Abstract
In Andalusia (Spain), there are different wine regions that have a great recognized tradition. In these regions, the cultivation of the vine is ancient and there are still vineyards planted with local varieties of Vitis vinifera L. that have not yet been identified. The aim of this research study was to identify 49 accessions of grapevine collected in the districts of four provinces in Andalusia (Spain). All samples were genotyped with 20 microsatellite markers in order to ascertain the identity and analyze the genetic diversity of the collected material. In total, 30 different genotypes were obtained, 22 of them which were identified with named, known varieties by comparison to the Spanish or European microsatellite databases, and eight which are referred to as new genotypes. All loci were polymorphic, and a total of 159 alleles were detected, ranging from 4 to 12 alleles per locus, with an average allele number of 7.95. The overall observed heterozygosity was 0.763 and was slightly higher than expected (0.715), while the gene diversity per locus varied between 0.167 (VVIN73) and 0.967 (VVMD5). A dendrogram representing the genetic similarities among cultivars was depicted using the UPGMA method to investigate their relationships. The eight new genotypes identified in this research work could represent ancient local varieties in danger of extinction. These new cultivars may be used to determine original wines.
1. Introduction
The Andalusia (Spain) region, in the south of the Iberian Peninsula, is one of the most ancient and important wine regions in Spain [1]. Archaeological, paleobotanical, and historical sources confirm that grapevines were spread and cultivated for a long time in this area. The presence of the species Vitis vinifera L. has been verified by pollen analysis performed in different Phoenician sites of Andalusia located in the provinces of Cádiz, Málaga and Almería [2]. In addition, numerous archaeological remains have been found at these sites, which may be associated with the existence of a wine industry [3,4,5]. The first evidence of planting techniques characteristic of protohistoric viticulture in the west has been documented in an archaeological site located in Huelva (Andalusia) dating back to the 1st millennium BC [6].
There are many citations that reference the diversity of grapevine (Vitis vinifera L.) varieties grown in Andalusia. Roxas Clemente [7], in his paper Essay of common grapevine varieties that are growing in Andalusia, includes 119 varieties grouped in two sections and 15 tribes. In 1831, James Busby, considered the “father of Australian viticulture” introduced 678 varieties in Australia [8]. These varieties originated in France and Spain. According to Morilla Critz [8], at least half of these varieties were from Andalusia.
Nevertheless, the genetic diversity of the Andalusia grapevine (Vitis vinifera L.) has been declining due to the phylloxera (Daktulosphaira vitifoliae) attack of the late 19th century [9], when severe regulations were approved, and the grapevine varieties authorized for wine production were restricted and the vineyard was restructured, frequently stimulated by subsidies. In Spain, previous to this vineyard restructuring, which begin in the 1970s, all vines were grafted in the field with mass-selected Vitis vinifera material from older vineyards, which often included different varieties [10]. With the aim of preserving grapevine phytogenetic resources, numerous studies on the surveying, localization, characterization, and maintaining of cultivars in germplasm banks are being carried out worldwide [11,12,13,14,15,16,17,18,19,20]. In Andalusia, a germplasm bank was established in 1940, and it was replanted between 1984 and 1987, and the number of accessions substantially increased [21]. Actually, this collection preserves 1417 accessions according to the Vitis International Variety Catalogue (VIVC, www.vivc.de accessed on 26 December 2022) [22].
The recovery of autochthonous or local varieties allows a genetic, ecological and agronomic enrichment capable of dealing with various diseases, improving the adaptation to edaphoclimatic conditions [23] or facilitating the adaptation in the face of future market changes [24]. For this reason, the accurate identification of local cultivars and their conservation could prevent their disappearance and preserve them for future needs. Traditionally, the identification of grape varieties has been based on the morphological features of vegetative and reproductive structures [25], but phenotypic traits are not sufficiently reliable for the classification of closely related varieties due to genotype–environment interactions [26]. Therefore, molecular characterization is the favoured technique for varietal identification. At the present time, there are different molecular markers available to carry out a molecular identification of a grape variety. However, microsatellites or Simple Sequence Repeats (SSRs) markers are the most used for this purpose [27,28]. In this sense, microsatellite markers have been widely used to identify and genotype grapevine cultivars collected in old vineyards of the Iberian Peninsula [29,30,31]. In addition, SSRs have been used for studies of genetic diversity and genetic relationships [32].
