Morpho-Physiological and Transcriptional Regulation of Root System under Saline Conditions in Nymphaea Plants
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Salt Treatment
2.2. Plant Growth and Root Measurement
2.3. Transcriptome Analysis of Roots
2.4. ICP-MS Analysis of Na+ and K+ Contents
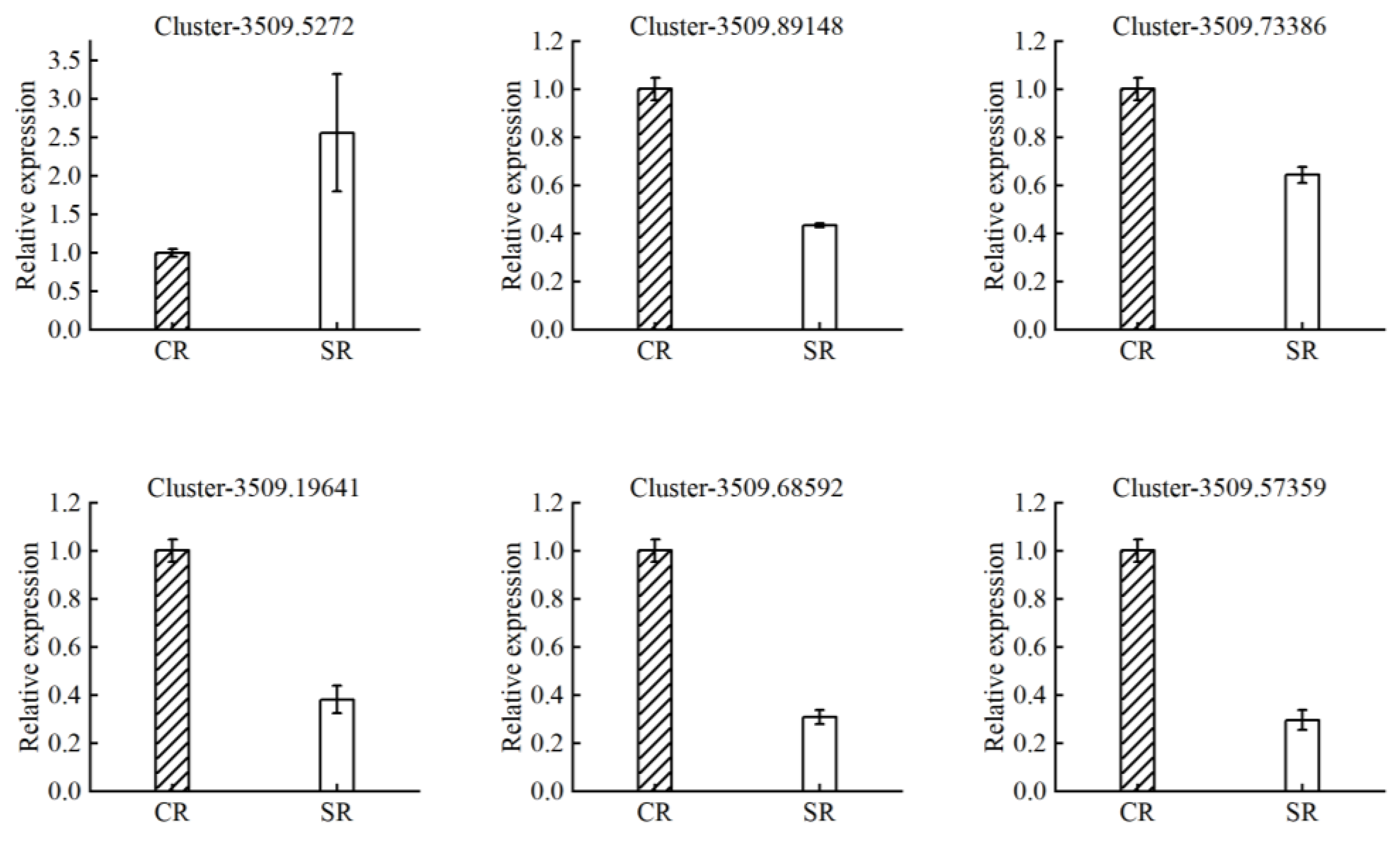
2.5. Quantitative Real-Time PCR
3. Results
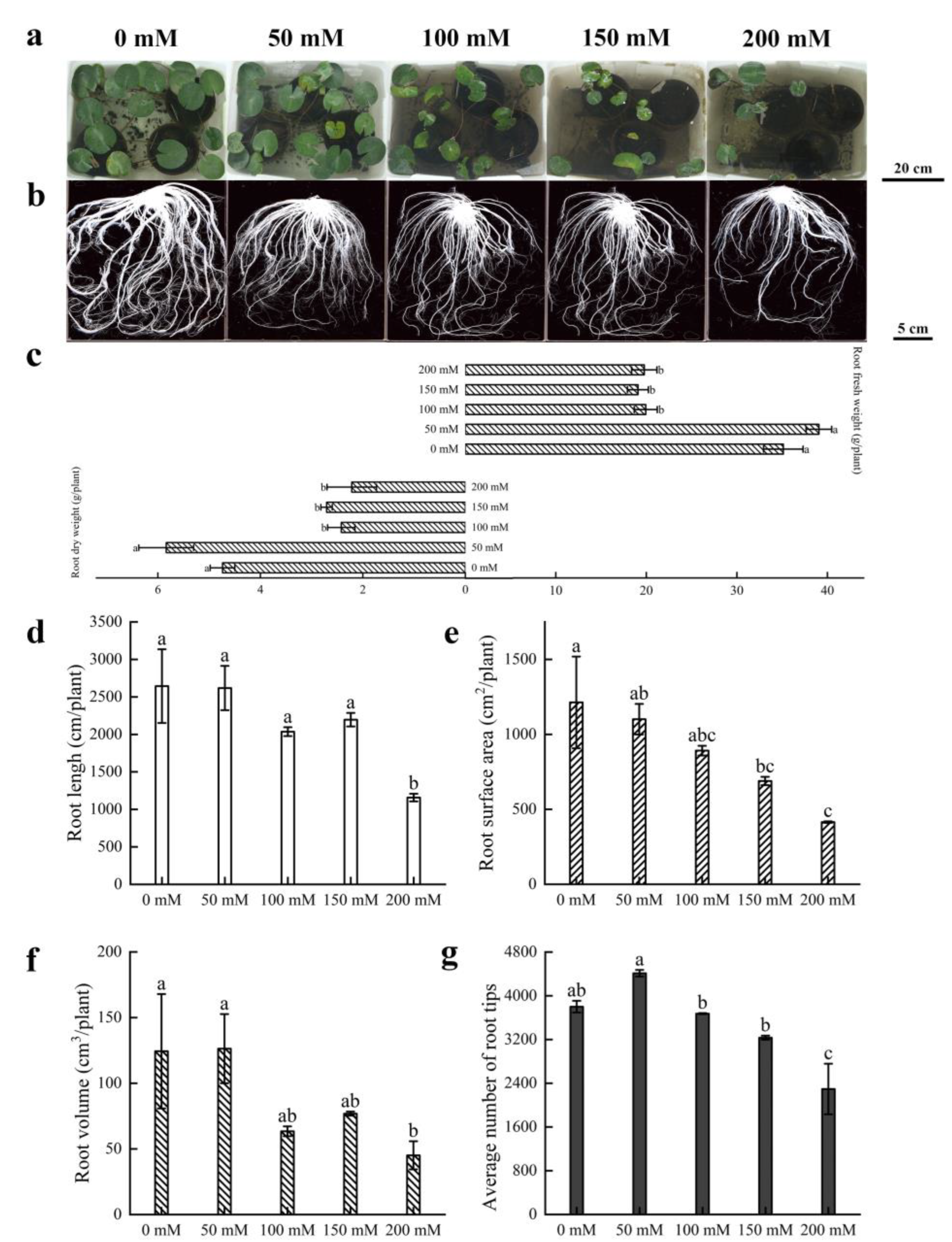
3.1. Morphological Characterization of Water Lily Plants
3.2. Analysis of RNA-Seq Data
3.3. Pathway Enrichment Analyses
3.4. Analysis of Genes Related to Ion Channels and Transporters
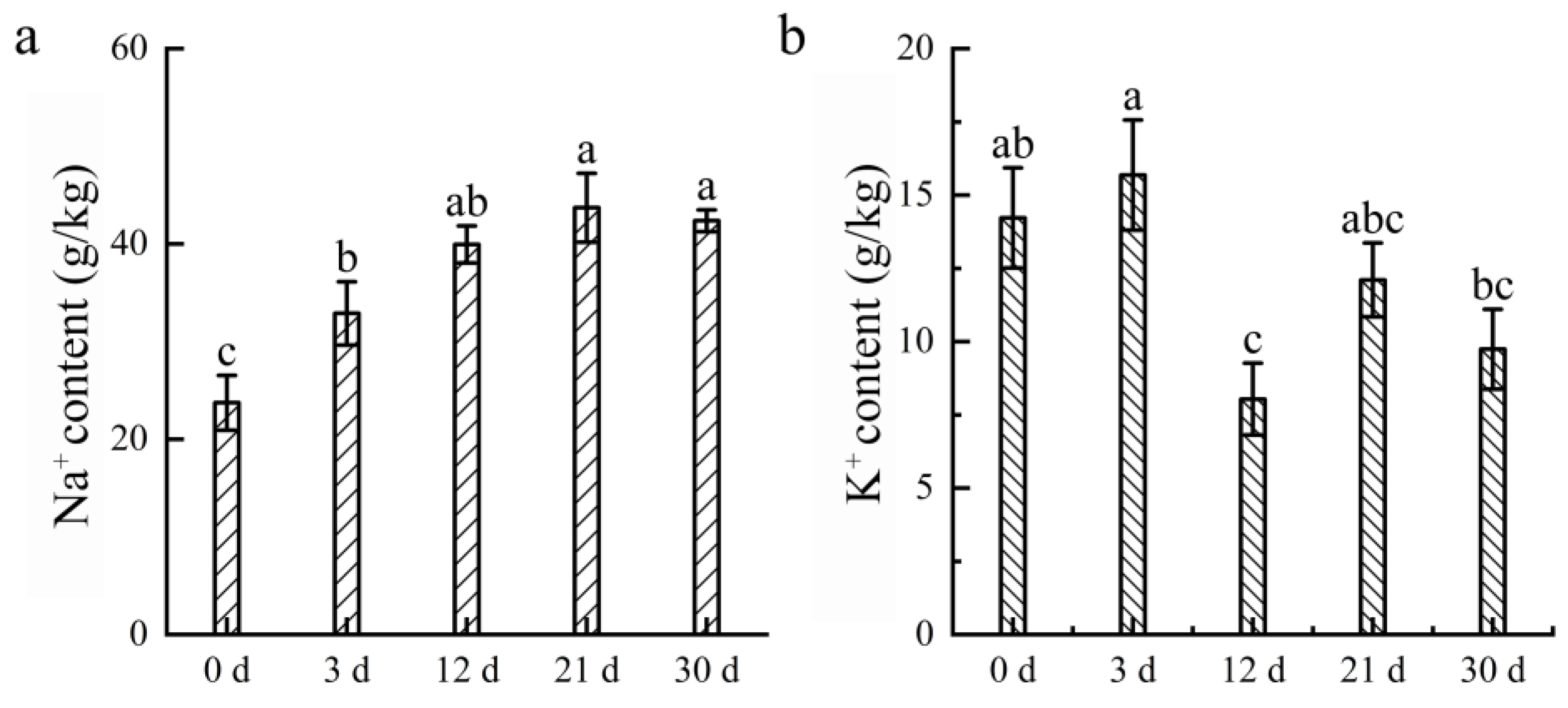
3.5. Analysis of Na+ and K+ in Water Lily Roots
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ha-Tran, D.M.; Nguyen, T.T.M.; Hung, S.-H.; Huang, E.; Huang, C.-C. Roles of Plant Growth-Promoting Rhizobacteria (PGPR) in Stimulating Salinity Stress Defense in Plants: A Review. Int. J. Mol. Sci. 2021, 22, 3154. [Google Scholar] [CrossRef]
- Parida, A.; Das, A. Salt Tolerance and Salinity Effects on Plants: A Review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef]
- Abiala, M.A.; Abdelrahman, M.; Burritt, D.J.; Tran, L.-S.P. Salt Stress Tolerance Mechanisms and Potential Applications of Legumes for Sustainable Reclamation of Salt-Degraded Soils. Land Degrad. Dev. 2018, 29, 3812–3822. [Google Scholar] [CrossRef]
- Mushtaq, Z.; Faizan, S.; Gulzar, B. Salt Stress, Its Impacts on Plants and the Strategies Plants Are Employing against It: A Review. J. Appl. Biol. Biotechnol. 2020, 8, 81–91. [Google Scholar] [CrossRef]
- Sunarpi, H.T.; Motoda, J.; Kubo, M.; Yang, H.; Yoda, K.; Horie, R.; Chan, W.; Leung, H.; Hattori, K. Enhanced Salt Tolerance Mediated by AtHKT1 Transporter-Induced Na+ Unloading from Xylem Vessels to Xylem Parenchyma Cells. Plant J. 2005, 44, 928–938. [Google Scholar] [CrossRef]
