Abstract
MIKCC-type MADS-box genes are involved in floral organ identity determination but remain less studied in the Malus lineage. Based on the conserved domains of this gene family, we identified 341 genes among 13 species. Classification results showed that the MIKCC-type were generated later than the M-type, after the formation of Chlamydomonas reinhardtii. By phylogenetic analysis, three different groups were divided among 12 plant species, and one group was an ancestral MIKCC-type MADS-box homologous gene cluster from lower moss to higher flowering plants. Comparative analysis of these genes in A. thaliana and Malus lineages revealed a similar pattern evolutionary relationship with the phylogenetic analysis. Three classes of genes of the ABC model in A. thaliana had orthologous genes in the Malus species, but they experienced different evolutionary events. Only a whole-genome duplication (WGD) event was considered to act on the expansion of ABC-model-related genes in the Malus lineage. Additionally, the expression pattern of genes showed to be involved in flowering development stages and anther development processes among different M. domestica cultivars. This study systematically traced the evolutionary history and expansion mechanism of the MIKCC-type MADS-box gene family in plants. The results also provided novel insights for ABC model research of flower development in the Malus lineage.
1. Introduction
MADS-box genes are an important class of transcription factors and play critical roles in diverse biological functions, particularly in plant vegetative growth and reproductive development [1,2]. The name MADS-box is derived from the four first letters of “MCM1” in Saccharomyces cerevisiae, “AGAMOUS” in Arabidopsis, “DEFICIENS” in Antirrhinum, and “SRF4” in humans [3,4]. Proteins encoded by these genes contain a highly conserved MADS domain containing approximately 60 amino acids, which act as cis-regulatory elements in promoters or enhancers of genes by binding to specific DNA sequences [5]. Based on their conserved protein domains, gene structures, and phylogenetic relationships with A. thaliana proteins, members of the MADS-box gene family are divided into two lineages, type I and type II [6]. Type I proteins consists of Mα, Mβ, and Mγ classes and possess only a MADS-box domain, while type II proteins contain additional domains such as the intervening (I) domain (responsible for protein dimerization), keratin-like (K) domain (involved in protein–protein interactions), and C-terminal (C) domain (required for interacting with other proteins and regulating transcriptional activation) [5,7,8,9]. According to structure divergence at the I domain, the MIKC-type genes are classified into two subtypes, MIKCC and MIKC* [10,11]. The majority of functionally known MADS-box genes are MIKCC-type genes [12].
MIKCC-type genes prominently present during all stages of flower development, from very early (floral transition) to very late (ovule and fruit development) [13,14]. The most primary members of the MIKCC-type MADS subfamilies are the floral organ identity genes, which establish the basic bauplan of angiosperm flowers [15]. On the basis of the ABC + DE model, A + E-, A + B + E-, B + C + E-, C + E-, and D + E-class genes determine sepal, petal, stamen, carpel, and ovule identities, respectively [14,16,17,18]. The MIKCC-type genes described in the ABC(DE) model include the following: A-class gene APETALA1 (AP1), B-class genes APETALA3 (AP3) and PISTILLATA (PI), C-class gene AGAMOUS (AG), D-class gene SEEDSTICK (STK), and E-class gene SEPALLATA (SEP, including SEP1, SEP2, SEP3, and SEP4) [19,20]. Based on the remarkable significance of MIKCC-type MADS-box genes in flower pattern formation, the MIKCC-type gene family has been reported in various plant species, such as Arabidopsis, rice, maize, soybean, petunia, Ziziphus jujuba, and Orchidaceae [21,22,23,24,25,26,27]. However, in the Malus lineage, MIKCC-type genes were only identified relative to their fleshy fruit development [28]. To date, limited genome-wide analyses of MIKCC-type MADS-box genes in Malus providing novel insights for the ABC(DE) model of flower development have been reported, except that the domesticated apple experienced a recent whole-genome duplication, and this will assist in knowing the expansion of the MIKCC-type MADS-box gene family in the Malus genus [29].
Here, we identified members of the MIKCC-type MADS-box gene family in 13 genome-sequenced species and used them to perform phylogenetic analysis among these different genomes, as well as conducted comparative analysis of MIKCC-type MADS-box genes between A. thaliana and Malus species. Additionally, we investigated the influences of whole-genome duplication (WGD) and tandem duplication (TD) events on MIKCC-type MADS-box gene family members in the Malus genus. The ABC model of flower-development-related genes in the MIKCC-type MADS-box gene family were also analyzed after including the entire gene family. Furthermore, we analyzed the expression patterns of different MIKCC-type MADS-box genes in different stages of flowering and anther development among different M. domestica cultivars and detected the expression divergence of the ABC model of flower-development-related genes among different tissues in two types of expression experiments in M. domestica. Our objectives were as follows: (1) tracing the evolutionary history and expansion mechanism of MIKCC-type MADS-box gene family in plants; (2) providing novel insights for the ABC model of flower development in the Malus lineage; and (3) offering a model for universal application for the study of gene families in plants.