The main objective of this research work is focused on the molecular identification of a total of 49 vine accessions collected in old Andalusian vineyards. The genotyping of these accessions could help to detect new local cultivars growing in Andalusia aiming to provide a solid basis to develop a regional germplasm collection to protect local biodiversity.
2. Materials and Methods
2.1. Plant Material
After prospecting more than 200 vineyards throughout the provinces of Almería, Cádiz, and Huelva y Málaga of the Andalusia region (Spain), those plants that were not visually identified as common varieties cultivated in Andalusia were sampled and placed in the germplasm bank at the Rancho de la Merced. This grapevine collection is located in Jerez de la Frontera (Cádiz, Spain) (36°41′10″ N; 6°08′10″ W; alt. 20 m). The list of the 49 accessions used in this study are shown in the Supplemental Table S1. Each accession was identified with a code of three letters and a number. The initials correspond to the name of the municipality where it was collected.
Two internationally known cultivars (‘Cabernet Sauvignon’ and ‘Syrah’) were also included to compare the genetic profiles obtained with the different published databases.
2.2. Molecular Analysis
DNA was extracted from young leaves collected from each accession and stored at −80 °C, using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). A genotypic characterization was performed for 20 nuclear microsatellite loci located in the 19 linkage groups of grapevine genome VMC1b11, VMC4F3-1 (Vitis Microsatellite Consortium); VVMD5, VVMD7, VVMD21, VVMD24, VVMD25, VVMD27, VVMD28, VVMD32 [33,34]; VVS2 [35]; VVIB01, VVIH54, VVIN16, VVIN73, VVIP31, VVIP60, VVIQ52, VVIV37, and VVIV67 [36]. Two multiplex PCR tests were set up to amplify the 20 microsatellite loci in a 20 µL reaction mix according to Vargas et al. [37]. PCR reactions were carried out in the 44 Applied Biosystems 9700 thermocycler.
Amplified products were separated by capillary electrophoresis using an automated sequencer (ABI Prism 3130, Applied Biosystems, Foster City, CA, USA). Fluorescently labelled fragments were detected and sized using GeneMapper v. 3.7 software (Applied Biosystems), and fragment lengths were determined with the help of internal size standards (GeneScan-500 LIZTM, Applied Biosystems, Foster City, CA, USA).
The identification of redundant genotypes was determined by comparing microsatellite genotypes with data contained in the Spanish microsatellite grapevine databases Rancho de la Merced [38,39,40] and the Vitis Germplasm Bank (BGV) at the Finca El Encín (IMIDRA, Alcalá de Henares, Spain) [41,42,43] and other European databases [22,44]. Genotype comparisons were carried out using the Microsatellite toolkit v. 9.0 software package [45].
2.3. Data Analysis
2.3.1. Genetic Diversity Analyses
For the calculation of the number of alleles (Na), expected (He) and observed (Ho) heterozygosity, frequency of null alleles (r) and probability of identity (PI), the GENALEX software [46] was used. The polymorphism information content (PIC) of each microsatellite loci was determined using an online tool [47].
2.3.2. Genetic Relationships among Cultivars
Genetic distances between grapevine genotypes were calculated as [-ln (proportion shared alleles)] using Microsat [48]. The obtained data was used for the construction of a dendrogram using the programs EXE from the PHYLIP package software [49] and MEGA version 7 [50].
3. Results and Discussion
3.1. Microsatellite Analysis and Genetic Diversity
The molecular analysis performed with the 49 studied accessions resulted in 30 non-redundant genotypes (Table 1). These genotypes were used for the calculation of genetic parameters (Table 2) in order avoid overestimation. A total of 159 alleles, ranging from 12 in VVMD7 and four in VVIN73, were detected, with an average of eight alleles per locus, similar to the mean Na attained by Fernández-González et al. [51]. The most frequent allele was VVIN73-264, which showed a frequency up to 90%, and 27 alleles were unique.

Table 1.
Thirty genotypes obtained for the 49 analyzed accessions at 20 microsatellite loci. Allele sizes are given in base pairs.

Table 2.
Characterization of 20 microsatellite markers in the 30 genotypes.