- Yang, T.; Zhang, S.; Hu, Y.; Wu, F.; Hu, Q.; Chen, G.; Cai, J.; Wu, T.; Moran, N.; Yu, L. The Role of a Potassium Transporter OsHAK5 in Potassium Acquisition and Transport from Roots to Shoots in Rice at Low Potassium Supply Levels. Plant Physiol. 2014, 166, 945-U757. [Google Scholar] [CrossRef]
- Zhang, K.; Tang, J.; Wang, Y.; Kang, H.; Zeng, J. The Tolerance to Saline-Alkaline Stress Was Dependent on the Roots in Wheat. Physiol. Mol. Biol. Plants 2020, 26, 947–954. [Google Scholar] [CrossRef]
- Chatenet, P.; Froissard, D.; Cook-Moreau, J.; Hourdin, P.; Ghestem, A.; Botineau, M.; Haury, J. Populations of Myriophyllum alterniflorum L. as Bioindicators of Pollution in Acidic to Neutral Rivers in the Limousin Region. Hydrobiologia 2006, 570, 61–65. [Google Scholar] [CrossRef]
- Bennett, S.; Barrett-Lennard, E.; Colmer, T. Salinity and Waterlogging as Constraints to Saltland Pasture Production: A Review. Agric. Ecosyst. Environ. 2009, 129, 349–360. [Google Scholar] [CrossRef]
- Setter, T.; Waters, I.; Sharma, S.; Singh, K.; Kulshreshtha, N.; Yaduvanshi, N.; Ram, P.; Singh, B.; Rane, J.; McDonald, G. Review of Wheat Improvement for Waterlogging Tolerance in Australia and India: The Importance of Anaerobiosis and Element Toxicities Associated with Different Soils. Ann. Bot. 2009, 103, 221–235. [Google Scholar] [CrossRef]
- Babourina, O.; Rengel, Z. Ion Transport in Aquatic Plants. In Waterlogging Signalling and Tolerance in Plants; Mancuso, S., Shabala, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 221–238. ISBN 978-3-642-10304-9. [Google Scholar]
