Abstract
Sansevieria Thunb. species are traditionally known as succulent ornamental plants worldwide. They are also cultivated for medicinal, fodder, soil conservation and fiber uses, and for their capacity to reduce environmental pollution. Sansevieria sexual propagation is limited by the lack of viable seeds, and reproduction is largely made via vegetative propagation by suckers or cuttings. For these reasons, genetic improvement by conventional breeding is limited. To overcome this problem and to address the increasing demand from customers for novel Sansevieria varieties, many commercial companies regularly use in vitro propagation, as is the case in the breeding process of several ornamental plants. In this paper, for the first time, we report a procedure for in vitro somatic embryogenesis and plant regeneration starting from three flower explants for seven different Sansevieria genotypes. Regeneration was attempted using stigmas/styles, anther/filament, and ovary which were cultured on a Murashige and Skoog solidified medium under three different plant growth regulator combinations. A good regeneration rate was obtained with all genotypes used under all culture conditions tested from every explant type, with percentages ranging from 0 to 73.3%. “Genetic stability” assessment of regenerated plants in respect to their mother plants was verified through flow cytometry analysis showing a high degree of uniformity, with only S. parva exhibiting a different level of DNA fluorescence among in vitro regenerated plants. This is an interesting achievement in the aim to produce true-to-type plants and new variants with desirable characteristics, both of which are desired features in ornamentals improvement.
1. Introduction
Sansevieria Thumb. is a genus of succulent plants with approximately 90 distinct species identified [1], originally found in tropical central Africa and drier parts of eastern and southern Africa [2]. Sansevieria plants are cultivated for medicinal, fodder, soil conservation and fiber uses [3]. Recently it has been reported that Sansevieria plants have also a high capacity to reduce environmental pollution produced by gases and heavy metals thanks to the ability to absorb hazardous pollutants such as VOCs and CO2 emissions [4,5]. Unquestionably, Sansevieria species are much better known as ornamental plants [6,7]. Sansevierias are liked for their shape, patterns and colors of leaves which varies from dark green, pale green, grayish green or a combination of green and white or yellow with several patterns of the lamina [8]. After the “green wave” in the 70’s, Sansevierias sales decreased but in recent years, the market has expanded and consumer demand has increased leading to an increase in the culture of Sansevieria. Nevertheless, intensive culture could not fulfil the demand due to the slow growing manner of plant propagation. Usually, Sansevierias are propagated by suckers and leaf cuttings but both methods need a great amount of starting plant material and a long period of time to produce a competitive number of new specimens [8]. Moreover, sexual propagation is limited by inviable seeds [9]. Due to these reasons, genetic improvement by conventional breeding is limited. Mass propagation through in vitro procedures has been investigated as a solution to address the increasing commercial demand. Recently, direct and indirect organogenesis from leaf explants have been reported for S. trifasciata and S. masoniana [3,10,11,12]. Here we report for the first time, to the best of our knowledge, a technique for plant regeneration starting from flower explants. These kinds of explants have been used to regenerate several crops, such as citrus [13], grapevine [14], caper [15] and fennel [16], with clear advantages since flower explants are easy to sterilize, due to the almost complete absence of contamination of their inner tissues. Further benefits of using of young floral tissues are inherent in the wide availability of young explants, which positively influence the regeneration potential [17,18] and in the preservation of the mother plants’ integrity (non-destructive procedure).
Ornamental plants are economically important for the horticulture industry and the growth of the sector depends on the creation of new cultivars with commercially attractive features [19]. Plant breeding studies are the main method to improve ornamental plants, and plant tissue culture is the tool used in both fundamental research and commercial applications. As tissue culture technologies sometimes can produce genetic variability, regenerants could bear new interesting features for the ornamental plants industry. For this reason, the exploitation of somaclonal variability has become part of the usual breeding activity of many commercial enterprises. Several ornamental cultivars derived from tissue culture via somaclonal variation have been described ([20] and further references are here reported). In this paper, we report for the first time a technique for plant regeneration starting from flower explants for seven different genotypes of Sansevieria. To detect the genetic fidelity of regenerated plants, flow cytometric analysis was performed.
