The Molecular Mechanism of Relatively Low-Temperature-Induced Broccoli Flower Bud Differentiation Revealed by Transcriptomic Profiling
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatment
2.2. Paraffin Sectioning
2.3. Sampling for Transcriptomic Analysis
2.4. RNA-Seq and Differential Expression Analysis
2.5. Motif Overrepresentation ESTIMATION
2.6. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
3. Results
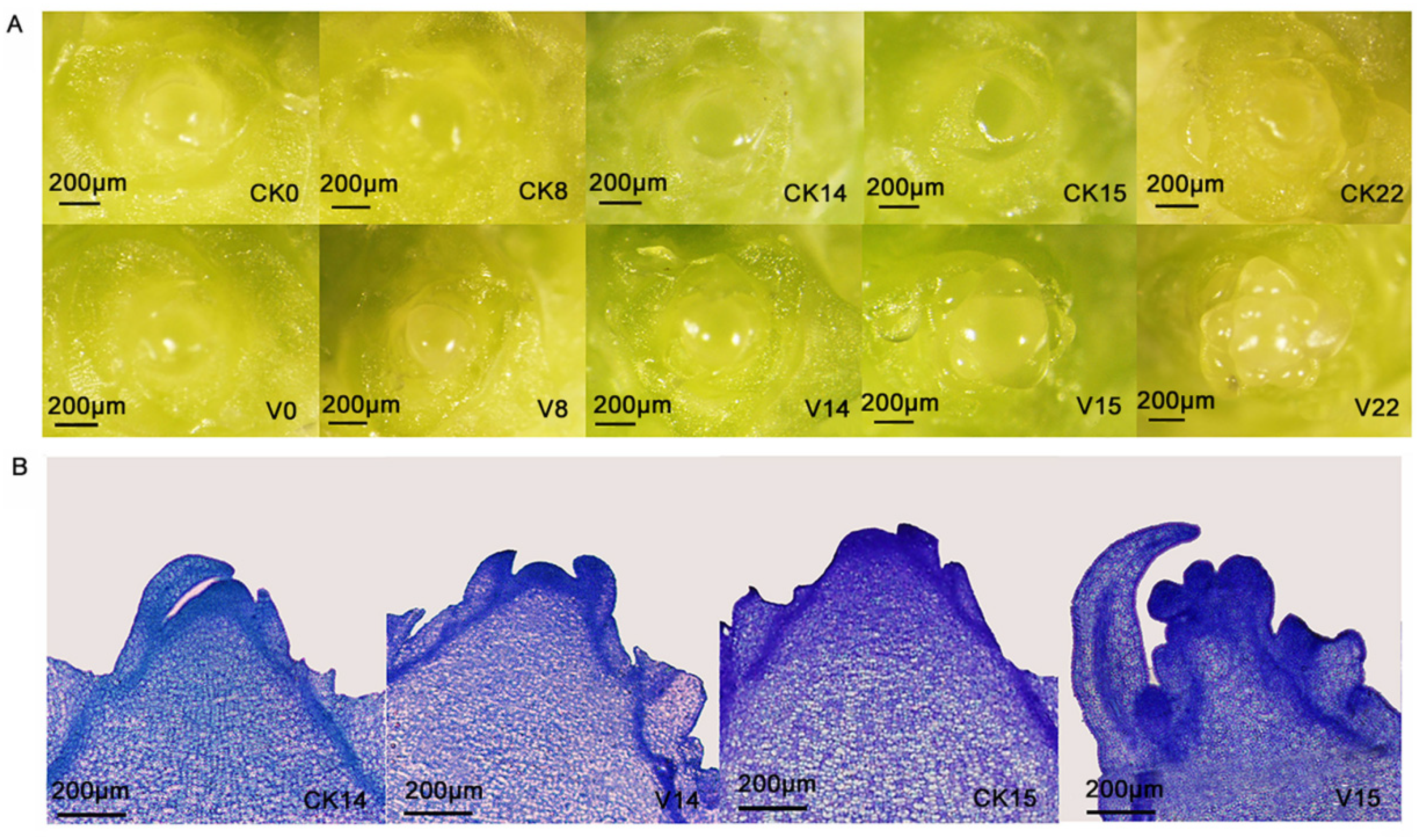
3.1. Morphological Changes at Different Stages of FBD
3.2. Evaluation of RNA-Seq Results
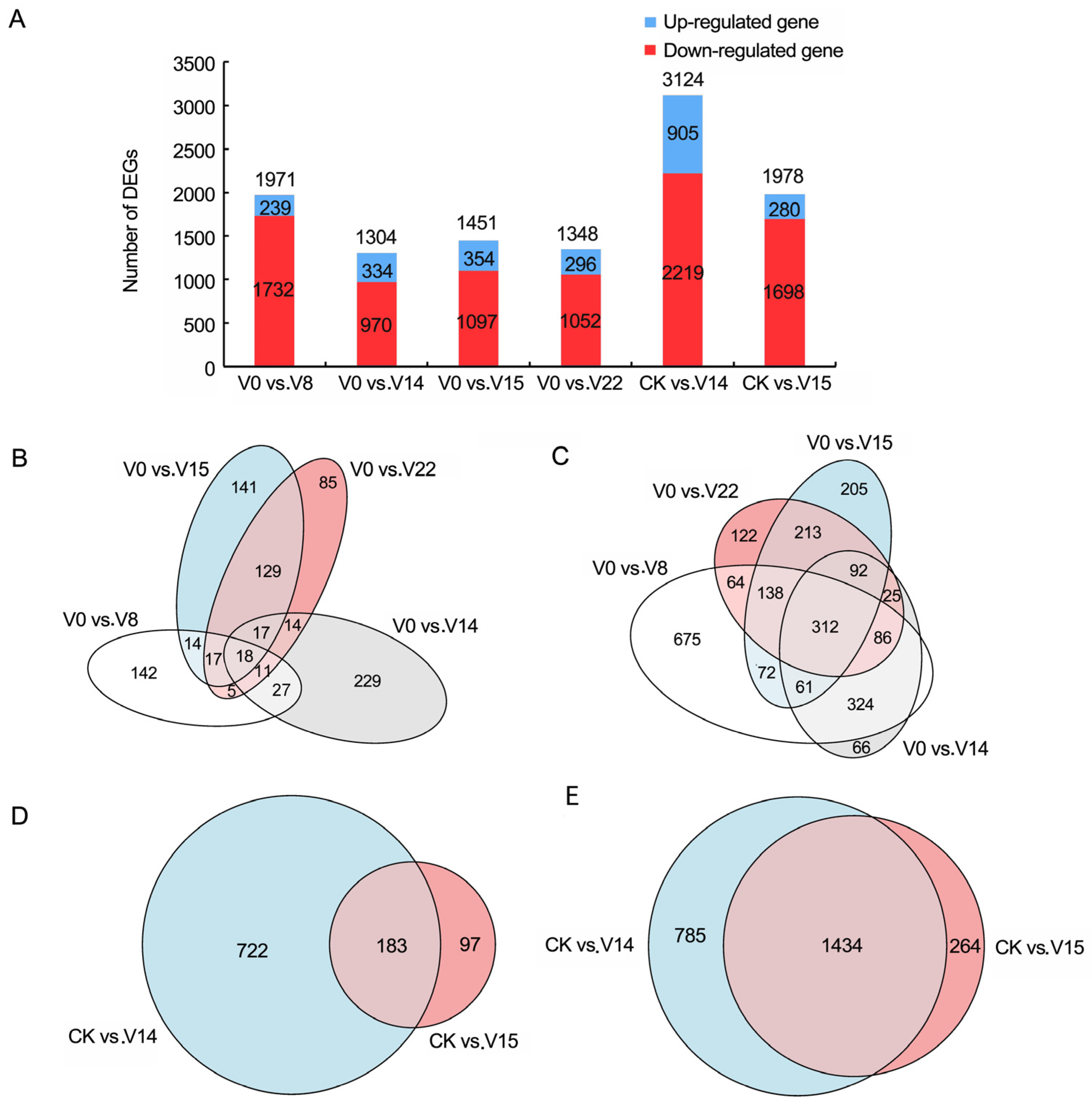
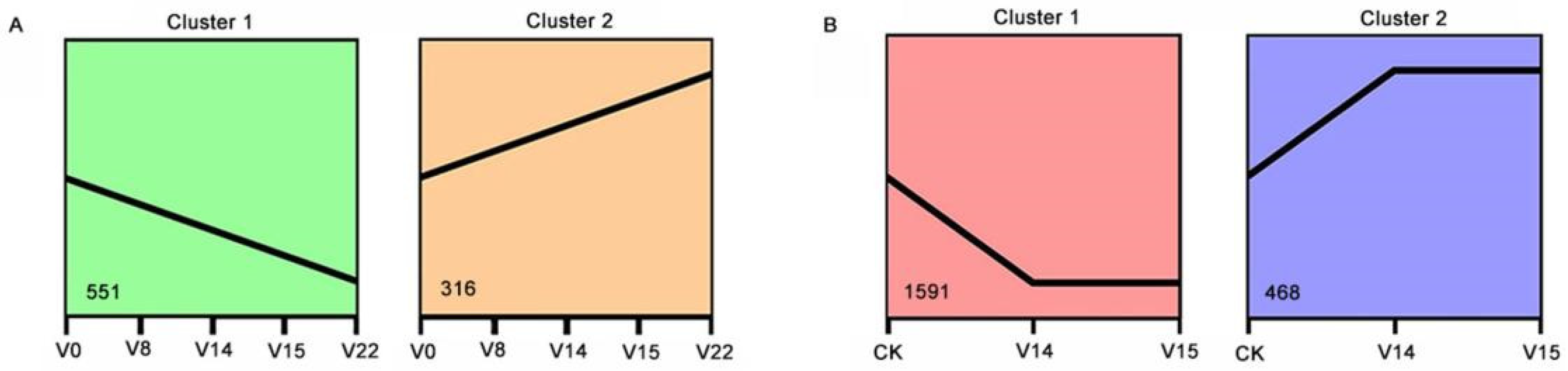
3.3. Screening and Analysis of DEGs
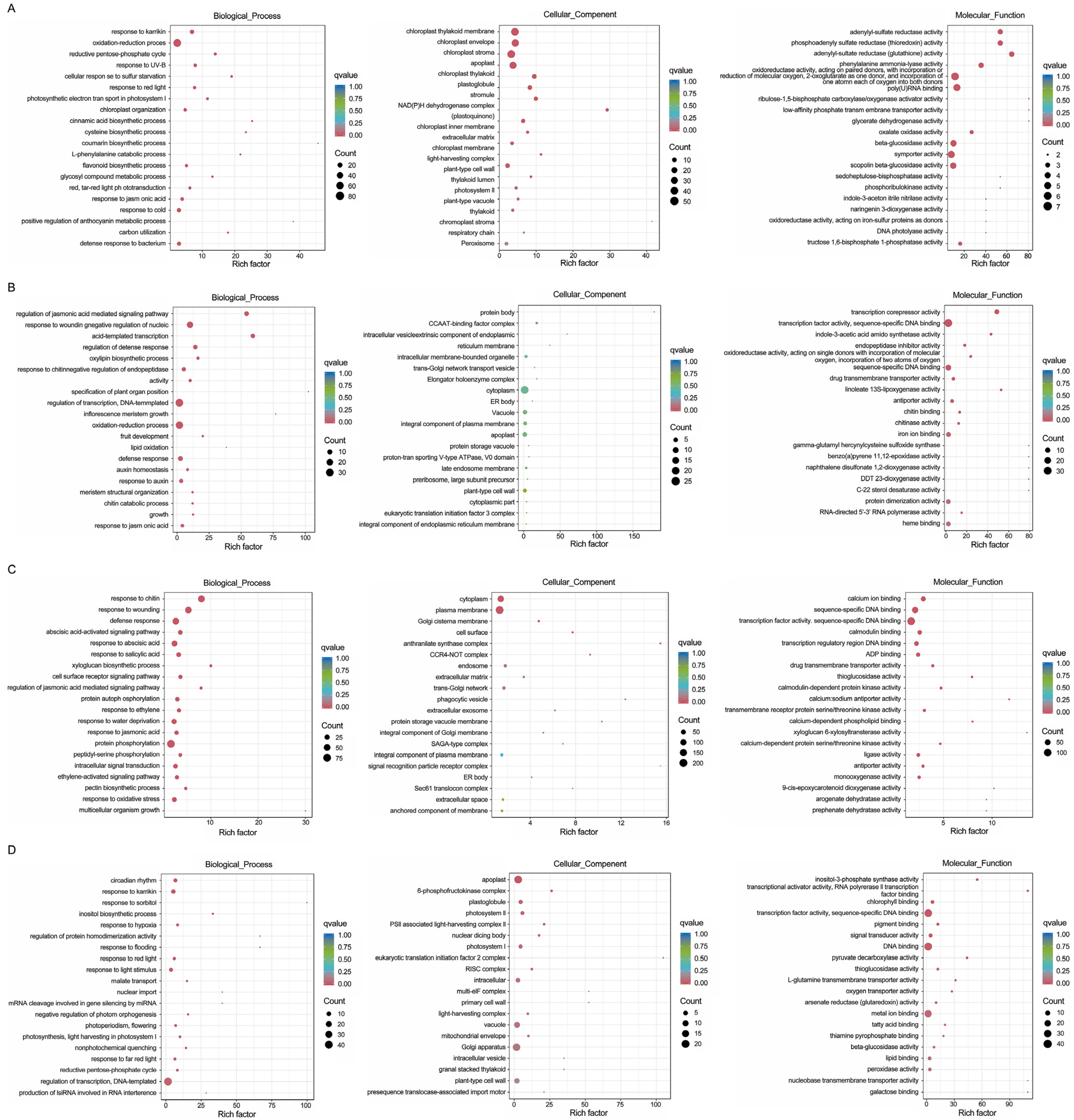
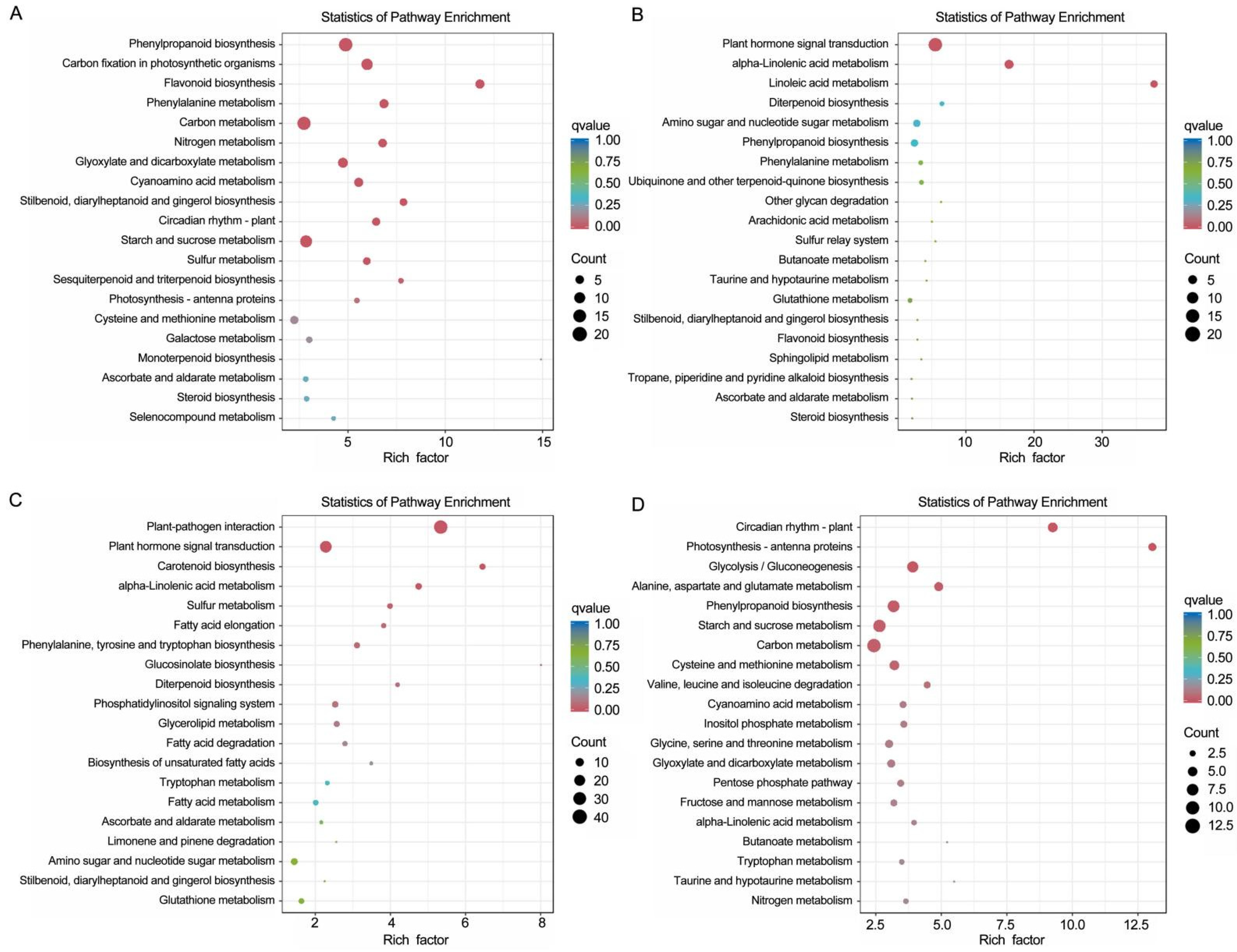
3.4. GO Enrichment Analysis and KEGG Pathway Analysis of Coexpressed DEGs
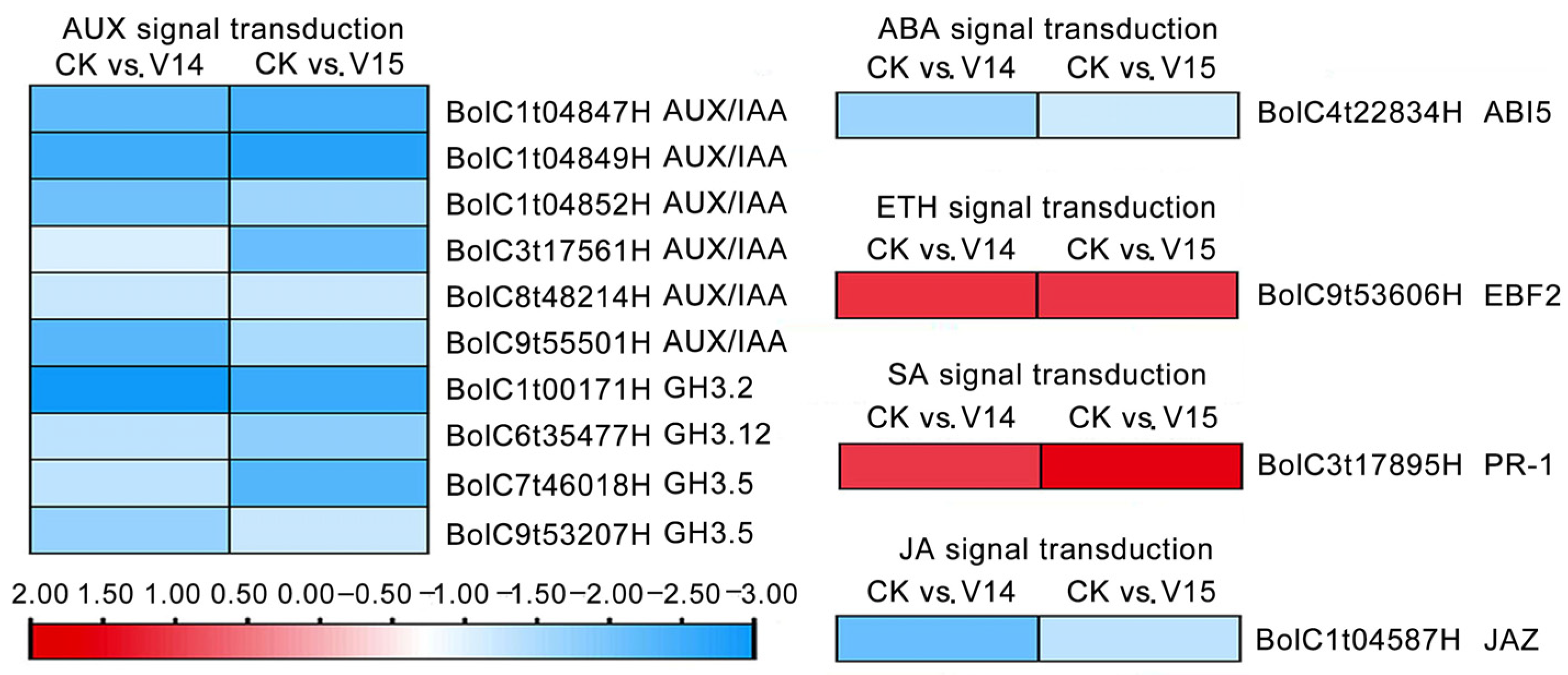
3.5. Identification of DEGs Involved in the Plant Hormone Signaling Pathways
3.6. Identification of Differentially Expressed TFs (DETFs)
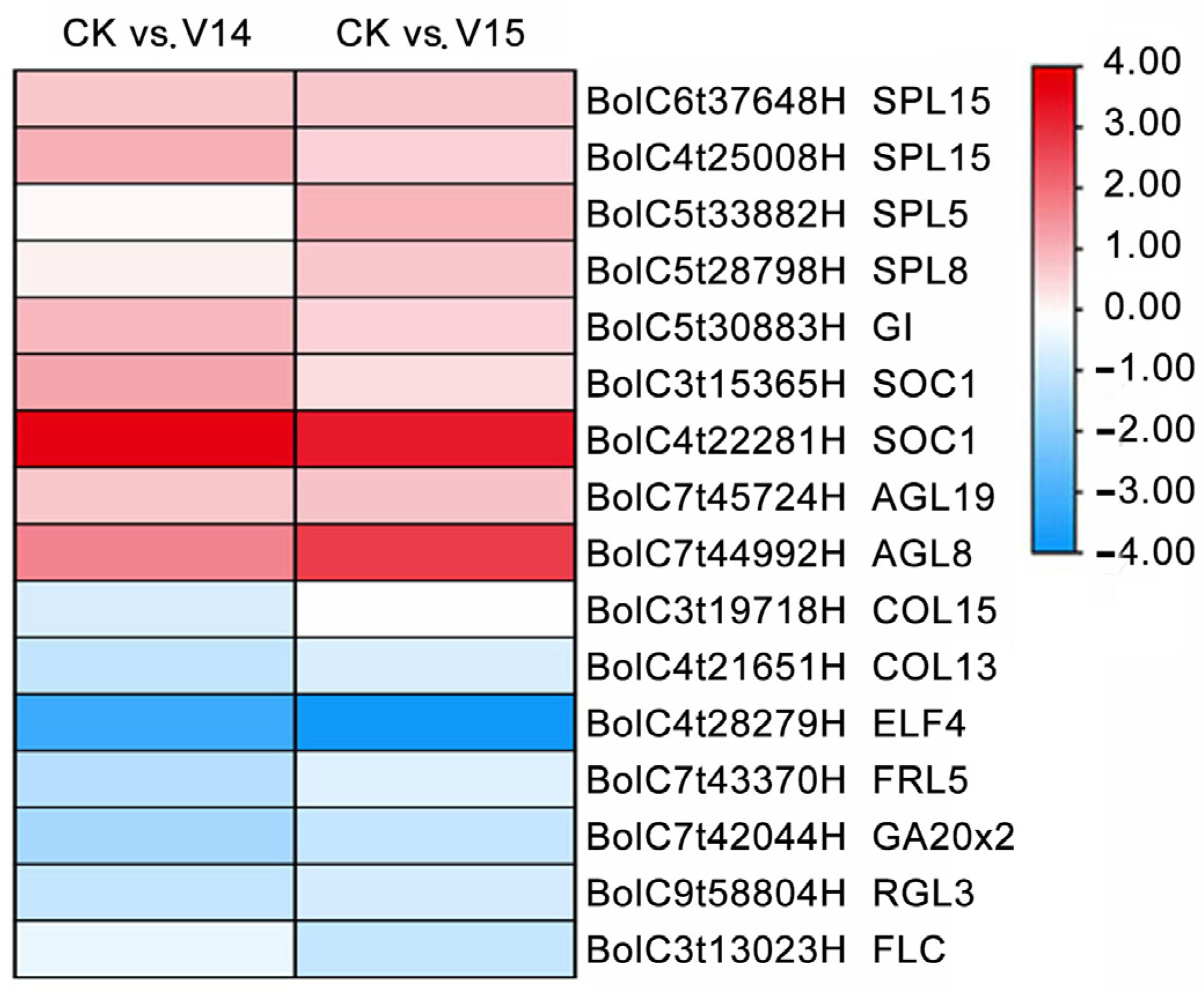
3.7. Identification of DEGs Involved in the FBD Pathways
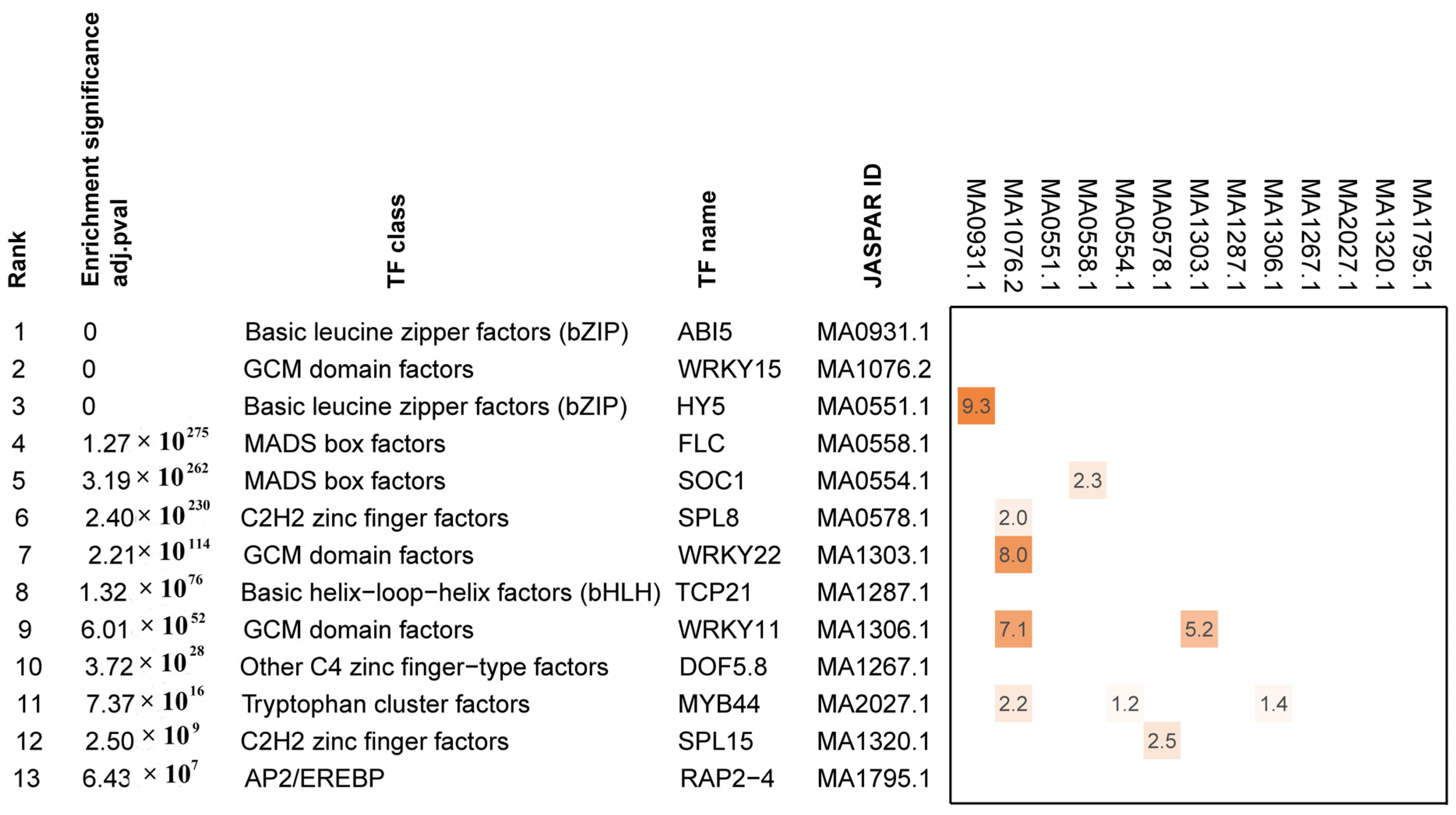