2. Materials and Methods
2.1. Resources
Genome sequences of Amborella trichopoda (v1.0), Chlamydomonas reinhardtii (v5.6), Citrus sinensis (v1.1), Fragaria vesca (v2.0.a2), Gossypium raimondii (v2.1), Malus × domestica (v1.1), Oryza sativa (v7.0), Physcomitrella patens (v3.3), Prunus persica (v2.1), Selaginella moellendorffii (v1.0), and Vitis vinifera (v2.1) were retrieved from Phytozome v12.1 (https://phytozome.jgi.doe.gov/pz/portal.html (accessed on 12 December 2022) [30]. Araport11 genome data of Arabidopsis thaliana were downloaded from TAIR10 (https://www.arabidopsis.org/ (accessed on 12 December 2022) [31], and the genome assembly and annotation data of Malus baccata (v1.0) were downloaded from CNGB with Project ID CNA0002537 (https://db.cngb.org/search/project/CNP0000421/ (accessed on 15 December 2022). For the conserved domains of MIKCC-type MADS-box transcription factor genes, the profile HMMs of the SRF-TF domain (PF00319.18) and K-box domain (PF01486.17) were retrieved from Pfam 32.0 (September 2018, 17,929 entries) (http://pfam.xfam.org/ (accessed on 20 December 2022) [32]. Two RNA-seq datasets of M. domestica were downloaded from the Sequence Read Archive (SRA) database with accession numbers: PRJNA302879 and PRJNA419119 [33].
2.2. Classification
First, the putative MIKCC-type MADS-box transcription factor genes of 13 algae, bryophytes, pteridophytes, and angiosperms were classified by the HMMER (v3.2.1-foss-2018b) program with ‘trusted cutoff’ as the threshold [34]. All putative MIKCC-type MADS-box transcription factor protein sequences were used to implement multiple sequence alignments through CLUSTALW2 with the protein weight matrix Gonnet [35]. Second, higher conserved MIKCC-type MADS-box transcription factor protein sequences were used to construct species-specific profile HMMs of SRF-TF and K-box domains among the different species using the ‘hmmbuild’ module by HMMER (v3.2.1-foss-2018b). Using the latest profile HMMs of MIKCC-type MADS-box transcription factor genes, the real target genes were identified from the 13 different species with HMMER (v3.2.1-foss-2018b) [34]. Finally, InterProScan was employed to investigate the validation of real MIKCC-type MADS-box transcription factor genes among the different species [36].
2.3. Phylogenetic Analysis of MIKCC-Type MADS-Box Genes
Using MIKCC-type MADS-box transcription factor genes, CLUSTALW2 was employed to perform multiple sequence alignments (MSA) with protein sequences of target genes from the 13 different species. Then, the MSA files with “meg” format were used to construct a phylogenetic tree through MEGA 7 with the maximum likelihood (ML) statistical method and 1000 bootstrap replications [37]. The phylogeny analysis of MIKCC-type MADS-box transcription factor genes in A. thaliana and Malus species followed the above procedures.
2.4. Analysis of Whole-Genome Duplication and Collinearity
All-against-all BLAST was implemented to M. baccata and M. domestica protein sequences for obtaining paralogous gene pairs in M. baccata and M. domestica genomes separately [38]. MCScanX toolkit was used to identify paralogous chromosomal regions using paralogous gene pairs within different Malus genomes with the parameters e = 1 × 10−20, u = 1, and s = 5 [39]. All paralogous chromosomal regions in separate M. baccata and M. domestica genomes constituted two subgenomes in two target Malus genomes, which were brought from a WGD event in the Malus lineage.
For collinear analysis among A. thaliana and Malus genomes, MCScanX toolkit was used to identify orthologous chromosomal regions using orthologous gene pairs with the above parameters, and then collinear gene pairs were obtained.
2.5. Analysis of Tandem Duplication in Malus Lineage
Following the identification of tandemly duplicated genes in PTGBase, all-against-all BLAST of protein sequences in Malus species was employed to identify the paralogous gene pairs in individual species with an E-value cutoff ≤ 1 × 10−20 [40]. After confirming that high similarity gene pairs anchored on closer location of same genomic regions, tandemly duplicated genes were obtained in different Malus species, and one unrelated gene was also allowed within one tandem array.
2.6. Expression Analysis of MIKCC-Type MADS-Box Transcription Factor Genes in M. domestica
Using the downloaded RNA-seq data of different samples from SRA in NCBI, all raw data were trimmed and cleaned by Trimmomatic (v0.33-Java-1.8.0_144) [41], and all clean short-reads were mapped to M. domestica reference genome sequences through STAR (v2.5.3a-foss-2016b) [42]. The featureCounts function in the Rsubread (http://www.bioconductor.org (accessed on 26 December 2022) software package (Bioc 3.16) was used to calculate the count numbers of mapped short-reads to target genes or transcripts in the M. domestica genome [43], and further the DESeq2 method (p < 0.05 and FoldChange ≥ 2) was performed for detecting differential analysis of count data of different samples in M. domestica [44].
3. Results
3.1. Identification and Distribution of MIKCC-Type MADS-Box Genes in 13 Genome-Released Species
MIKCC-type MADS-box proteins contain SRF-TF and K-box conserved domains, representing important structure characterization of MIKCC-type MADS-box genes in the plant kingdom. In this study, based on the profile Hidden Markov Models (HMMs) of SRF-TF and K-box domains in the Pfam v32.0 database, species-specific profile HMMs of the 13 genome-sequenced species (C. reinhardtii, P. patens, S. moellendorffii, A. trichopoda, A. thaliana, C. sinensis, F. vesca, G. raimondii, M. baccata, M. domestica, P. persica, V. vinifera, and O. sativa) were rebuilt by HMMER v3.2.1 software. Using their corresponding species-specific profile HMMs, a total of 341 MIKCC-type MADS-box genes were identified, and were unevenly distributed in Chlorophyta (1), Bryophyta (1), Lycopodiophyta (1), and Magnoliophyta (basal: 1, dicot: 8, and monocot: 1) (Table 1). After curation, we finally obtained 6 MIKCC-type MADS-box genes in P. patens (0.018%), 3 in S. moellendorffii (0.013%), 15 in A. trichopoda (0.056%), 37 in A. thaliana (0.134%), 19 in C. sinensis (0.075%), 32 in F. vesca (0.095%), 48 in G. raimondii (0.128%), 36 in M. baccata (0.078%), 50 in M. domestica (0.111%), 32 in P. persica (0.119%), 28 in V. vinifera (0.088%), and 35 in O. sativa genomes (0.063%), respectively. No MIKCC-type MADS-box genes were identified in the C. reinhardtii genome (Table S1). For MIKCC-type MADS-box gene classification in C. reinhardtii, we identified two genes (Cre11.g467577.t1.1 and Cre06.g253250.t1.1) containing the SRF-TF conserved domain and one gene (Cre01.g011600.t1.2) containing the K-box conserved domain, but these three genes had the incomplete conserved domain feature of MIKCC-type MADS-box genes.