The expected heterozygosity (He, gene diversity) ranged from 0.185 at locus VVIN73 to 0.866 at locus VVIP31, with a mean value of 0.715. The observed heterozygosity (Ho) varied between 0.167 at locus VVIN73 and 0.967 at locus VVMD5. For 16 loci, Ho was higher than He, and the probability of null alleles was always negative, except for VMC4F31, VVMD21, VVMD25, VVMD28, VVIN73 and VVIP60. Samples in which only one single allele per locus was detected were considered as homozygous genotypes instead of heterozygous with a null allele. The VVIN73 and VVIP31 markers displayed the minimum (0.1769) and maximum (0.8522) PIC values, respectively. The 20 microsatellite loci showed a mean PIC value of 0.67241.
The 20 microsatellite loci used reflected a high discrimination power and a low probability that two randomly chosen individuals had identical genotypes using the 20 loci (PI. 1.74 × 10−19). This indicates the probability that two of the 30 varieties analyzed randomly were chosen to share the same genotype using the set of these 20 microsatellite loci.
The values obtained from the statistical characterization of the 20 microsatellite loci used in this research study (Table 2) are similar to those obtained in other studies on the genetic characterization of local grapevine cultivars using microsatellite markers [51,52,53]. Nevertheless, the percentage of new accessions recovered (16.3%) is higher than that obtained by Balda et al. [10] for 45 accessions recovered in Rioja (Spain) (4.4%), Fort et al. for 223 accessions in recovered in Lanzarote (Canary Islands, Spain) (3.6%) [20], and Augusto et al. for 310 accessions recovered in northeast Portugal [32]. This suggests that the grapevine richness of the Andalusian region has not been prospected with the same degree of intensity.
3.2. Cultivar Analysis
Most of the analyzed accessions were identified with known grapevine cultivars. The varietal names were assigned based on the comparison with Spanish [38,39,40,41,42,43] and European [22,44] microsatellite databases and using the genetic profile of reference varieties for adapting the allele sizes. Allele sizes of genotypes obtained for the twenty SSRs loci analyzed are shown in Table 1 and Table 2, and the prime names of the identified cultivars according to VIVC [22], indicating the code of sampled accession for each cultivar (Table 3). Thirty-nine accessions corresponded to 22 known varieties and the ten accessions remaining (Can-1, Comp-3, Lau-3, Lau-4, Lau-16, Lau-11, Man-5, Ron-3, Ron-5 and Ron-6) to the eight unidentified cultivars. These cultivars showed genotypes that did not match any of the published cultivars in the Spanish and European microsatellite databases consulted in this research. Half of the identified accessions are of Spanish origin according to the VIVC database [22] (Table 3), and the country of origin of the rest was France (four accessions), Portugal (three accessions), the United States (one accession), Italy (one accession), Greece (one accession), Algeria (one accession) and Lebanon (one accession). The accessions coded as Lau-3, Lau-4 and Lau-16 showed the same genotype, and they were collected in the same location (Laujar de Andarax, Almería, Spain).

Table 3.
Grapevine material studied with SSR identification, utilization and country of origin of the variety are according to VIVC [21].
The identified accessions include table and wine grapevine varieties. The table grape varieties, identified by ‘Molinera’ (Ins-1), ‘Imperial Napoleon’ (Ins-3) and ‘Attika seedless’ (Pla-2), have been collected in different regions of the province of Almería (Spain). In this province the cultivation of table grapes was predominant until the 1960s [54]. Furthermore, one hybrid interspecific (‘Jacquez’) was identified (Table 2). This hybrid was used for the reconstitution of European vineyards [55]. It is currently prohibited from use in Europe.
One genotype (Com-4) was identified as ‘Rome Tinto’, after comparing it with the microsatellite database from Rancho de la Merced [38,39,56]. This variety was only conserved in the Rancho de la Merced Germplasm bank according to the VIVC database and presents a different genotype to the ‘Rome’ cultivar published by Ibáñez et al. [41].
Four of the varieties identified, ‘Beba’, ‘Jaén negro’, ‘Pedro Ximenez’ and ‘Rome Tinto’, were already mentioned by Rojas Clemente [6] as being present in the Andalusian region. This shows the antiquity of the cultivation of these varieties in this region. ‘Jaén negro’ and ‘Rome Tinto’ are two red grapevine cultivars that have already been identified in old vineyards in the province of Málaga [57].
Currently, most of the cultivars identified in this work have disappeared from the Andalusia vineyards, and the unique cultivar that is growing in the commercial vineyards is ‘Pedro Ximenez’. All of this vegetal material recovered from old vineyards could be interesting for the wine industry in Andalusia or regions with similar agroclimatic conditions. However, many of these identified cultivars are not included in the official register of Spanish grapevine varieties for the community of Andalusia, which would make their cultivation difficult. Recently, ‘Beba’ has been included as an authorized variety in the regulation of wines of the Protected Designation of Origin “Jerez-Xérès-Sherry” (Spain) [58], as there is some interest in increasing the diversity of wines [59].