3.8. Analysis of Enrichment with Binding Sites for DETFs Involved in the FBD Pathway
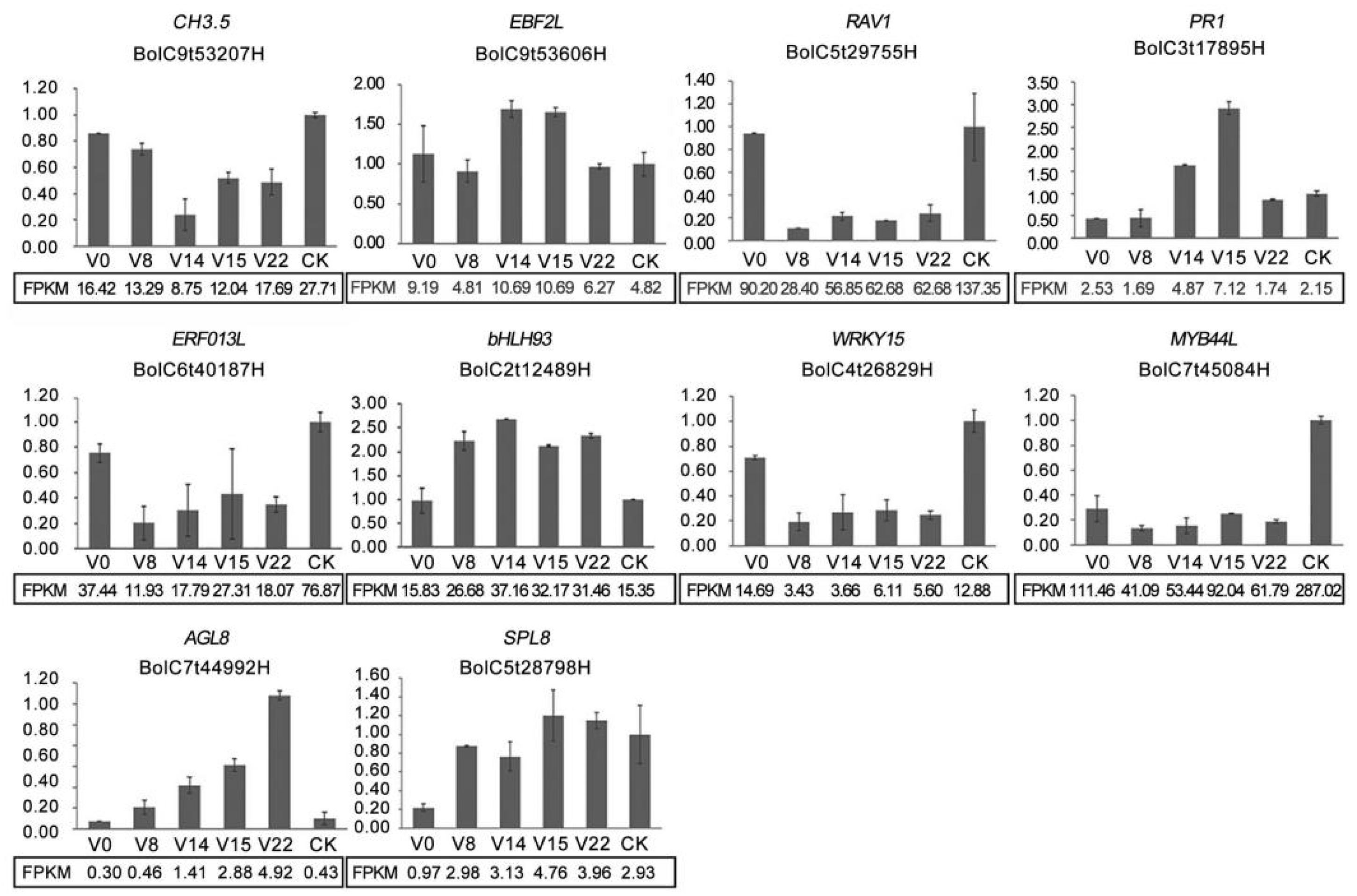
3.9. qRT-PCR Analysis Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Grabowska, A.; Kunicki, E.; Kalisz, A.; Wojciechowska, R.; Leja, M.; Sękara, A. Chilling stress applied to broccoli transplants of different age affects yield of the plants cultivated in summer. Hortic. Sci. 2014, 41, 71–79. [Google Scholar] [CrossRef]
- Thiruvengadam, M.; Shih, C.F.; Yang, C.H. Expression of an antisense Brassica oleracea GIGANTEA (BoGI) gene in transgenic broccoli causes delayed flowering, leaf senescence, and post-harvest yellowing retardation. Plant Mol. Biol. Rep. 2015, 33, 1499–1509. [Google Scholar] [CrossRef]
- Fahey, J.W.; Zhang, Y.; Talalay, P. Broccoli sprouts: An exceptionally rich source of inducers of enzymes that protect against chemical carcinogens. Proc. Natl. Acad. Sci. USA 1997, 94, 10367–10372. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.W.; Fu, S.F.; Liu, Y.J.; Chen, C.C.; Chang, C.H.; Yang, Y.W.; Huang, H.J. Analysis of ambient temperature-responsive transcriptome in shoot apical meristem of heat-tolerant and heat-sensitive broccoli inbred lines during floral head formation. BMC Plant Biol. 2019, 19, 3. [Google Scholar] [CrossRef] [PubMed]