Table 1.
Summary of MIKCC-type MADS-box genes in different genome-released species.
3.2. Phylogeny of MIKCC-Type MADS-Box Genes among Different Species
To investigate the evolutionary relationships of the above-mentioned 341 target MADS-box genes, a phylogenetic tree was constructed based on their protein sequences (Figure 1). All MIKCC-type MADS-box genes present in 12 plant species were classified into three different groups: I, II, and III. Group I contained 182 (53.37%) MIKCC-type MADS-box genes that were only distributed in 10 plant species in angiosperms without bryophyta and lycopodiophyte, indicating specific MIKCC-type MADS-box genes in angiosperms. Group II contained 89 (26.10%) MIKCC-type MADS-box genes, and similar to those in group I, genes in group II only existed in angiosperms without bryophyta and lycopodiophyta, and they also belonged to the angiosperm-specific MIKCC-type MADS-box genes, which were identified in 10 plant species. Genes distributed in I and II groups belonged to the same type in angiosperms but clustered into different groups. Group III contained 70 (20.53%) MIKCC-type MADS-box genes, and compared with those in groups I and II, the target genes in bryophyta and lycopodiophyta were clustered in group III. These results illustrate that these genes were ancestral and existed in lower plant species.
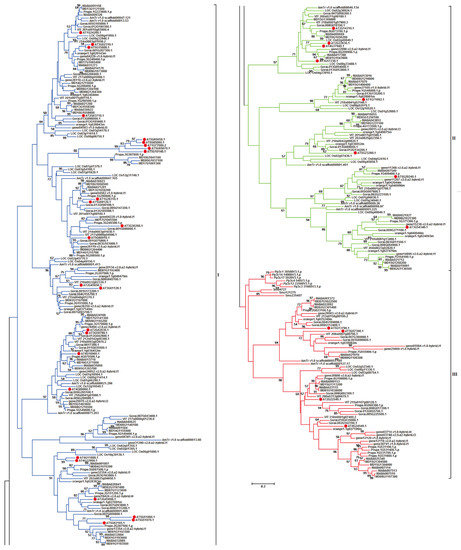
Figure 1.
Phylogenetic tree of MIKCC-type MADS-box genes in genome-released species. The phylogenetic tree has been truncated, and evolutionary branches with blue, green, and red colors represent I, II, and III groups in phylogenetic tree of MIKCC-type MADS-box genes among different species. Solid red circle represents the MIKCC-type MADS-box genes in A. thaliana.
3.3. Comparative Analysis of MIKCC-type MADS-Box Genes in A. thaliana and Malus Species
A. thaliana is an important model plant species, and its genome was released more than ten years ago, creating an excellent opportunity for studying MIKCC-type MADS-box genes within the whole genomes [45]. A. thaliana contained two A-class genes (AT1G69120, AP1 and AT4G36920, AP2), two B-class genes (AT5G20240, PI and AT3G54340, AP3), and a single C-class gene (AT4G18960, AG), although AP2 was not a MIKCC-type MADS-box gene. In order to provide molecular evidence for studying the floral organ identity determination in Malus species, we performed a comparative genomics analysis of MIKCC-type MADS-box genes between A. thaliana and Malus genomes. All MIKCC-type MADS-box genes in A. thaliana, M. domestica, and M. baccata were clustered in two different groups, results consistent with what was observed in the phylogenetic analysis of MIKCC-type MADS-box genes of the 12 plant species (Figure 2A). Furthermore, the orthologous relationship of the ABC genetic model related genes between A. thaliana and Malus species were traced (Table 2). We found that the A-class gene AT1G69120 (AP1) had orthologous genes in Malus genomes. AT1G69120 (AP1) and AT1G26310 (CAL1, AGL10, and CAL) genes were clustered in one subgroup, while MD16G1058500 and MD13G1059200 genes in M. domestica and MABA030923 and MABA021281 genes in M. baccata were clustered in another subgroup. From the plant genome duplication database (PGDD), we retrieved AT1G69120 (AP1) and AT1G26310 (CAL1, AGL10, and CAL) from syntenic genome blocks in the A. thaliana genome, which revealed that the AP1 gene in Arabidopsis and Malus lineages experienced a separate evolutionary event (Figure 2B). For B-class genes, we found that AT5G20240 (PI) and MABA007549 in M. baccata were grouped in one group, and with AT3G54340 (AP3), three genes (MABA021927, MABA021712, and MABA015091) in M. baccata and three genes (MD08G1021300, MD15G1250200, and MD02G1136500) in M. domestica were grouped in another subgroup. We only identified one orthologous gene in M. baccata not in M. domestica for AT5G20240 (PI), but for AT3G54340 (AP3), we attained three orthologous genes in M. baccata and M. domestica separately, indicating the expansion of gene copy number of AP3 in the Malus lineage (Figure 2C). A. thaliana only had a single C-class gene: AT4G18960 (AG). Through phylogenetic analysis, we found that AT4G18960 (AG) might have two orthologous genes in M. baccata and M. domestica separately, which meant that AG also experienced the expansion of gene copy numbers in the Malus lineage (Figure 2D).