2. Materials and Methods
2.1. Plant Material
Flowers used in this work were collected from seven different genotypes of Sansevieria: S. concinna N.E.Br. (S-CNR-014; Figure 1(A1,A2)), S. fasciata Cornu ex Gérôme & Labroy, × forskaoliana (Schult.f.) Hepper & J.R.I. Wood (S-CNR-074; Figure 1(B1,B2)), S. forskaoliana (Schult.f.) Hepper & J.R.I.Wood (S-CNR-036; Figure 1(C1,C2)), S. elliptica (Chiovenda) Cudofontis (S-CNR-103; Figure 1(D1,D2)), S. parva N.E.Br. (S-CNR-054; Figure 1(E1,E2)), S. pearsonii N.E.Br. (S-CNR-058; Figure 1(F1,F2)) and S. caulescens N.E.Br. (S-CNR-088; Figure 1(G1,G2)). All specimens utilized in the present study come from the former UK National Collection of Sansevierias recognized by the Royal Horticultural Society and held by Alan Butler, Brookside Nursery. Other information about area of origin and accession names are reported in Table S1. The plant material was collected from the germplasm repository for perennial plants at the Institute of Biosciences and BioResources of the National Research Council of Italy (CNR-IBBR) located in Collesano Palermo, Italy (38° N, 14° E). Three different floral explants (stigmas/styles (Sti/Sty), anther/filament (Ant/Fil), and ovary (Ov)) used for culture initiation were dissected from flowers harvested 15 days before anthesis. Whole flowers were rinsed with tap water, surface sterilized by immersion for 1 min in 70% ethanol, 7 min in 2% (w/v) sodium hypochlorite, followed by three 5 min rinses in sterile distilled water. With the aid of a stereo-microscope, the flowers were cut under sterile conditions, and Ov, Ant/Fil and Sti/Sty explants (Figure 2A) were excised from the flowers and plated as single explants under 3 different culture conditions in contact with the medium.

Figure 1.
Sansevieria plants and inflorescences: (A1,A2) Sansevieria concinna (S-CNR-014), Bars 8 and 10 cm; (B1,B2) S. fasciata x forskaoliana (S-CNR-074), Bars 15 and 10 cm; (C1,C2) S. forskaoliana (S-CNR-036), Bars 5 and 3 cm; (D1,D2) S. elliptica (S-CNR-103), Bars 10 cm; (E1,E2) S. parva (S-CNR-054), Bars 6 cm; (F1,F2) S. pearsonii (S-CNR-058), Bars 5 cm; (G1,G2) S. caulescens (S-CNR-088), Bars 3.5 cm.

Figure 2.
Plant regeneration in Sansevieria through somatic embryogenesis. (A) Sansevieria flower collected before opening and cut open under sterile conditions. From left to right: whole flower; flower with two petals removed; flower with all petals removed and stigma/style (Sti/Sty) and ovary (Ov) emphasized; anther and filament (Ant/Fil) explants dissected from Sansevieria flowers collected before opening (bar = 4 mm); (B) Creamy-white callus from the Sti/Sty explants (S-CNR-054) 90 days after culture initiation (bar = 1.5 cm); (C) Somatic embryos (S-CNR-054) generated after 120 days of culture initiation at the surface of anther explant-derived callus (bar = 2.5 mm); (D) Somatic embryos (S-CNR-103) generated after 150 days of culture at the surface of Ov explant-derived callus (bar = 2 mm); (E) Somatic embryos (S-CNR-103) generated after 200 days of culture initiation at the surface of Sti/Sty explant-derived callus (bar = 1.5 mm); (F) Plant (S-CNR-036) derived from germinated somatic embryos growing in Magenta jars for 150 days on MS medium (bar = 2.5 cm); (G,H) Somatic embryo-derived plant of S-CNR-074 transferred to sterilized soil and incubated in a growth chamber, respectively 14 and 60 days after acclimatization (bar = 3 cm); (I) Regenerated plants after two months of growth under greenhouse conditions (bar = 10 cm).
2.2. Media and Culture Conditions
Explants were cultured on Murashige and Skoog [21] solidified (6 g L−1 Plantagar) medium (MS) supplemented with 88 mM sucrose as carbon source under three different plant growth regulator (PGR) combinations: (1) T4 medium, 5 µM N-(2-chloro-4-pyridyl)-N-phenylurea (4-CPPU) + 5 µM 2,4-dichlorophenoxy acetic acid (2,4-D); (2) T5 medium, 20 µM β-naphthoxyacetic acid (NOA) + 4 µM 1-phenyl-3-(1,2,3-thiadiazol-5-yl) urea (TDZ); and (3) T16 medium, 10 µM NOA + 4.4 µM N6-benzylaminopurine (BA). The pH of the media was adjusted to 5.7–5.8 with 1 N NaOH before autoclaving. All chemicals were purchased from Duchefa Biochemie, The Netherlands. Explants were incubated in a climatic chamber at 25 ± 1°C with a 16 h photoperiod (40 µmol m−2s−1 at shelf level, provided by Osram Cool White18 W fluorescent lamps) and subcultured in the same culture medium at 60 d intervals until embryogenic callus was produced (Figure 2B). Usually embryogenic callus was produced after two transfers. Among the explants initially incubated on culture medium, only those showing embryonic response were transferred to basal MS-medium deprived of PGRs, supplemented with 88 mM sucrose and cultured for four more weeks under the same culture conditions described above, to allow embryo proliferation and development (Figure 2C,D). Embryogenic calli were individually transferred to Petri dishes containing 20 mL of basal solid MS medium deprived of PGRs and incubated under the same light and temperature conditions as described above, to allow further growth (Figure 2E).
