390/horticulturae9040446
Chicago/Turabian StyleLin, Jinsheng, Shaoxuan Qu, Xianyi Chen, Huiping Li, Lijuan Hou, Shi-En Lu, Ping Xu, Ning Jiang, and Lin Ma. 2023. "Systematic Analysis of Two Tandem GGDEF/EAL Domain Genes Regulating Antifungal Activities in Pseudomonas glycinae MS82" Horticulturae 9, no. 4: 446. https://doi.org/10.3390/horticulturae9040446
APA StyleLin, J., Qu, S., Chen, X., Li, H., Hou, L., Lu, S.-E., Xu, P., Jiang, N., & Ma, L. (2023). Systematic Analysis of Two Tandem GGDEF/EAL Domain Genes Regulating Antifungal Activities in Pseudomonas glycinae MS82. Horticulturae, 9(4), 446. https://doi.org/10.3390/horticulturae9040446