- Garcia-Sanchez, M.; Jaime, M.; Ramos, A.; Sanders, D.; Fernandez, J. Sodium-Dependent Nitrate Transport at the Plasma Membrane of Leaf Cells of the Marine Higher Plant Zostera marina L. Plant Physiol. 2000, 122, 879–885. [Google Scholar] [CrossRef] [PubMed]
- Rubio, L.; Linares-Rueda, A.; Garcia-Sanchez, M.; Fernandez, J. Physiological Evidence for a Sodium-Dependent High-Affinity Phosphate and Nitrate Transport at the Plasma Membrane of Leaf and Root Cells of Zostera marina L. J. Exp. Bot. 2005, 56, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Ishitani, M.; Kim, C.; Zhu, J.-K. The Arabidopsis thaliana Salt Tolerance Gene SOS1 Encodes a Putative Na+/H+ Antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [PubMed]
- Garciadeblas, B.; Haro, R.; Benito, B. Cloning of Two SOS1 Transporters from the Seagrass Cymodocea nodosa. SOS1 Transporters from Cymodocea and Arabidopsis Mediate Potassium Uptake in Bacteria. Plant Mol. Biol. 2007, 63, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Qiao, G.; Qiu, W.; Yu, D.; Zhou, S.; Shen, Y.; Yu, G.; Jiang, J.; Han, X.; Liu, M. Molecular Breeding of Water Lily: Engineering Cold Stress Tolerance into Tropical Water Lily. Hortic. Res. 2018, 5, 73. [Google Scholar] [CrossRef]
- Luo, H.; Chen, S.; Jiang, J.; Chen, Y.; Chen, F.; Teng, N.; Yin, D.; Huang, C. The Expression of Floral Organ Identity Genes in Contrasting Water Lily Cultivars. Plant Cell Rep. 2011, 30, 1909–1918. [Google Scholar] [CrossRef]
- Hinojosa-Garro, D.; Mason, C.; Underwood, G. Macrophyte Assemblages in Ditches of Coastal Marshes in Relation to Land-Use, Salinity and Water Quality. Fundam. Appl. Limnol. 2008, 172, 325–337. [Google Scholar] [CrossRef]
- Julkowska, M.; Koevoets, I.; Mol, S.; Hoefsloot, H.; Feron, R.; Tester, M.; Keurentjes, J.; Korte, A.; Haring, M.; de Boer, G. Genetic Components of Root Architecture Remodeling in Response to Salt Stress. Plant Cell 2017, 29, 3198–3213. [Google Scholar] [CrossRef]