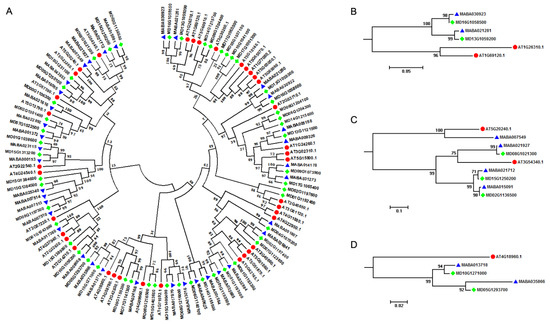
Figure 2.
Phylogenetic analysis of MIKCC-type MADS-box genes among A. thaliana and Malus species. (A) Phylogenetic tree. (B) A-class MIKCC-type MADS-box genes. (C) B-class MIKCC-type MADS-box. (D) C-class MIKCC-type MADS-box genes. Solid red circles, blue triangle, and green rhombus represent MIKCC-type MADS-box genes in A. thaliana, M. baccata, and M. domestica, respectively.

Table 2.
MIKCC-type MADS-box orthologous genes between A. thaliana and Malus species.
3.4. Analysis of WGD Events for MIKCC-Type MADS-Box Genes in Malus Lineage
Using the MCScanX toolkit with default parameters, we identified 1225 and 877 collinear genomic regions containing 22,457 and 9589 paralogous gene pairs in M. domestica and M. baccata genomes, respectively (Table S2). Through collinear analysis, we determined that 46 of 50 (representing 92%) MIKCC-type MADS-box genes from 23 paralogous gene pairs were located on syntenic genomic regions in the M. domestica genome. For ABC genetic model related MIKCC-type MADS-box genes in the A. thaliana genome, AT1G69120 (AP1) and AT1G26310 (CAL1, AGL10, and CAL) for A-class genes had two orthologous genes (MD16G1058500 and MD13G1059200) in the M. domestica genome, and we further explored that these two genes were generated by a WGD event in the M. domestica genome. AT3G54340 (AP3) for B-class genes had three orthologous genes (MD08G1021300, MD15G1250200, and MD02G1136500) in the M. domestica genome. By syntenic analysis in the M. domestica genome, MD08G1021300 and MD15G1250200, and MD08G1021300 and MD02G1136500 were both paralogous gene pairs in the M. domestica genome. AT4G18960 (AG) for C-class genes had two orthologous genes (MD10G1271000 and MD05G1293700) in the M. domestica genome, which was a paralogous gene pair generated from a WGD event.
In the M. baccata genome, we found 27 MIKCC-type MADS-box genes locating on M. baccata syntenic genomic regions representing 75% of the total target genes in its genome. As mentioned above, AT3G54340 (AP3) for B-class genes had three orthologous genes (MABA021927, MABA021712, and MABA015091) in the M. baccata genome, and we concluded that MABA021712 and MABA015091 were a paralogous gene pair generated by a WGD event in the M. baccata genome. AT4G18960 (AG) for C-class genes in the ABC genetic model had two orthologous genes (MABA013718 and MABA035866), and they all were generated from a WGD event in M. baccata genome.
3.5. Analysis of TD Events for MIKCC-Type MADS-Box Genes in Malus Lineage
A tandem duplication event is a critical mechanism in plants resulting in the increase in gene copy numbers leading to gene family expansion of main traits, phenotype, and genome size [46,47,48]. With the method implemented in PTGBase, we identified 5797 tandemly duplicated genes distributed in 2431 tandem arrays, representing 12.85% of the total protein-coding genes in the M. domestica genome (Table S3). Out of 50 MIKCC-type MADS-box genes in the M. domestica genome, 15 were generated from a TD event, and were distributed in one- and six-, three-, and two-copy tandem arrays (Table 3). In M. baccata, we identified 5379 tandemly duplicated genes distributed in 2378 tandem arrays, and the proportion of tandemly duplicated genes in the M. baccata genome indicated lower (11.71%) than that in the M. domestica genome. For 36 MIKCC-type MADS-box genes, only 8 target genes, distributed in four two-copy tandem arrays, were generated from a TD event in the M. baccata genome.

Table 3.
Tandemly duplicated MIKCC-type MADS-box genes in Malus species.
Comparative analysis of MIKCC-type MADS-box genes in A. thaliana and Malus species indicated that most of the A. thaliana MIKCC-type MADS-box genes had orthologous genes in Malus species. For instance, AT4G24540 (AGAMOUS-like 24, AGL24) had three orthologous genes (MD08G1197300, MD15G1384500, and MD15G1384600) in M. domestica genome, with four (MABA007510, MABA007513, MABA007514, and MABA025340) existing in the M. baccata genome (Figure 3A). Through TD analysis, we found that two of three M. domestica MIKCC-type MADS-box genes (MD15G1384500 and MD15G1384600) were generated by a TD event, and two of four MIKCC-type MADS-box genes (MABA007513 and MABA007514) originated from a two-copy tandem array in the M. baccata genome. However, this illustrated a different orthologous relationship between A. thaliana and Malus species (Figure 3B). Three MIKCC-type MADS-box genes (AT5G62165, AGL42, AT5G51860, AGL70, and AT5G51870, AGL69) were clustered together, and two of these three genes (AT5G51860, AGL70 and AT5G51870, AGL69) were from a two-copy tandem array in the A. thaliana genome. Two M. baccata (MABA032884 and MABA032885) and three M. domestica (MD01G1193300, MD01G1193500, and MD01G1193600) MIKCC-type MADS-box genes were clustered in different subgroups, with two M. baccata genes from a two-copy tandem array and three M. domestica genes from a three-copy tandem array. These results showed that the AGL42, AGL69, and AGL70 groups in A. thaliana, Arabidopsis, and Malus lineages experienced separate TD events after splitting from their ancestor. It is noteworthy to mention that the Malus ancestor experienced this event first to generate a two-copy tandem array, which was retained in M. domestica and M. baccata genomes after species divergence or formation.