- Berndtsson, E.; Andersson, R.; Johansson, E.; Olsson, M.E. Side streams of broccoli leaves: A climate smart and healthy food ingredient. Int. J. Environ. Res. Public Health 2020, 17, 2406. [Google Scholar] [CrossRef]
- Cai, G.Q.; Yang, Q.Y.; Chen, H.; Yang, Q.; Zhang, C.Y.; Fan, C.C.; Zhou, Y.M. Genetic dissection of plant architecture and yield-related traits in Brassica napus. Sci. Rep. 2016, 6, 21625. [Google Scholar] [CrossRef]
- Li, H.G.; Zhang, L.P.; Hu, J.H.; Zhang, F.G.; Chen, B.Y.; Xu, K.; Gao, G.Z.; Li, H.; Zhang, T.Y.; Li, Z.Y.; et al. Genome-wide association mapping reveals the genetic control underlying branch angle in rapeseed (Brassica napus L.). Front. Plant Sci. 2017, 8, 1054. [Google Scholar] [CrossRef]
- Fujime, Y. A difference of response to low temperature between cauliflower and broccoli. Acta Hortic. 1988, 218, 141–152. [Google Scholar] [CrossRef]
- Grevsen, K.; Olesen, J.E. Modeling development of broccoli (Brassica oleracea L. var. italica) from transplanting to head initiation. J. Hortic. Sci. Biotechnol. 1999, 74, 698–705. [Google Scholar] [CrossRef]
- Kałużewicz, A.; Krzesiński, W.; Knaflewski, M.; Lisiecka, J.; Spiżewski, T.; Frąszczak, B. The effect of temperature on the broccoli yield and length of the period from head initiation to harvest. Acta Sci. Pol. Hortorum Cultus 2010, 9, 167–174. [Google Scholar]
- Grabowska, A.; Sękara, A.; Bieniasz, M.; Kunicki, E.; Kalisz, A. Dark-chilling of seedlings affects initiation and morphology of broccoli inflorescence. Not. Bot. Horti Agrobot. Cluj-Napoca 2013, 41, 213–218. [Google Scholar] [CrossRef]
- Huang, B.Y.; Qian, P.P.; Gao, N.; Shen, J.; Hou, S.W. Fackel interacts with gibberellic acid signaling and vernalization to mediate flowering in Arabidopsis. Planta 2017, 245, 939–950. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; He, Y.H. Experiencing winter for spring flowering: A molecular epigenetic perspective on vernalization. J. Integr. Plant Biol. 2020, 62, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Mouradov, A.; Cremer, F.; Coupland, G. Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell 2002, 14, S111–S130. [Google Scholar] [CrossRef]