2.3. Embryo Germination, Plant Development and Acclimatization
Individual somatic embryos were isolated from callus and germination was attempted in Magenta vessels (Figure 2F) containing hormone free MS solidified medium (7 g/L plantagar, Duchefa) supplemented with 88 mM sucrose as carbon source. Magenta vessels were maintained in a climate chamber at 25 ± 1C under the same culture conditions as described above. Embryos were considered as germinated when there was root extension. Plantlets, 2–4 cm tall and with well-developed roots were collected and washed with tap water in order to remove the medium before being transplanted individually into plastic pots 70 mm × 70 mm containing sterile soil. The potted plants, covered with transparent polyethylene bags to maintain temperature and high humidity (Figure 2G), were placed in a climate chamber at 25 ± 1 °C under a 16 h day length and a photosynthetic photon flux of 50 µmol m−2s−1 provided by Osram cool-white 18 W fluorescent lamps. After 20–30 days plantlets were exposed to gradual reduction of humidity obtained by gradually perforating the polyethylene bags (Figure 2H) and after 40 days plants were transferred outdoors under natural daylight conditions (Figure 2I). The survival rates were recorded after 2 months.
2.4. Flow Cytometry Analysis
The analyzed plant material consisted of seven different Sansevieria genotypes (mother plants: MP) and 16 in vitro regenerated plants (regenerants: R) (Table 1) obtained from different explant types under different culture conditions.

Table 1.
The table summarizes the relationship between Sansevieria’s genotypes used, the number of the accession, the ratio between the G1 peak of regenerants (R) and corresponding mother plant (MP), the type of explant and the regeneration medium used.
Flow Cytometry was carried on Sansevieria nuclei in suspension obtained from 2–4 mg leaf tissue, which was homogenized into 200 µL of LB01 extraction buffer [22] and stained with DAPI (4,6-diamidino-2-phenylindole) at the final concentration of 2 µg/mL.
To obtain the nuclei suspensions, mechanical disruption was performed for 12 s at 9500 rpm with a Mini-Turrax T8 with a S10N-5G generator (IKA, Staufen, Germany). Samples were filtered through a 36 µm nylon mesh and their integrity and concentration were preliminarily determined on a Nikon Eclipse TE2000-S epifluorescence microscope (Nikon Corp., Tokyo, Japan).
All analyses were run on a CytoFLEX S flow analyzer (Beckman Coulter Flow Cytometry, Milan, Italy), using as internal reference standard (IRS) 2.5 µm polystyrene microspheres (Alignflow Beads for UV lasers cod. A16502, ThermoFisher, Milan, Italy) to monitor instrument stability and ensuring a true experimental comparison. Genetic stability analysis was estimated as fluorescence DNA emission on 5000 DAPI stained nuclei/sample, excited by a violet laser (ext 405 nm) with a main DAPI fluorescence emission collected at 450/45 nm. In order to prevent staining instability and ensure reliable measurements, all samples were diluted to get a similar concentration of nuclei and a constant number of standard beads (an amount of 10% of beads was added after a preliminary FCM check done sample by sample).
DNA fluorescence histograms were acquired and processed using CytExpert software v. 2.3 (Beckman Coulter Flow Cytometry, Milan, Italy) and the Internal synthetic Reference Standard fluorescence peak (IRS) was carefully set at the same channel value for all acquisitions (Figure 3). The IRS CV was of 3.04 ± 0.05% through all the experiments and its mean fluorescence peak was set at channel 210,436.4 ± 0.25% (on a basis of 16.8 × 106 instrumental channels resolution).
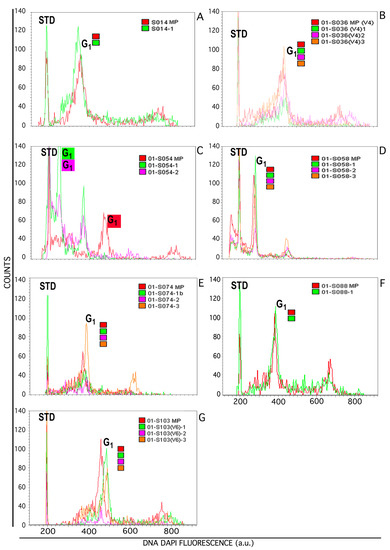
Figure 3.
FCM analysis of genetic stability of Sansevieria spp. mother plant (MP) and their regenerants. DAPI DNA fluorescence histogram overlapping among MP and respective regenerants are presented. Fluorescence histograms of DAPI stained nuclei in suspension show an IRS (STD) region that contains reference beads to allow histogram comparison among several nuclei isolations from Sansevieria individuals. The G1 region shows basic DNA content for each sample; the use of color coding refers to MP and their Rs per line/species. In the abscissa for DAPI fluorescent emission (arbitrary units) captured by a 450–45 band pass filter after violet laser excitation is shown. The panels show: (A) S. concinna S-CNR-014, (B) S. forskaoliana S-CNR-036, (C) S. parva S-CNR-054, (D) S. pearsonii S-CNR-058, (E) S. fasciata × forskaoliana S-CNR-074, (F). S. caulescens S-CNR-088, (G) S. elliptica S-CNR-103.