- Korver, R.A.; van den Berg, T.; Meyer, A.J.; Galvan-Ampudia, C.S.; ten Tusscher, K.H.W.J.; Testerink, C. Halotropism Requires Phospholipase D Zeta 1-Mediated Modulation of Cellular Polarity of Auxin Transport Carriers. Plant Cell Environ. 2020, 43, 143–158. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Yang, X.; Wang, H.; Pan, T.; Wang, Y.; Xu, Y.; Xu, C.; Yang, Z. Genetic Control of Root Plasticity in Response to Salt Stress in Maize. Theor. Appl. Genet. 2021, 134, 1475–1492. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, T.; Zhang, W.; Li, X. SOS3 Mediates Lateral Root Development under Low Salt Stress through Regulation of Auxin Redistribution and Maxima in Arabidopsis. New Phytol. 2011, 189, 1122–1134. [Google Scholar] [CrossRef] [PubMed]
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Saqib, M.; Zorb, C.; Rengel, Z.; Schubert, S. The Expression of the Endogenous Vacuolar Na+/H+ Antiporters in Roots and Shoots Correlates Positively with the Salt Resistance of Wheat (Triticum aestivum L.). PLANT Sci. 2005, 169, 959–965. [Google Scholar] [CrossRef]
- Sarasketa, A.; Gonzalez-Moro, M.; Gonzalez-Murua, C.; Marino, D. Exploring Ammonium Tolerance in a Large Panel of Arabidopsis thaliana Natural Accessions. J. Exp. Bot. 2014, 65, 6023–6033. [Google Scholar] [CrossRef]
- Tolay, I. The Impact of Different Zinc (Zn) Levels on Growth and Nutrient Uptake of Basil (Ocimum basilicum L.) Grown under Salinity Stress. PLoS ONE 2021, 16, e0246493. [Google Scholar] [CrossRef] [PubMed]