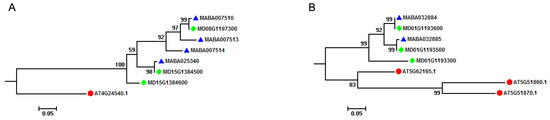
Figure 3.
Orthologous relationship of MIKCC-type MADS-box genes among A. thaliana and Malus species. (A) Phylogenetic tree subgroup of AT4G24540 MIKCC-type MADS-box gene indicating the effect of tandem duplication event on the generation of MIKCC-type MADS-box genes in Malus species. (B) Phylogenetic tree subgroup of AT5G62165, AT5G51860, and AT5G51879 MIKCC-type MADS-box genes indicating the effect of tandem duplication event on the generation of MIKCC-type MADS-box genes in A. thaliana and Malus species. These phylogenetic tree subgroups are from the phylogenetic analysis of MIKCC-type MADS-box genes among A. thaliana and Malus species in Figure 3. Solid red circles, blue triangle, and green rhombus represent MIKCC-type MADS-box genes in A. thaliana, M. baccata, and M. domestica, respectively.
3.6. Expression Analysis of MIKCC-Type MADS-Box Genes in M. domestica
To investigate the expression differences of M. domestica MIKCC-type MADS-box genes in different stages of flowering development, we first analyzed these genes from FPKM values of transcripts in 10 tissues, including dormant apical bud, dormant spur bud, initial pink flower, full pink flower, full flower, petal fall, fruit, silver tip, green tip, and tight cluster from the NCBI-SRA database with accession number PRJNA302879. The transcript abundance analysis of MIKCC-type MADS-box genes showed that 50 MIKCC-type MADS-box genes were expressed across the 10 different tissues. Through hierarchical clustering analysis with the complete linkage method of MIKCC-type MADS-box gene expression in M. domestica, this gene expression profiling was grouped in two different groups, indicating different expression patterns among the 10 tissue types (Figure 4A). MD16G1058500 and MD13G1059200 were orthologous genes of AT1G69120 (AP1) and AT1G26310 (CAL1, AGL10, and CAL) in M. domestica, which belonged to the A-class genes of the ABC model. These two genes were up-regulated in initial pink flower, full pink flower, full flower, green tip, fruit, petal fall, and tight cluster tissues but were down-regulated in dormant apical bud and dormant spur bud tissues, with no expression in silver tip tissue. For B-class genes of the ABC model, AT3G54340 (AP3) had three orthologous genes (MD08G1021300, MD15G1250200, and MD02G1136500) in M. domestica. Expression analysis among the different tissues showed that these genes were down-regulated in dormant apical bud, dormant spur bud, fruit, and tight cluster tissues and up-regulated in initial pink flower, full pink flower, full flower, and green tip tissues, with a lower expression in silver tip and petal fall tissues. The two orthologous genes (MD05G1293700 and MD10G1271000) in M. domestica of the C-class gene AT4G18960 (AG) showed different expression patterns, although these two genes were up-regulated in initial pink flower, full pink flower, and full flower tissues and down-regulated in dormant apical bud and dormant spur bud tissues. MD05G1293700 was up-regulated in green tip tissue and down-regulated in fruit tissue, but MD10G1271000 was not found to be expressed in these two tissues.
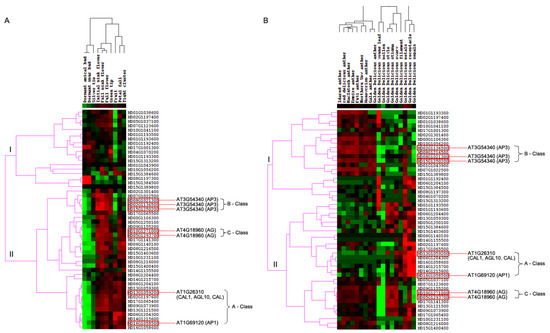
Figure 4.
Expression heatmap of MIKCC-type MADS-box genes in M. domestica. (A) Expression heatmap of M. domestica MIKCC-type MADS-box genes in different flowering development stages. (B) Expression heatmap of M. domestica MIKCC-type MADS-box genes in different M. domestica cultivars. Green color indicates that the expression levels of genes are low, while red color indicates that the levels are high.