The relative stability in DAPI-DNA fluorescence level, which is correlated to the nuclear DNA amount for each mother plant and its regenerants, was calculated on cell cycle histograms and defined in relation to the ratio: G1 mean peak position of its Rs /G1 mean peak position of MP (Table 1).
2.5. Data Analyses
All experiments were carried out in a randomized complete block design. Each treatment comprised ten replicates. Five explants each for stigma/style and ovary, and thirty explants for anther/filament were used per Petri plate representing a replicate, making a total of 8400 explants in the experiment. The number of embryogenic explants was recorded after 6 months from culture initiation.
Embryo germination data were collected 2 months from the incubation of somatic embryos on PGR-free medium. Effects of genotype, PGR combination and type of explant on percentages of embryogenic explants, and percentages of embryo germination were tested by ANOVA (p ≤ 0.05) and the differences among means were tested by Tukey’s test. Prior to analysis, percentage data were arcsin-square root transformed. Statistical analysis was performed using SigmaStat 3.5 for Windows.
3. Results
3.1. Plant Regeneration
Callus formation was observed after 60 days from the beginning of the culture under all the PGR combinations tested. Regeneration was obtained from all genotypes with differences according to explant type and medium used. Embryos arose from callus after 4–5 months from culture initiation (Figure 2C,D; Figure 4). Table 2 reports the embryogenic response after 6 months from the culture initiation. The percentages of success varied greatly (0–73.3%) depending on genotypes, explants and PGR combinations. Differences were observed among genotypes in the efficiency of embryogenesis. Percentage of embryogenic explants of the different cultivars calculated across the media and explant types, ranged between 9.7 and 31.1% (Table 3). The capacity of producing new individuals was greater in S. parva (31.1%) and in S. forskaoliana (28.9%) even if all genotypes produced somatic embryos at remarkable percentages (Table 3). Somatic embryogenesis occurred under all the PGR combinations tested but with significant differences in the response of explants and cultivars according to the PGR combination used. The percentage of embryogenic explants, calculated across the genotypes and explant types ranged between 8.8 and 19.1% (T4 and T16 media, respectively) (Table 4). The best response was obtained with the T16 (19.1%). T5 medium was slightly less effective than T16 while T4 was significantly less effective with the lowest embryogenic potential of 8.8% (Table 4). In our experimental conditions the explant type plays an important role in embryogenic response. When calculated across the cultivars and PGR combinations, the response of different explants tested ranged between 4.08% for style/stigma and 13.75% for anthers/filament, respectively (Table 3). When embryos produced roots, germination was considered to have occurred. About 90% of explants germinated and developed into plantlets when transferred onto MS basal medium. Plantlets reached about 10 cm in height in about 60 days (Figure 2F); these well-developed plantlets were transferred into plastic pots 70 × 70 × 100 mm containing sterile soil and covered with a polythene bag to maintain humidity (Figure 2G). After about 30 days, plantlets were transferred to the greenhouse and exposed to natural daylight conditions at 22/27 °C, night/day. In order to reduce humidity levels holes of increasing size were made in the polyethylene bags. When plantlets were transferred into greenhouse conditions, the percentage of acclimatized plants was about 80%.
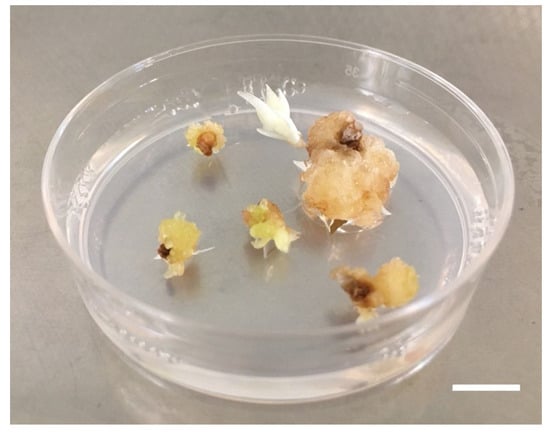
Figure 4.
Somatic embryos appeared after 4 months of culture. Somatic embryo (S-CNR-103) generated from Ov explant-derived callus (bar = 1 mm).

Table 2.
Embryogenic response of seven Sansevieria genotypes using three explant types and three PGR combinations. Data were collected 6 months after culture initiation and each treatment comprised 300 explants for Ant/Fil and 50 explants for Ov and Sti/Sty. Means + SE, values followed by the same letter are not significantly different at p ≤ 0.05 level (Tukey’s test).

Table 3.
Genotype specificity of the embryogenic response in different Sansevieria genotypes. Percentages of embryogenic explants of seven Sansevieria varieties. Data were collected 6 months after the beginning of the experiment; each treatment comprises 1200 explants. Means ± SE, values followed by the same letter are not significantly different at p ≤ 0.05 level (Tukey’s test).

Table 4.
Effect of three PGR combinations (T4, T5, T16) and three explant types (Ov, Sti/Sty, Ant/Fil) on embryogenic response. Data were collected 6 months after the beginning of the experiment. Means ± SE, in each column values followed by the same letter are not significantly different at p ≤ 0.05 level (Tukey’s test).