- Coelho, D.G.; Miranda, R.D.; Paula-Marinho, S.D.; de Carvalho, H.H.; Prisco, J.T.; Gomes, E. Ammonium Nutrition Modulates K+ and N Uptake, Transport and Accumulation during Salt Stress Acclimation of Sorghum Plants. Arch. Agron. Soil Sci. 2020, 66, 1991–2004. [Google Scholar] [CrossRef]
- Chebbi, M.; Amdouni, T.; Ben, A.S.; Msilini, N.; Lachaal, M.; Ouerghi, Z. Does the Source of Nitrogen Affect the Response of Fenugreek Plants to Saline Stress? Agrochimica. 2017, 61, 296–316. [Google Scholar] [CrossRef]
- Hessini, K.; Jeddi, K.; Siddique, K.; Cruz, C. Drought and Salinity: A Comparison of Their Effects on the Ammonium-Preferring Species Spartina Alterniflora. Physiol. Plant 2021, 172, 431–440. [Google Scholar] [CrossRef]


| Sample Name | Raw Reads | Clean Reads | Clean Bases | Error Rate (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|---|
| CR1 | 22,947,598 | 22,451,618 | 6.7G | 0.03 | 97.58 | 93.25 | 48.55 |
| CR2 | 22,741,980 | 22,228,034 | 6.7G | 0.03 | 97.72 | 93.55 | 47.91 |
| CR3 | 22,409,365 | 21,936,861 | 6.6G | 0.03 | 97.63 | 93.25 | 48.17 |
| SR1 | 23,281,962 | 21,788,747 | 6.5G | 0.03 | 97.86 | 93.75 | 46.52 |
| SR2 | 22,502,300 | 22,344,183 | 6.7G | 0.03 | 97.89 | 93.92 | 47.56 |
| SR3 | 20,460,810 | 19,022,924 | 5.7G | 0.03 | 97.99 | 94.03 | 43.76 |
| Gene ID | log2FC | p-Value | Description |
|---|---|---|---|
| Downregulated | |||
| Cluster-3509.89148 | −3.11 | 2.67 × 10−3 | sodium transporter HKT1 |
| Cluster-3509.60818 | −4.40 | 9.59 × 10−3 | potassium transporter 5 |
| Cluster-3509.73386 | −2.68 | 5.13 × 10−3 | potassium transporter 5-like |
| Cluster-3509.79079 | −1.72 | 4.51 × 10−2 | potassium transporter 5-like |
| Cluster-3509.78332 | −1.58 | 3.22 × 10−2 | K+ uptake permease 4 |
| Cluster-3509.41717 | −1.34 | 1.31 × 10−2 | boron transporter 2 |
| Cluster-3509.64063 | −1.47 | 2.36 × 10−4 | boron transporter 2 |