- Komeda, Y. Genetic regulation of time to flower in Arabidopsis thaliana. Annu. Rev. Plant Biol. 2004, 55, 521–535. [Google Scholar] [CrossRef]
- Kim, D.H. Current understanding of flowering pathways in plants: Focusing on the vernalization pathway in Arabidopsis and several vegetable crop plants. Hortic. Environ. Biotechnol. 2020, 61, 209–227. [Google Scholar] [CrossRef]
- Li, H.J.; Fan, Y.H.; Yu, J.Y.; Chai, L.; Zhang, J.F.; Jiang, J.; Cui, C.; Zheng, B.C.; Jiang, L.C.; Lu, K. Genome-wide identification of flowering-time genes in Brassica species and reveals a correlation between selective pressure and expression patterns of vernalization-pathway genes in Brassica napus. Int. J. Mol. Sci. 2018, 19, 3632. [Google Scholar] [CrossRef]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef]
- Sheldon, C.C.; Rouse, D.T.; Finnegan, E.J.; Peacock, W.J.; Dennis, E.S. The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. USA 2000, 97, 3753–3758. [Google Scholar] [CrossRef]
- Searle, I.; He, Y.H.; Turck, F.; Vincent, C.; Fornara, F.; Kröber, S.; Amasino, R.A.; Coupland, G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006, 20, 898–912. [Google Scholar] [CrossRef]