3.2. Flow Cytometric Analyses
The Sansevieria species used for cytofluorimetric analysis consist of fresh leaves of adult regenerated plants on which homogenization was demonstrated to be effective in disrupting tissues to release nuclei in suspension. LB01 extraction buffer proved to be superior for the isolation of Sansevieria nuclei in suspension in respect to other buffers we have tested such as Otto and Catalano buffers [23,24]; LB01 gave a larger number of nuclei per mg of tissue with a stronger fluorescent emission (Supplemental Figure S1).
Regenerated plants’ clones were compared to their MP to verify “genetic stability” in term of similar levels of DNA fluorescence emission, which is correlated to DNA content. In Figure 3, overlapping of histogram of MP and respective Rs are shown.
Sansevieria explants were characterized by a variable effectiveness of the isolation procedure in terms of nuclei release and debris amount. To generate effective DNA fluorescence histograms, debris and other contaminating particles were removed using the analytical tools from CytExpert software (Supplemental Figure S2).
It was observed that on six of the seven groups of different genotypes analyzed, (1) S-CNR-014: Mother Plant, Regenerant 1; 2) S-CNR-074: Mother Plant, Regenerants 1, 2, 3; 3) S-CNR-036: Mother Plant, Regenerants 1, 2, 3; 4) S-CNR-103: Mother Plant, Regenerants 1, 2, 3; 5) S-CNR-058: Mother Plant, Regenerants-1, 2, 3; 6) S-CNR-088: Mother Plant, Regenerant 1), the alignment of the position of the respective IRS makes the histograms superimposable, demonstrating homogeneity in DNA fluorescence emission from regenerants in respect to their mother plant, independently from explant type used and culture conditions (Figure 3 and Table 1).
S-CNR-054/1 and CNR-054/2 instead, exhibited a fluorescence content of the G1 peak lower than that of the mother plant (Figure 3C).
4. Discussion
Ornamental plants play a significant role in the human environment. They are intentionally grown for decoration in a variety of places, including gardens, floriculture plantations, specialised garden collections, open land and indoors. Since ancient times, humans have been lured by ornamental plants and this trend is increasing year by year. Ornamental plants possess important attributes that influence the buying decision and for this reason the ornamental plant industry is under constant pressure from customers for novel varieties with new intriguing traits [25].
In order to fulfil market needs, it is necessary to develop a genetic improvement program oriented to consumers’ demands and aimed at the discovery of commercially attractive plants competitive with the ones currently available [19]. Nowadays, many commercial companies regularly use in vitro-biotechnology in the breeding process of ornamental plants.
In vitro regeneration starting from flower explants is a well-documented procedure with high regenerative potential used for several species [13,26,27,28,29,30] but so far, never described, to the best of our knowledge, for Sansevieria. The inflorescence of Sansevieria is a many-flowered raceme and the flowers are stalkless, white, cream-colored or greenish white to pale mauve. A single inflorescence gives dozens of flowers and one single inflorescence gives hundreds of explants with limited or no effects for the plant. A good regeneration rate was obtained with all genotypes used under all culture conditions tested from every explant type. Nevertheless, regeneration potential is influenced from several factors as reported also for other species [14,31,32]. The best combination of genotype, medium and explant in terms of regeneration rate was ovary of S. fasciata × S. forskaoliana (S-CNR-074) cultured in T5 medium. All rooted plants obtained after transfer under ex vitro conditions are of good quality and fully functional.
In vitro tissue culture is a rapid method for production and commercialization of valuable species, and it is of most relevance to ensure a true-to-type regeneration to assuring a final product uniformity. However, it is well known that in vitro tissue culture can generate somaclonal variations [33] when cells are forced to a certain differentiation pathway. Among the most influential factors in inducing somaclonal variation are the PGRs. It is well known that, among others, 2,4-D and TDZ have disrupting action causing abnormalities in somatic embryos [34] even if, in our experimental procedure, modifications have been obtained with NOA and BA, as reported also for grapevine [24]. The most important modification consists of changes in chromosome number, generating polyploids or aneuploids [35]. Somaclonal variation is somehow uncontrollable, but in some situations, it could be a valuable source of variability in order to generate and select new genotypes while it is a disadvantage if the goal is the preservation of elite genotypes and their micropropagation [36]. Among the several methods to study the genetic stability of regenerants during plant tissue culture [37], flow cytometry, more than the long and laborious cytogenetic methodologies, represents a precise and fast method to identify changes in ploidy and DNA content of a species, thus measuring the genetic variability and somaclonal variation in various species [38,39,40]. Flow cytometry measures fluorescence intensity of nuclei DNA stained with DNA specific fluorochromes, and for DNA ploidy and nuclei content identification the use of a known internal standard is recommended [41]. In the present study the ploidy level of the Sansevieria mother plants is not defined; therefore, the analyses tend to evaluate the relative genetic stability in terms of DNA fluorescence variations of the nuclei extracted from plant materials in our particular growth conditions. In particular, our experiments entailed the comparison among MP and their Rs to assess the fluorescence emission, which is related to nuclei DNA amount. The present analysis aims to highlight any variations witnessed in the potential genetic instability of Sansevieria’s spp. clones, regenerated from several types of explants, grown in different culture media.