| Cluster-3509.65281 | −2.94 | 2.04 × 10−4 | anion transporter 3, chloroplastic |
| Cluster-3509.82188 | −1.25 | 3.10 × 10−2 | organic cation/carnitine transporter 7-like |
| Cluster-3509.72310 | −1.05 | 4.78 × 10−2 | cation-transporting P-type ATPase |
| Cluster-3509.19641 | −2.02 | 7.55 × 10−3 | ABC transporter G family member 36 |
| Cluster-3509.96046 | −3.61 | 9.84 × 10−4 | ABC transporter G family member |
| Cluster-3509.61142 | −1.33 | 3.79 × 10−2 | ABC transporter G family member 29 |
| Cluster-3509.60854 | −5.93 | 8.54 × 10−3 | ABC transporter G family member 21 |
| Cluster-3509.98196 | −2.31 | 1.13 × 10−2 | ABC transporter I family member 17-like |
| Cluster-3509.82719 | −1.14 | 3.20 × 10−2 | ABC transporter I family member 6, chloroplastic |
| Cluster-3509.68592 | −2.68 | 1.59 × 10−2 | ammonium transporter 1 member 1-like |
| Cluster-3509.60927 | −2.84 | 2.33 × 10−2 | ammonium transporter 2-like |
| Cluster-3509.57872 | −2.78 | 1.36 × 10−3 | ammonium transporter 1 member 1-like |
| Cluster-3509.78326 | −4.61 | 4.17 × 10−3 | ammonium transporter 3 member 1-like |
| Cluster-3509.57359 | −3.13 | 8.47 × 10−5 | high-affinity nitrate transporter 2.1 |
| Cluster-3509.63374 | −3.31 | 1.47 × 10−4 | high-affinity nitrate transporter-activating protein 2.1-like |
| Cluster-3509.66911 | −3.23 | 6.24 × 10−25 | urea-proton symporter DUR3 |
| Cluster-3509.51839 | −2.80 | 6.07 × 10−3 | urea-proton symporter DUR3 |
| Cluster-3509.81777 | −1.57 | 3.48 × 10−2 | proton-dependent oligopeptide transporter family |
| Cluster-3509.18628 | −6.08 | 5.55 × 10−3 | proton-dependent oligopeptide transporter family |
| Cluster-3509.40878 | −1.70 | 7.53 × 10−3 | oligopeptide transporter 4 |
| Cluster-3509.77846 | −1.43 | 4.91 × 10−2 | peptide transporter |