In addition, of the identified varieties, the eight new genotypes should be studied and evaluated in order to make their oenological potential and adaptation climate change known among the wine sector. Furthermore, these cultivars could be important genetic resources for future breeding programs.
3.3. Genetic Relationships among Cultivars
Based on the results of the analysis of the microsatellites, the distance matrix was used to carry out a grouping using UPGMA. To characterize the genetic structure of different genotypes obtained and two references varieties (‘Cabernet Sauvignon’ and ‘Syrah’), a dendrogram based on the proportion of shared alleles was constructed. Figure 1 shows the resulting dendrogram of the 30 non-redundant genotypes found in this study.
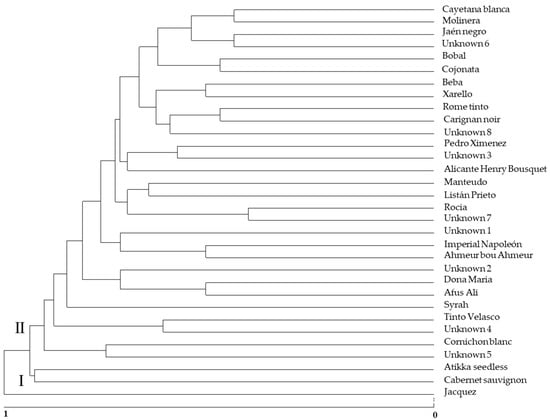
Figure 1.
Genetic relationships among the thirty genotypes obtained and two reference cultivars.
SSR analysis allowed for the evaluation of the genetic relationships among European cultivars and unknown accessions recollected in different regions of Andalusia. The dendrogram in Figure 1 shows the existence of two defined groups. Group I includes only one cultivar identified with ‘Jacquez’, which is a hybrid interspecific of the cross between Vitis aestivalis × Vitis vinifera [22]. All of the rest of the identified and unknown cultivars are included in group II and are cultivars of the Vitis vinifera species. The formation of these two groups may be related to the pedigree of the cultivars. In group II, there is no clear separation of different subgroups in relation to regions of origin as found in other published research papers on Sicilian varieties [60]. Varieties with different countries of origin are grouped in this cluster II (Table 3).
Two varieties, ‘Cabernet Sauvignon’ and ‘Atikka seedless’ are markedly distant from the rest of the cultivars in Group II, probably because of a different origin and use. ‘Atikka seedless’ is considered a seedless variety of Greek origin according to VIVC [22].
Phylogenetic distances of the subgroup where variety “Unknown 6” is included indicates that it could be a wine and table grape, since it is grouped with other grapes that are used as wine and table grapes, such as ‘Cayetana Blanca’, ‘Molinera’ and ‘Jaén Tinto’ (Table 3). The same behavior could be said for the variety “Unknown 7” and the ‘Roal’, “Unknown 4” and ‘Tinto Velasco’ or “Unknown 5” and ‘Cornichon Blanc’.
4. Conclusions
Forty-nine accessions collected in Andalusia have been described by molecular methods. A total of 83.7% of these accessions analyzed have been identified by comparison to Spanish or European microsatellite databases with known cultivars. However, eight genotypes have not yet been identified and could represent old local cultivars in danger of extinction. All of these genotypes have been preserved within the Rancho de la Merced germplasm bank (Andalusia, Spain).
This study indicates an important biodiversity within the old vineyards from the Andalusia region that provides interesting information for the wine industry and that points out the wide genetic diversity of grapevines which are still unexploited. Our efforts should lead to the protection and study of local grape natural richness.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae9030316/s1, Table S1: List of the 49 accessions collected and their place of origin in the Andalusia region (Spain).