The greater variation in DNA amount has been observed in plants regenerated from S-CNR-054; both regenerants in fact showed a different level of DNA fluorescence compared to MP, probably due to the in vitro culture conditions, while a high degree of uniformity has been observed among other Sansevieria lines. In contrast, comparison within mother plants showed differences in relative DNA fluorescence, and this is not surprising since they belong to several species. Few indications are given in the literature regarding the DNA content in Sansevieria spp. [42,43]. Additional studies are needed to better characterize the group of regenerants belonging to S. parva species (group MP S-CNR-054) by flow cytometric analysis of the absolute DNA content, using the appropriate internal biological reference standard with a known DNA amount. A more precise response regarding inner variability could be achieved also by classic cytogenetic studies on metaphases spreads of regenerants compared to S. parva, known to have a chromosome complement of 2n = 40 [44].
The presence of genetic variability in regenerated plants of S. parva is an interesting aspect from the perspective of producing new variants for commercial purposes. Nevertheless, the high degree of uniformity in DNA relative content, verified in the other samples, validates the regeneration system starting from floral explants. These results are in agreement with those reported for other species [14,15,35,45] about stability of regenerated plants.
We can conclude that the fast assessment of the flow DNA content stability of in vitro regenerated plants is essential both for valuation of true-to-type clones to be used for production of homogeneous plants and for selection of individuals bearing genetically stable variations, useful in the improvement of ornamental plants.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae9020138/s1, Figure S1: Flow cytometry evaluation of three extraction buffers for Sansevieria nuclei isolation in suspension from 2 mg leaf tissue. Figure S2: An example of “clearing up” FCM analysis from debris and unnecessary particles. Table S1: List of seven Sansevieria genotypes used for the regeneration of somatic embryos in vitro.
Author Contributions
Conceptualization, C.C., A.C., F.C., M.S., A.B., S.L. and L.A.; methodology, C.C., A.C., D.G., A.F. and L.A.; validation, F.C., M.S. and S.L.; investigation, C.C., A.C., A.M., D.G., A.F. and L.A.; data curation, C.C., A.C., D.G., A.F. and L.A; writing—original draft preparation, C.C., A.C., F.C., D.G., A.F. and L.A.; writing—review and editing, C.C., A.C., F.C., A.M., M.S., A.B., S.L., D.G., A.F. and L.A.; supervision, C.C., A.C., F.C., M.S., S.L. and L.A.; funding acquisition, M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Fondo Finalizzato alla Ricerca di Ateneo dell’Università di Palermo, FFR-D15-005355 to Maurizio Sajeva.
Data Availability Statement
Data are available from the corresponding author upon request.
Acknowledgments
We would like to thank Francesca La Bella for technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Webb, R.H.; Newton, L.E. The Genus Sansevieria: A Pictorial Guide to the Species; Arid Lands Press: Tucson, Arizona, 2022; p. 200. [Google Scholar]
- Brown, N.E. Sansevieria. A monograph of all the known species. Bull. Misc. Inform. Kew. 1915, 5, 185–261. [Google Scholar] [CrossRef]
- García-Hernández, E.; Loera-Quezada, M.M.; Morán-Velázquez, D.C.; López, M.G.; Chable-Vega, M.A.; Santillán-Fernández, A.; Zavaleta-Mancera, H.A.; Tang, J.Z.; Azadi, P.; Ibarra-Laclette, E.; et al. Indirect organogenesis for high frequency shoot regeneration of two cultivars of Sansevieria trifasciata Prain differing in fiber production. Sci. Rep. 2022, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bandehali, S.; Miri, T.; Onyeaka, H.; Kumar, P. Current state of indoor air phytoremediation using potted plants and green walls. Atmosphere 2021, 12, 473. [Google Scholar] [CrossRef]
- Chatakul, P.; Janpathompong, S. Interior plants: Trends, species, and their benefits. Build Environ. 2022, 222, 109325. [Google Scholar] [CrossRef]
- Suberi, I.; Noor, N.; Darnis, D.; Mukai, Y.; Usup, G. The Potential of Ornamental Plant, Sansevieria trifasciata to Inhibit the Growth of Harmful Algal Bloom Species; Isals Publishing: Yazd, Iran, 2014; p. 42. [Google Scholar]
- Takawira-Nyenya, R.; Newton, L.E.; Wabuyele, E.; Stedje, B. Ethnobotanical uses of Sansevieria Thunb (Asparagaceae) in Coast Province of Kenya. Ethnobot Res. App. 2014, l12, 51–69. [Google Scholar]
- Yusnita, Y.; Pungkastiani, W.; Hapsoro, D. In vitro organogenesis of two Sansevieria trifasciata cultivars on different concentrations of benzyladenine (BA). AGRIVITA J. Agric. Sci. 2011, 33, 147–153. [Google Scholar] [CrossRef]
- Henley, R.W.; Chase, A.R.; Osborne, L.S. Sansevieria Production Guide. USA: University of Florida. 1991. Available online: https://mrec.ifas.uf.edu/foliage/folnotes/sansevie.htm (accessed on 10 November 2022).