| Cluster-3509.97247 | −1.53 | 3.45 × 10−2 | amino acid transporter |
| Cluster-3509.81032 | −1.55 | 6.73 × 10−3 | amino acid/polyamine transporter I |
| Cluster-3509.22635 | −1.91 | 3.95 × 10−2 | cationic amino acid transporter 1 |
| Upregulated | |||
| Cluster-3509.5272 | 3.56 | 6.35 × 10−3 | aluminum-activated malate transporter |
| Cluster-3509.5272 | 3.56 | 6.35 × 10−3 | aluminum-activated malate transporter |
| Cluster-3509.69281 | 2.29 | 7.37 × 10−3 | NRT1/PTR FAMILY 7.2 |
| Cluster-3509.41673 | 1.51 | 2.56 × 10−2 | amino acid transporter AVT6A-like |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Du, F.; Huang, Q.; Gao, X.; Zhang, Z.; Cui, J.; Chang, Y.; Liu, X.; Yao, D. Morpho-Physiological and Transcriptional Regulation of Root System under Saline Conditions in Nymphaea Plants. Horticulturae 2023, 9, 132. https://doi.org/10.3390/horticulturae9020132
Chen S, Du F, Huang Q, Gao X, Zhang Z, Cui J, Chang Y, Liu X, Yao D. Morpho-Physiological and Transcriptional Regulation of Root System under Saline Conditions in Nymphaea Plants. Horticulturae. 2023; 9(2):132. https://doi.org/10.3390/horticulturae9020132
Chicago/Turabian StyleChen, Shaozhou, Fengfeng Du, Qianhao Huang, Xiaojing Gao, Zhiyuan Zhang, Jian Cui, Yajun Chang, Xiaojing Liu, and Dongrui Yao. 2023. "Morpho-Physiological and Transcriptional Regulation of Root System under Saline Conditions in Nymphaea Plants" Horticulturae 9, no. 2: 132. https://doi.org/10.3390/horticulturae9020132
APA StyleChen, S., Du, F., Huang, Q., Gao, X., Zhang, Z., Cui, J., Chang, Y., Liu, X., & Yao, D. (2023). Morpho-Physiological and Transcriptional Regulation of Root System under Saline Conditions in Nymphaea Plants. Horticulturae, 9(2), 132. https://doi.org/10.3390/horticulturae9020132