Author Contributions
Conceptualization, A.J.-C., A.P.-P. and R.A.-G.; methodology, A.J.-C., A.P.-P. and R.A.-G.; software, A.J.-C. and R.A.-G.; validation, A.J.-C. and R.A.-G.; formal analysis, A.J.-C.; investigation, A.J.-C., A.P.-P. and R.A.-G.; resources, A.J.-C.; data curation, A.J.-C.; writing—original draft preparation, A.J.-C., A.P.-P. and R.A.-G.; writing—review and editing, A.J.-C., A.P.-P. and R.A.-G.; visualization, A.J.-C., A.P.-P. and R.A.-G.; supervision, A.J.-C., A.P.-P. and R.A.-G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Ministerio de Ciencia y Tecnología (INIA-Spain), grants number RF2004-00014-00-00, VIN00-036-C6-5X, RF2006-00011-00-00 and RF2007-00017-00-00.
Acknowledgments
The authors would like to thank the IFAPA for facilitating the material conserved in the Rancho de la Merced germplasm bank for the study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pozo-Bayón, M.Á.; Moreno-Arribas, M.V. Sherry wines. In Advances in Food and Nutrition Research; Jackson, R.S., Ed.; Academic Press: Cambridge, MA, USA, 2011; Volume 63, pp. 17–40. [Google Scholar] [CrossRef]
- Prados Martínez, F. La producción vinícola en el mundo fenicio-púnico. Apuntes sobre cultivo de la vid y consumo del vino a través de las fuentes arqueológicas y literarias. Gerión 2011, 29, 9–35. [Google Scholar] [CrossRef]
- González Rodríguez, R. Excavaciones de Urgencia en el Cerro Naranja (Jerez de la Frontera, Cádiz). In Anuario Arqueológico de Andalucía, III; Junta de Andalucía: Sevilla, Spain, 1985; pp. 90–96. [Google Scholar]
- Torres, J.R. Las Ánforas Fenicio-Púnicas del Mediterráneo Central y Occidental. Ph.D. Thesis, University of Barcelona, Barcelona, Spain, 1991. [Google Scholar]
- Aubet Semmler, M.E.; Buxó Capdevila, R. Los recursos y la economía colonial. En Aubet Semmler, M.E.; et al. ii Cerro del Villar I. El Asentamiento fenicio en la desembocadura del río Guadalhorce y su interacción con el hinterland. Arqueol. Monogr. 1999, 5, 334–339. [Google Scholar]
- Vera, J.C.; Echevarría, A. Sistemas agrícolas del I milenio a.C. en el yacimiento de La Orden-Seminario de Huelva: Viticultura protohistórica a partir del análisis arqueológico de las huellas de cultivo. In Proceedings of the Patrimonio Cultura de la Vid y el Vino, Madrid, Spain, 2013; pp. 95–106. [Google Scholar]
- De Rojas Clemente y Rubio, S. Ensayo Sobre las Variedades de vid Común que Vegetan en Andalucía; Imprenta Villalpando: Madrid, Spain, 1807; p. 324. [Google Scholar]
- Morilla Critz, J. La viticulture de Andalucía en 1831 vista por James Busby, padre de la viticultura Australiana. Estud. Reg. 1997, 49, 261–298. [Google Scholar]
- Fernández de Bobadilla, G. Viníferas Jerezanas y de Andalucía Occidental; Instituto Nacional de Investigaciones Agronómicas: Madrid, Spain, 1956. [Google Scholar]
- Balda, P.; Ibáñez, J.; Sancha, J.C.; Martínez de Toda, F. Characterization and identification of minority red grape varieties recovered in Rioja, Spain. Am. J. Enol. Vitic. 2014, 65, 148–152. [Google Scholar] [CrossRef]
- Fatahi, R.; Ebadi, A.; Bassil, N.; Mehlenbacher, S.A.; Zamani, Z. Characterization of Iranian grapevine cultivars using microsatellite markers. Vitis 2003, 42, 185–192. [Google Scholar]
- Martínez, L.E.; Cavagnaro, P.F.; Masuelli, R.W.; Zuñiga., M. SSR-based assessment of genetic diversity in South American Vitis vinifera varieties. Plant Sci. 2006, 170, 1036–1044. [Google Scholar] [CrossRef]
- De Mattia, F.; Imazio, S.; Grassi, F.; Lovicu, G.; Tardaguila, J.; Failla, O.; Maitt, C.; Scienza, A.; Labra, M. Genetic characterization of Sardinia grapevine cultivars by SSR markers analysis. J. Int. Sci. Vigne et du Vin. 2007, 41, 175–184. [Google Scholar] [CrossRef]
- Lacombe, T.; Boursiquot, J.M.; Laucou, V.; Dechesne, F.; Varès, D.; This, P. Relationships and genetic diversity within the accessions related to Malvasia held in the Domaine de Vassal grape germplasm repository. Am. J. Enol. Vitic. 2007, 58, 124–131. [Google Scholar] [CrossRef]