- Putterill, J.; Robson, F.; Lee, K.; Simon, R.; Coupland, G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 1995, 80, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Samach, A.; Onouchi, H.; Gold, S.E.; Ditta, G.S.; Schwarz-Sommer, Z.; Yanofsky, M.F.; Coupland, G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 2000, 288, 1613–1616. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.W.; Ma, J.H.; Han, Y.Z.; Chen, X.J.; Fu, Y.F. Cloning and expression analysis of the soybean CO-Like gene GmCOL9. Plant Mol. Biol. Rep. 2011, 29, 352–359. [Google Scholar] [CrossRef]
- Cheng, X.F.; Wang, Z.Y. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 2005, 43, 758–768. [Google Scholar] [CrossRef] [PubMed]
- Steinbach, Y. The Arabidopsis thaliana CONSTANS-LIKE 4 (COL4)–A modulator of flowering time. Front. Plant Sci. 2019, 10, 651. [Google Scholar] [CrossRef] [PubMed]
- Carr, S.M.; Irish, V.F. Floral homeotic gene expression defines developmental arrest stages in Brassica oleraeea L. vars botrytis and italica. Planta 1997, 201, 179–188. [Google Scholar] [CrossRef]
- Duclos, D.V.; Björkman, T. Meristem identity gene expression during curd proliferation and flower initiation in Brassica oleracea. J. Exp. Bot. 2008, 59, 421–433. [Google Scholar] [CrossRef]
- Ridge, S.; Brown, P.H.; Hecht, V.; Driessen, R.G.; Weller, J.L. The role of BoFLC2 in cauli-flower (Brassica oleracea var. botrytis L.) reproductive development. J. Exp. Bot. 2015, 66, 125–135. [Google Scholar] [CrossRef]
- Tyagi, S.; Sri, T.; Singh, A.; Mayee, P.; Shivaraj, S.M.; Sharma, P.; Singh, A. SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 influences flowering time, lateral branching, oil quality, and seed yield in Brassica juncea cv. Varuna. Funct. Integr. Genom. 2019, 19, 43–60. [Google Scholar] [CrossRef]
- Fujime, Y.; Okuda, N. The physiology of flowering in Brassicas, especially about cauliflower and broccoli. Acta Hortic. 1996, 407, 247–254. [Google Scholar] [CrossRef]
- Grabowska, A.; Kunicki, E.; Libik, A. Effects of age and cold storage of transplants on the growth and quality of broccoli heads. J. Fruit Ornam. Plant Res. 2007, 66, 31–38. [Google Scholar] [CrossRef]