- Kaur, J.; Mudgal, G. An efficient and quick protocol for in vitro multiplication of snake plant, Sansevieria trifasciata var. Laurentii [Prain]. Plant Cell Tissue Organ Cult. 2021, 147, 405–411. [Google Scholar] [CrossRef]
- Sarmast, M.K.; Salehi, M.; Salehi, H. The potential of different parts of Sansevieria trifasciata L. leaf for meristemoids production. Aust. J. Basic Appl. Sci. 2009, 3, 2506–2509. [Google Scholar]
- Agrotan, J.; Inderiati, S. Indirect organogenesis and induction of morphogenic callus for in vitro propagation of Sansevieria masoniana. Jurnal Agrotan 2015, 1, 1–8. [Google Scholar]
- Carra, A.; De Pasquale, F.; Ricci, A.; Carimi, F. Diphenylurea derivatives induce somatic embryogenesis in Citrus. Plant Cell Tissue Organ Cult. 2006, 87, 41–48. [Google Scholar] [CrossRef]
- Carra, A.; Sajeva, M.; Abbate, L.; Siragusa, M.; Pathirana, R.; Carimi, F. Factors affecting somatic embryogenesis in eight Italian grapevine cultivars and the genetic stability of embryo-derived regenerants as assessed by molecular markers. Sci. Hortic. 2016, 204, 123–127. [Google Scholar] [CrossRef]
- Carra, A.; Sajeva, M.; Abbate, L.; Siragusa, M.; Sottile, F.; Carimi, F. In vitro plant regeneration of caper (Capparis spinosa L.) from floral explants and genetic stability of regenerants. Plant Cell Tissue Organ Cult. 2012, 109, 373–381. [Google Scholar] [CrossRef]
- Fiore, M.C.; Carimi, F.; Carra, A.; Sunseri, F. Efficient plant regeneration via somatic embryogenesis in bulbing fennel using immature flower explants. In Vitro Cell. Dev. Biol. Plant. 2012, 48, 440–445. [Google Scholar] [CrossRef]
- Lou, Q.; Liu, H.; Luo, W.; Chen, K.; Liu, Y. Creating a novel petal regeneration system for function identification of colour gene of grape hyacinth. Plant Methods 2021, 17, 1–10. [Google Scholar] [CrossRef]
- de Almeida, N.V.; Rivas, E.B.; Cardoso, J.C. Somatic embryogenesis from flower tepals of Hippeastrum aiming regeneration of virus-free plants. Plant Sci. 2022, 317, 111191. [Google Scholar] [CrossRef]
- Botelho, F.B.S.; Rodrigues, C.S.; Bruzi, A.T. Ornamental plant breeding. Ornam. Hortic. 2015, 21, 9–16. [Google Scholar] [CrossRef]
- Datta, S.K. Breeding of ornamentals: Success and technological status. Nucleus 2022, 65, 107–128. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassay with tobacco tissue culture. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Dolezel, J.; Binarova, P.; Lucretti, S. Analysis of nuclear-DNA content in plant-cells by flow-cytometry. Biol. Plant. 1989, 31, 113–120. [Google Scholar] [CrossRef]
- Otto, F. Preparation and staining of cells for high-resolution DNA analysis. In Flow Cytometry and Cell Sorting; Radbruch, A., Ed.; Springer: Berlin, Germany, 1992; pp. 101–104. [Google Scholar] [CrossRef]
- Catalano, C.; Abbate, L.; Motisi, A.; Crucitti, D.; Cangelosi, V.; Pisciotta, A.; Di Lorenzo, R.; Carimi, F.; Carra, A. Autotetraploid emergence via somatic embryogenesis in Vitis vinifera induces marked morphological changes in shoots, mature leaves, and stomata. Cells 2021, 10, 1336. [Google Scholar] [CrossRef]
- Palma, M.A.; Hall, C.R.; Collart, A.J. Repeat buying behavior for ornamental plants: A consumer profile. J. Food Distrib. Res. 2011, 42, 67–77. [Google Scholar] [CrossRef]
- Carimi, F.; Barizza, E.; Gardiman, M.; Schiavo, F.L. Somatic embryogenesis from stigmas and styles of grapevine. In Vitro Cell Dev. Biol. Plant 2005, 41, 249–252. [Google Scholar] [CrossRef]
- Sajeva, M.; Carra, A.; de Pasquale, F.; Carimi, F. Somatic embryogenesis and plant regeneration from pistil transverse thin cell layers of lemon (Citrus limon). Plant Biosyst. 2008, 142, 199–203. [Google Scholar] [CrossRef]
- Wojciechowicz, M.K. Organogenesis and somatic embryogenesis induced in petal cultures of Sedum species. Acta Biol. Cracov. Bot. 2009, 51, 83–90. [Google Scholar]