Furthermore, we detected the anther expression profiling of 50 M. domestica MIKCC-type MADS-box genes in 18 different M. domestica cultivars, including ‘Idared’, ‘MacIntosh’, ‘Northern Spy’, ‘red delicious’, ‘Honeycrisp’, ‘Empire’, ‘Fuji’, ‘Gala’, and ‘Golden Delicious’, as well as different floral organs of M. domestica ‘Golden Delicious’, with all anther tissues retrieved from buds 1–3 days prior to flower opening. Using all downloaded RNA-seq short-read data of different M. domestica tissues, we implemented the expression profile of total protein-coding genes in the M. domestica genome and retrieved expression levels of 50 M. domestica MIKCC-type MADS-box genes in different M. domestica cultivars (Figure 4B). For A-class genes of the ABC model in M. domestica, MD13G1059200 and MD16G1058500 showed significant up-regulation in receptacle and sepal tissues of ‘Golden Delicious’ and different expression divergences in anther tissues of different cultivars and the rest of the tissues of ‘Golden Delicious’, indicating significant functional divergence in different M. domestica cultivars. The three AP3 orthologous genes in M. domestica were up-regulated in anther tissues of different M. domestica cultivars and petal tissues of ‘Golden Delicious’. Notably, these genes were slightly up-regulated in the remaining ‘Golden Delicious’ tissues. MD05G1293700 and MD10G1271000, orthologous genes of AT4G18960 (AG) belonging to C-class genes for the ABC model, displayed a significant down-regulation in young leaf, pollen tissues, petals, and sepals, and higher up-regulation in style, stigma, and ovary tissues in ‘Golden Delicious’. These genes were slightly up-regulated in anther tissues of different M. domestica cultivars.
4. Discussion
In this study, we collected 13 genome-sequenced species arranged from lower to higher organisms to trace the evolutionary history of the MIKCC-type MADS-box gene family. We found that the M-type MADS-box genes emerged in the lower single-cell green alga but were generated after the formation of C. reinhardti. Qu et al. bioinformatics analyses showed that the MIKC*-type MADS-box genes were a transition between the M-type and MIKC-type MADS-box genes in the Salix suchowensis genome [49]. Our evolutionary relationships of the target MADS-box genes showed that all genes within group I and II belonged to the angiosperm-specific MIKCC-type MADS-box genes. However, in group III the target genes were clustered in bryophyta and lycopodiophyta. These MIKCC-type genes were ancestral and existed in lower plant species. We presumed that MIKCC-type MADS-box genes in angiosperm species belonging to group III were inherited from an ancestor or lower plant species and were retained in angiosperm species.
The MADS-box genes are thought to originate from gene duplication that initially occurred in the most recent common ancestor of living eukaryote lineages [50]. A previous study illustrated that a relatively recent genome-wide duplication occurred in the M. domestica genome approximately more than 50 MYA [29]. The analysis of 4dTv distribution suggested that M. baccata have experienced a recent WGD event that brought two copies of protein-coding genes in each of the M. domestica and M. baccata genome, leading to MIKCC-type MADS-box gene family expansion in the Malus lineage [28]. In our analysis, both WGD and TD events affected the expansion of the MIKCC-type MADS-box gene family in the Malus lineage. Our results illustrated that MIKCC-type MADS-box genes in the M. domestica genome (46 of 50, ~92%) experienced more influences of WGD events than those in the M. baccata genome (27 of 36, 75%). For the ABC-model-related genes, the three-class genes were all influenced by a WGD event in the M. domestica genome, but only B-class and C-class genes in the M. baccata genome were influenced by a WGD event. For the TD event, MIKCC-type MADS-box genes in the M. domestica genome (15 of 50, ~30%) also experienced more influences of TD events than that in M. baccata genome (7 of 36, ~19%). For ABC-model-related genes, only A-class genes in M. domestica (MD13G1059200 and MD16G1058500) and M. baccata (MABA030923) genomes were influenced by a TD event, but these genes were all located in a two-copy tandem array and other members of the tandem array did not belong to the ABC-model-related genes in Malus species. We surmised that the WGD events affected the expansion of the ABC-model-related genes without the influence of TD events on these in Malus species. However, an analysis by Arora et al. revealed that the different number of the total M-type MADS-box genes in rice and Arabidopsis was mainly due to duplication events derived from TD [51]. Research found that MADS-box genes seem to have evolved mainly through gene duplication events, and then the form of neofunctionalization, subfunctionalization, and pseudogenization [52].
MIKCC-type MADS-box genes have been thought to be mainly involved in the control of flowering time and floral organ identity in plants [53]. The present study analyzed the transcript abundance of 50 MIKCC-type MADS-box genes in different stages of M. domestica flowering development across 10 varied tissues. Furthermore, the expression profiles of 50 MIKCC-type MADS-box genes were also detected in 18 different M. domestica cultivars in anther tissues. Altogether, most MIKCC-type MADS-box genes of ABC-model -related genes showed a trend of up-regulated expression in different flowering developmental stages and varieties. These results confirmed that MIKCC-type MADS-box genes were indeed involved in flowering and anther development processes. Co-expression network analysis of 58 MIKCC-type MADS-box genes found them to be composed of central hubs in a regulatory network of flowering development and floral organogenesis in rose [53]. In Adonis amurensis transcriptomes, 38 MIKCC-type MADS-box genes were discovered in flower organs and may play a regulatory role in floral organogenesis and flowering time [54]. Further, 34 MIKCC-type MADS-box genes were identified from the barley genome, and transcript analysis further confirmed their potential roles involved in processes of inflorescence meristem initiation, floral meristem identity, and floral organ determination [55]. Considering the above results, it is prudent to assume that the MIKCC-type MADS-box genes play an important regulatory role in the process of flower development.