- Dzhambazova, T.; Tsvetkov, I.; Atanassov, I.; Rusanos, K.; Martínez Zapater, J.M.; Atanassov, A.; Hvarleva, T. Genetic diversity in native Bulgarian grapevine germplasm (Vitis vinifera L.) based on nuclear and chloroplast microsatellite polymorphisms. Vitis 2009, 48, 115–121. [Google Scholar]
- De Olivera, G.L.; De Souza, A.P.; De Oliver, F.A.; Zucchi, M.I.; De Souza, L.M.; Moura, M.F. Genetic structure and molecular diversity of Brazilian grapevine germplasm: Management and use in breeding programs. PLoS ONE 2020, 15, e0240665. [Google Scholar] [CrossRef]
- Upadhayay, A.; Aher, L.B.; Shinde, M.P.; Mundankar, K.Y.; Datre, A.; Karibasappa, G.S. Microsatellite analysis to rationalize grape germplasm in India and development of a molecular database. Plant Genet. Resour. 2013, 11, 225–233. [Google Scholar] [CrossRef]
- Popescu, C.F.; Maul, E.; Dejeu, L.C.; Dinu, D.; Gheorge, R.N.; Laucou, V.; Lacombe, T.; Migliaro, D.; Crespan, M. Identification and characterization of Romanian grapevine genetic resources. Vitis 2017, 56, 173–180. [Google Scholar]
- Marsal, G.; Méndez, J.J.; Mateo, J.M.; Ferrer, S.; Canals, J.M.; Zamora, F.; Fort, F. Molecular characterization of Vitis vinifera L. local cultivars from volcanic areas (Canary Islands and Madeira) using SSR markers. OENO One 2019, 53, 667–680. [Google Scholar] [CrossRef]
- Fort, F.; Marsal, G.; Mateo-Sanz, J.M.; Pena, V.; Canals, J.M.; Zamora, F. Molecular characterisation of the current cultivars of Vitis vinifera L. in Lanzarote (Canary Islands, Spain) reveals nine individuals which correspond to eight new varieties and two new sports. OENO One 2022, 56, 281–295. [Google Scholar] [CrossRef]
- García de Luján, A.; Lara Benítez, M. La colección de vides del Rancho de la Merced; Junta de Andalucía, Consejería de Agricultura y Pesca: Sevilla, Spain, 1997; p. 157.
- Vitis International Variety Catalogue. Available online: www.vivc.de (accessed on 26 December 2022).
- Pastor, I.; Rovira, J.M. La Investigación arqueológica en los procesos de recuperación de variedades de uva autóctonas tradicionales. In Proceedings of the Patrimonio Cultura de la Vid y el Vino, Madrid, Spain, 2013; pp. 71–79. [Google Scholar]
- Sancho-Galán, P.; Amores-Arrocha, A.; Palacios, V.; Jiménez-Cantizano, A. Identification and characterization of white grape varieties autochthonous of a warm climate region (Andalusia, Spain). Agronomy 2020, 10, 205. [Google Scholar] [CrossRef]
- Boursiquot, J.M.; This, P. Les nouvelles techniques utilisées en ampélographie: Informatique et marquage. J. Int. Sci. Vigne Vin 1996, 610, 13–23. [Google Scholar]
- Emanuelli, F.; Lorenzi, S.; Grzeskowiak, L.; Catalano, V.; Stefanini, M.; Troggio, M.; Myles, S.; Martínez-Zapater, J.M.; Zyprian, E.; Moreira, F.M.; et al. Genetic diversity and population structure assessed by SSR and SNP markers in large germplasm collection of grape. BMC Plant Biol. 2013, 13, 39. [Google Scholar] [CrossRef]
- Sefc, K.M.; Lefort, F.; Grando, M.S.; Scott, K.D.; Steinkellner, H.; Thomas, M.R. Microsatellite markers for grapevine: A state of the art. In Molecular Biology and Biotechnology of Grapevine; Roubelakis-Angelakis, K.A., Ed.; Kluwer Academic: Dordrecht, The Netherlands, 2001; pp. 433–463. [Google Scholar]
- This, P.; Jung, A.; Boccacci, P.; Borrego, J.; Botta, R.; Costantini, L.; Crespan, M.; Dangl, G.S.; Eisenheld, C.; Ferreira-Monteiro, F.; et al. Development of a standard set of microsatellite reference alleles for identification of grape cultivars. Theor. Appl. Genet. 2004, 109, 1448–1458. [Google Scholar] [CrossRef]
- González-Andrés, F.; Martín, J.P.; Yuste, J.; Rubio, J.A.; Arranz, C.; Ortiz, J.M. Identification and molecular biodiversity of autochthonous grapevine cultivars in the “Comarca del Bierzo”, León, Spain. Vitis 2007, 46, 71–76. [Google Scholar]