- Vidal, J.R.; Rama, J.; Taboada, L.; Martin, C.; Ibañez, M.; Segura, A.; González-Benito, M.E. Improved somatic embryogenesis of grapevine (Vitis vinifera) with focus on induction parameters and efficient plant regeneration. Plant Cell Tissue Organ Cult. 2009, 96, 85–94. [Google Scholar] [CrossRef]
- Prado, M.J.; Grueiro, M.P.; González, M.V.; Testillano, P.S.; Domínguez, C.; López, M.; Rey, M. Efficient plant regeneration through somatic embryogenesis from anthers and ovaries of six autochthonous grapevine cultivars from Galicia (Spain). Sci. Hortic 2010, 125, 342–352. [Google Scholar] [CrossRef][Green Version]
- Akin-Idowu, P.E.; Ibitoye, D.O.; Ademoyegun, O.T. Tissue culture as a plant production technique for horticultural crops. Afr. J. Biotechnol. 2009, 8, 3782–3788. [Google Scholar]
- Shumilina, D.; Kornyukhin, D.; Domblides, E.; Soldatenko, A.; Artemyeva, A. Effects of genotype and culture conditions on microspore embryogenesis and plant regeneration in Brassica rapa ssp. rapa L. Plants 2020, 9, 278. [Google Scholar] [CrossRef]
- Bednarek, P.T.; Orłowska, R.; Koebner, R.M.D.; Zimny, J. Quantification of the tissue-culture induced variation in barley (Hordeum vulgare L.). BMC Plant Biol. 2007, 7, 10. [Google Scholar] [CrossRef]
- Garcia, C.; Furtado de Almeida, A.A.; Costa, M.; Britto, D.; Valle, R.; Royaert, S.; Marelli, J.P. Abnormalities in somatic embryogenesis caused by 2, 4-D: An overview. Plant Cell Tissue Organ Cult. 2019, 137, 193–212. [Google Scholar] [CrossRef]
- De-la-Peña, C.; Nic-Can, G.; Galaz-Ávalos, R.; Avilez-Montalvo, R.; Loyola-Vargas, V. The role of chromatin modifications in somatic embryogenesis in plant. Front. Plant Sci. 2015, 6, 635. [Google Scholar] [CrossRef]
- Krishna, H.; Alizadeh, M.; Singh, D.; Singh, U.; Chauhan, N.; Eftekhari, M.; Sadh, R.K. Somaclonal variations and their applications in horticultural crops improvement. 3 Biotech 2016, 6, 1–18. [Google Scholar] [CrossRef]
- Bairu, M.W.; Aremu, A.O.; Van Staden, J. Somaclonal variation in plants: Causes and detection methods. Plant Growth Regul. 2011, 63, 147–173. [Google Scholar] [CrossRef]
- Loureiro, J.; Pinto, G.; Lopes, T.; Doležel, J.; Santos, C. Assessment of ploidy stability of the somatic embryogenesis process in Quercus suber L. using flow cytometry. Planta 2005, 221, 815–822. [Google Scholar] [CrossRef]
- Sharma, S.; Bryan, G.; Winfield, M.; Millam, S. Stability of potato (Solanum tuberosum L.) plants regenerated via somatic embryos, axillary bud proliferated shoots, microtubers and true potato seeds: A comparative phenotypic, cytogenetic and molecular assessment. Planta 2007, 226, 1449–1458. [Google Scholar] [CrossRef]
- Grosso, V.; Farina, A.; Giorgi, D.; Nardi, L.; Diretto, G.; Lucretti, S. A high-throughput flow cytometry system for early screening of in vitro made polyploids in Dendrobium hybrids. Plant Cell Tissue Organ Cult. 2018, 132, 57–70. [Google Scholar] [CrossRef]
- Dolezel, J.; Bartoš, J. Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. 2005, 95, 99–110. [Google Scholar] [CrossRef]
- Zonneveld, B.J.M.; Leitch, I.J.; Bennett, M.D. First nuclear DNA amounts in more than 300 Angiosperms. Ann. Bot. 2005, 96, 229–244. [Google Scholar] [CrossRef]
- Smarda, P.; Bure, P.; Horová, L.; Leitch, I.J.; Mucina, L.; Pacini, E.; Tichý, L.; Grulich, V.; Rotreklová, O. Ecological and evolutionary significance of genomic GC content diversity in monocots. Proc. Natl. Acad. Sci. USA 2014, 111, E4096–E4102. [Google Scholar] [CrossRef]
- Nazeer, M.A.; Khoshoo, T.N. Cytology of Some Species of Sansevieria Thunb. Cytologia 1984, 49, 325–332. [Google Scholar] [CrossRef]
- Catalano, C.; Abbate, L.; Fatta Del Bosco, S.; Motisi, A.; Carimi, F.; De Michele, R.; Mercati, F.; D’Onghia, A.M.; Carra, A. Different cell types affect the transition from juvenile to mature phase in Citrus plants regenerated through somatic embryogenesis. Plants 2022, 11, 1811. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).