5. Conclusions
Here, we identified 341 MIKCC-type MADS-box genes among 13 genome-released species. Our results showed that MIKCC-type genes were generated later than M-type genes in the MADS-box gene family after the formation of Chlamydomonas reinhardtii. Phylogenetic analysis revealed that the MIKCC-type MADS-box genes belonged to group III in angiosperm species, which were inherited from an ancestor or lower plant species. Comparative analysis of MIKCC-type MADS-box genes between A. thaliana and Malus lineages exhibited a similar pattern of evolutionary relationship with the phylogenetic analysis. For orthologous analysis of ABC-model-related genes, three classes of genes in Malus species existed in orthologous genes compared with A. thaliana. Both WGD and TD evolutionary events affected the expansion of the MIKCC-type MADS-box gene family in the Malus lineage; among them, only the WGD event was considered to have acted on the expansion of the ABC-model-related genes. The expression profile of 50 MIKCC-type MADS-box genes in different M. domestica developmental stages and cultivars confirmed that these genes were indeed involved in flowering and anther development processes. These results provided novel insights for the ABC model research of flower development in the Malus lineage and a model for widespread application for studying gene families in plants.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae9030373/s1, Table S1: Protein sequences of MIKCC type MADS-Box genes from 12 species; Table S2: Collinear genomic regions in M. domestica and M. baccata genomes; Table S3: Tandemly duplicated genes distributed in M. domestica and M. baccata genomes.
Author Contributions
K.N. and T.Z. conceived the project, prepared the data analysis, and wrote the manuscript. T.Z. supervised the project. W.Z., D.Z. and Y.A.E.-K. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Natural Science Foundation of Jiangsu Province (BK20220751), the National Natural Science Foundation of China (32201618), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (20KJB220001), and the Scientific Research Programs for High-level Talents Start-up Fund of the Jinling Institute of Technology (jit-b-202008).
Data Availability Statement
All data generated or analyzed are included in this article and its Supplementary Information files.
Acknowledgments
We thank anonymous reviewers for their critical reading, stimulating discussions, and helpful comments and suggestions, which allowed us to improve the quality of this manuscript.
Conflicts of Interest
The authors declare that they have no competing interests.
Abbreviations
| WGD | whole-genome duplication |
| TF | tandem duplication |
| HMM | Hidden Markov model |
| ML | maximum likelihood |
References
- Theißen, G.; Gramzow, L. Structure and evolution of plant MADS domain transcription factors. In Plant Transcription Factors: Evolutionary, Structural and Functional Aspects; Gonzalez, D.H., Ed.; Elsevier: Philadelphia, PA, USA, 2016; pp. 127–138. [Google Scholar]
- Zhu, P.; Dong, T.; Xu, T.; Kang, H. Identification, characterisation and expression analysis of MADS-box genes in sweetpotato wild relative Ipomoea trifida. Acta Physiol. Plant. 2020, 42, 163. [Google Scholar] [CrossRef]
- Medard, N.; Martin, F.Y. Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2001, 2, 186–195. [Google Scholar]
- Ghorbani, M.; Bagheri, H.; Gholami, M. Genome-wide study of flowering-related MADS-box genes family in Cardamine hirsuta. 3 Biotech. 2020, 10, 518. [Google Scholar] [CrossRef] [PubMed]
- Smaczniak, C.; Immink, R.G.H.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef]
- Parenicova, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef] [PubMed]
- Honma, T.; Goto, K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 2001, 409, 525–529. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Krizek, B.A.; Meyerowitz, E.M. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 1996, 93, 4793–4798. [Google Scholar] [CrossRef]
- Xu, Z.; Zhang, Q.; Sun, L.; Du, D.; Cheng, T.; Pan, H.; Yang, W.; Wang, J. Genome-wide identification, characterization and expression analysis of the MADS-box gene family in Prunus mume. Mol. Genet. Genom. 2014, 289, 903–920. [Google Scholar] [CrossRef]
- Henschel, K.; Kofuji, R.; Hasebe, M.; Saedler, H.; Munster, T.; Theissen, G. Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol. Biol. Evol. 2002, 19, 801–814. [Google Scholar] [CrossRef]
- Kwantes, M.; Liebsch, D.; Verelst, W. How MIKC* MADS-box genes originated and evidence for their conserved function throughout the evolution of vascular plant gametophytes. Mol. Biol. Evol. 2012, 29, 293–302. [Google Scholar] [CrossRef]
- Aceto, S.; Gaudio, L. The MADS and the beauty: Genes involved in the development of Orchid flowers. Curr. Genom. 2011, 12, 342–356. [Google Scholar] [CrossRef]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef]
- Theißen, G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001, 4, 75–85. [Google Scholar] [CrossRef]
- Kaufmann, K.; Melzer, R.; Theißen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Ditta, G.; Pinyopich, A.; Robles, P.; Pelaz, S.; Yanofsky, M.F. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004, 14, 1935–1940. [Google Scholar] [CrossRef]
- Krishnamurthy, K.V.; Bahadur, B. Genetics of flower development. In Plant Biology and Biotechnology; Bahadur, B., Venkat Rajam, M., Sahijram, L., Krishnamurthy, K., Eds.; Springer: New Delhi, India, 2015; pp. 385–407. [Google Scholar]
- Grimplet, J.; Martinez-Zapater, J.M.; Carmona, M.J. Structural and functional annotation of the MADS-box transcription factor family in grapevine. BMC Genom. 2016, 17, 80. [Google Scholar] [CrossRef]