- Jiménez-Cantizano, A.; Lara, M.; Ocete, M.E.; Ocete, R. Short communication: Characterization of the relic Almuñécar grapevine cultivar. Span. J. Agric. Res. 2012, 10, 454–460. [Google Scholar] [CrossRef]
- Mena, A.; Martínez, M.; Fernández-González, M. Recovery, identification and relationships by microsatellite analysis of ancient grapevine cultivars from Castilla-La Mancha: The largest wine growing region in the world. Genet. Resour. Crop Evol. 2014, 61, 625–637. [Google Scholar] [CrossRef]
- Augusto, D.; Ibáñez, J.; Pinto-Sintra, A.L.; Falco, V.; Leal, F.; Martínez-Zapater, J.M.; Oliveira, A.A.; Castro, I. Grapevine Diversity and Genetic Relationships in Northeast Portugal Old Vineyards. Plants 2021, 10, 2755. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.E.; Dangl, G.S.; Vignani, R.; Meredith, C.P. Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.). Genome 1996, 39, 628–633. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.E.; Dangl, G.S.; Meredith, C.P. Development and characterization of additional microsatellite markers for grape. Am. J. Enol. Vitic. 1999, 50, 243–246. [Google Scholar] [CrossRef]
- Thomas, M.R.; Scott, N.S. Microsatellite repeats in grapevine reveal DNA polymorphisms when analyzed as sequence-tagged sites (STSs). Theor. Appl. Genet. 1993, 86, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Merdinoglu, D.; Butterlin, G.; Bevilacqua, L.; Chiquet, V.; Adam-Blondon, A.F.; Decroocq, S. Development and characterization of a large set of microsatellite markers in grapevine (Vitis vinifera L.) suitable for multiplex PCR. Mol. Breed. 2005, 15, 349–366. [Google Scholar] [CrossRef]
- Vargas, A.M.; Velez, M.D.; De Andrés, M.T.; Laucou, V.; Lacombe, T.; Boursiquot, J.M.; Borrego, J.; Ibáñez, J. Corinto bianco: A seedless mutant of Pedro Ximenes. Am. J. Enol. Vitic. 2007, 58, 540–543. [Google Scholar] [CrossRef]
- Jiménez-Cantizano, A. Caracterización Molecular del Banco de Germoplasma de vid del Rancho de la Merced. Ph.D. Thesis, University of Cádiz, Cádiz, Spain, 2014. Available online: http://hdl.handle.net/10498/17919 (accessed on 12 November 2022).
- Jiménez-Cantizano, A.; García De Luján, A.; Arroyo-García, R. Molecular characterization of table grape varieties preserved in the Rancho de la Merced Grapevine Germplasm Bank (Spain). Vitis 2018, 57, 93–101. [Google Scholar]
- Cretazzo, E.; Moreno Sanz, P.; Lorenzi, S.; Benítez, M.L.; Velasco, L.; Emanuelli, F. Genetic Characterization by SSR Markers of a Comprehensive Wine Grape Collection Conserved at Rancho de la Merced (Andalusia, Spain). Plants 2022, 11, 1088. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez, J.; Vargas, A.M.; Palancar, M.; Borrego, J.; De Andrés, M.T. Genetic Relationships among Table-Grape Varieties. Am. J. Enol. Vitic. 2009, 60, 35–42. [Google Scholar] [CrossRef]
- Vargas, A.M.; De Andrés, M.T.; Borrego, J.; Ibáñez, J. Pedigrees of fifty tables-grape cultivars. Am. J. Enol. Vitic. 2009, 60, 525–531. [Google Scholar] [CrossRef]
- De Andrés, M.T.; Benito, A.; Pérez-Rivera, G.; Ocete, R.; López, M.A.; Gaforio, L.; Muñoz, G.; Cabello, F.; Martínez-Zapater, J.M.; Arroyo-García, R. Genetic diversity of wild grapevine populations in Spain and their genetic relationships with cultivated grapevines. Mol. Ecol. 2012, 21, 800–816. [Google Scholar] [CrossRef]
- Lacombe, T.; Boursiquot, J.M.; Laucou, V.; Di Vecchi-Staraz, M.; Péros, J.P.; This, P. Large-scale parentage analysis in an extended set of grapevine cultivars (Vitis vinifera L.). Theo. Appl. Genet. 2013, 126, 401–414. [Google Scholar] [CrossRef]
- Park, S.D.E. Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection. Ph. D. Thesis, University of Dublin, Dublin, Ireland, 2001. [Google Scholar]
- Peakall, R.; Smouse, P. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Res. 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Bi ’nkowski, J.; Miks, S. Gene-Calc. 2018. Available online: www.gene-calc.pl. (accessed on 20 February 2023).
- Minch, E.; Ruiz-Linares, A.; Goldstein, D.; Feldman, M.; Cavalli-Sforza, M. Microsat 1.5d: A Computer Program for Calculating Various Statistics on Microsatellite Allele Date; Washington State University: Pullman, WA, USA, 1997. [Google Scholar]
- Felsenstein, J. Phylogeny inference package. Cladistics 1989, 5, 164–166. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA 7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2015, 33, 1870–1874. [Google Scholar] [CrossRef]
- Fenández-González, M.; Mena, A.; Izquierdo, P.; Martínez, J. Genetic characterization of grapevine (Vitis vinífera L.) cultivars from Castilla La Mancha (Spain) using microsatellite markers. Vitis 2007, 46, 126–130. [Google Scholar]
- Žulj Mihaljević, M.; Šimon, S.; Pejić, I.; Carka, F.; Sevo, R.; Kojić, A.; Gaši, F.; Tomić, L.; Jovanović Cvetković, T.; Maletić, E.; et al. Molecular characterization of old local grapevine varieties from South East European countries. Vitis 2013, 52, 69–76. [Google Scholar]
- Basheer-Salimia, R.; Lorenzi, S.; Batarseh, F.; Moreno-Sanz, P.; Emanuelli, F.; Grando, M.S. Molecular identification and genetic relationships of Palestinian grapevine cultivars. Mol. Biotechnol. 2014, 56, 546–556. [Google Scholar] [CrossRef]
- Gómez-Espín, J.M.; Gil Meseguer, E. La exportación española de uva de mesa en la segunda mitad del siglo XX. Papeles Geografía 1987, 13, 87–104. [Google Scholar]
- Santiago, J.L.; González, I.; Gago, P.; Alonso-Villaverde, V.; Boso, S.; Martínez, M.C. Identification of and relationships among a number of teinturier grapevines that expanded across Europe in the early 20th century. Aust. J. Grape and Wine Res. 2008, 14, 223–229. [Google Scholar] [CrossRef]
- Jiménez-Cantizano, A.; Amores-Arrocha, A.; Gutiérrez-Escobar, R.; Palacios, V. Identification and relationship of the autochthonous ‘Romé’ and ‘Rome Tinto’ grapevine cultivars. Span. J. Agric. Res. 2018, 16, e07SC02. [Google Scholar] [CrossRef]
- Jiménez-Cantizano, A.; Muñoz-Martín, A.; Amores-Arrocha, A.; Sancho-Galán, P.; Palacios, V. Identification of Red Grapevine Cultivars (Vitis vinifera L.) Preserved in Ancient Vineyards in Axarquía (Andalusia, Spain). Plants 2020, 9, 1572. [Google Scholar] [CrossRef] [PubMed]
- Orden de 4 de Octubre de 2022, Boletín Oficial de la Junta de Andalucía (BOJA). Available online: https://www.juntadeandalucia.es/eboja/2022/195/s54.html (accessed on 3 January 2023).
- Sancho-Galán, P.; Amores-Arrocha, A.; Palacios, V.; Jiménez-Cantizano, A. Genetical, Morphological and Physicochemical Characterization of the Autochthonous Cultivar ‘Uva Rey’ (Vitis vinifera L.). Agronomy 2019, 9, 563. [Google Scholar] [CrossRef]
- Carimi, F.; Mercati, F.; Abbate, L.; Sunseri, F. Microsatellite analyses for evaluation of genetic diversity among Sicilian grapevine cultivars. Genet. Resour. Crop Evol. 2010, 57, 703–719. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).