- Theißen, G.; Rumpler, F.; Gramzow, L. Array of MADS-box genes: Facilitator for rapid adaptation? Trends Plant Sci. 2018, 23, 563–576. [Google Scholar] [CrossRef]
- Won, S.Y.; Jung, J.A.; Kim, J.S. Genome-wide analysis of the MADS-Box gene family in Chrysanthemum. Comput. Biol. Chem. 2021, 90, 107424. [Google Scholar] [CrossRef] [PubMed]
- Ohmori, S.; Kimizu, M.; Sugita, M.; Miyao, A.; Hirochika, H.; Uchida, E.; Nagato, Y.; Yoshida, H. MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 2009, 21, 3008–3025. [Google Scholar] [CrossRef] [PubMed]
- Thompson, B.E.; Bartling, L.; Whipple, C.; Hall, D.H.; Sakai, H.; Schmidt, R.; Hake, S. Bearded-ear encodes a MADS box transcription factor critical for maize floral development. Plant Cell 2009, 21, 2578–2590. [Google Scholar] [CrossRef]
- Rijpkema, A.S.; Zethof, J.; Gerats, T.; Vandenbussche, M. The petunia AGL6 gene has a SEPALLATA-like function in floral patterning. Plant J. 2009, 60, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, K.; Wellmer, F.; Muino, J.M.; Ferrier, T.; Wuest, S.E.; Kumar, V.; Serrano-Mislata, A.; Madueno, F.; Krajewski, P.; Meyerowitz, E.M. Orchestration of floral initiation by APETALA1. Science 2010, 328, 85–89. [Google Scholar] [CrossRef]
- Wong, C.E.; Singh, M.B.; Bhalla, P.L. Novel members of the AGAMOUS LIKE 6 subfamily of MIKCC-type MADS-box genes in soybean. BMC Plant Biol. 2013, 13, 105. [Google Scholar] [CrossRef] [PubMed]
- Acri-Nunes-Miranda, R.; Mondragon-Palomino, M. Expression of paralogous SEP-, FUL-, AG- and STK-like MADS-box genes in wild-type and peloric Phalaenopsis flowers. Front. Plant Sci. 2014, 5, 76. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jin, Z.; Feng, C.; Liu, M.; Wang, J.; Hu, Y. Genome-wide identification, characterization of the MADS-box gene family in Chinese jujube and their involvement in flower development. Sci. Rep. 2017, 7, 1025. [Google Scholar] [CrossRef]
- Tian, Y.; Dong, Q.; Ji, Z.; Chi, F.; Cong, P.; Zhou, Z. Genome-wide identification and analysis of the MADS-box gene family in apple. Gene 2014, 555, 277–290. [Google Scholar] [CrossRef]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.K.; Troggio, M.; Pruss, D.; et al. The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant Genom. Nucleic Acids Res. 2012, D1, D1178–D1186. [Google Scholar] [CrossRef]
- Cheng, C.; Krishnakumar, V.; Chan, A.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef]
- Sara, E.G.; Jaina, M.; Alex, B.; Eddy, S.R.; Aurélien, L.; Potter, S.C.; Matloob, Q.; Richardson, L.J.; Salazar, G.A.; Alfredo, S. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar]
- Leinonen, R.; Sugawara, H.; Shumway, M. The sequence read archive. Nucleic Acids Res. 2010, 39, D19–D21. [Google Scholar] [CrossRef]
- Potter, S.C.; Aurélien, L.; Eddy, S.R.; Youngmi, P.; Rodrigo, L.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; Mcwilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Philip, J.; David, B.; Chang, H.; Matthew, F.; Li, W.; Craig, M.; Hamish, M.; John, M.; Alex, M.; Gift, N. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for 16 bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Tae-ho, L.; Jin, H.; Barry, M.; Guo, H. CScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Yu, J.; Ke, T.; Tehrim, S.; Sun, F.; Liao, B.; Hua, W. PTGBase: An integrated database to study tandem duplicated genes in plants. Database 2015, 2015, bav017. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Gingeras, T.R. Mapping RNA-seq reads with STAR. Curr. Protoc. Bioinform. 2015, 51, 11–14. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kaul, S.; Koo, H.L.; Jenkins, J.; Rizzo, M.; Rooney, T.; Tallon, L.J. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Maher, C. Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 2006, 16, 510–519. [Google Scholar] [CrossRef]
- Shang, Q.; Liang, L.; Dong, C. Multiple tandem duplication of the phenylalanine ammonia-lyase genes in Cucumis sativus L. Planta 2012, 236, 1093–1105. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Bi, C.; He, B.; Ye, N.; Yin, T.; Xu, L. Genome-wide identification and characterization of the MADS-box gene family in Salix suchowensis. Peer J. 2019, 7, e8019. [Google Scholar] [CrossRef] [PubMed]
- Gramzow, L.; Ritz, M.S.; Theißen, G. On the origin of MADS-domain transcription factors. Trends Genet. 2010, 26, 149–153. [Google Scholar] [CrossRef]
- Arora, R.; Agarwal, P.; Ray, S.; Singh, A.K.; Singh, V.P.; Tyagi, A.K.; Kapoor, S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007, 8, 242. [Google Scholar] [CrossRef]
- Irish, V.F.; Litt, A. Flower development and evolution: Gene duplication, diversification and redeployment. Curr. Opin. Genet. Dev. 2005, 15, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Fu, X.; Dong, Y.; Lu, J.; Ren, M.; Zhou, N.; Wang, C. MIKCC-type MADS-box genes in Rosa chinensis: The remarkable expansion of ABCDE model genes and their roles in floral organogenesis. Hortic. Res. 2018, 5, 25. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Sun, H.; Dai, S.; Feng, S.; Qiao, K.; Wang, J.; Gong, S.; Zhou, A. Identification and characterization of MIKCc-type MADS-Box genes in the flower organs of Adonis amurensis. Int. J. Mol. Sci. 2021, 22, 9362. [Google Scholar] [CrossRef] [PubMed]
- Kuijer, H.N.J.; Shirley, N.J.; Khor, S.F.; Shi, J.; Schwerdt, J.; Zhang, D.; Li, G.; Burton, R.A. Transcript profiling of MIKCc MADS-Box genes reveals conserved and novel roles in barley inflorescence development. Front. Plant Sci. 2021, 12, 705286. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).