- Kalisz, A.; Cebula, S.; Kunicki, E.; Gil, J.; Sekara, A.; Grabowska, A. Effect of chilling stress before transplanting on morphological parameters of broccoli heads. Acta Sci. Pol. Hortorum Cultus 2014, 13, 129–139. [Google Scholar]
- Garcês, H.; Sinha, N. Fixing and sectioning tissue from the plant Kalanchoë daigremontiana. Cold Spring Harb. Protoc. 2009, 10, pdb.prot5301. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Oshchepkov, D.; Chadaeva, I.; Kozhemyakina, R.; Shikhevich, S.; Sharypova, E.; Savinkova, L.; Klimova, N.V.; Tsukanov, A.; Levitsky, V.G.; Markel, A.L. Transcription factors as important regulators of changes in behavior through domestication of gray rats: Quantitative data from RNA sequencing. Int. J. Mol. Sci. 2022, 23, 12269. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kazan, K.; Lyons, R. The link between flowering time and stress tolerance. J. Exp. Bot. 2016, 67, 47–60. [Google Scholar] [CrossRef]
- Bao, S.J.; Hua, C.M.; Shen, L.S.; Yu, H. New insights into gibberellin signaling in regulating flowering in Arabidopsis thaliana. J. Integr. Plant Biol. 2020, 62, 118–131. [Google Scholar] [CrossRef]
- Liu, F.; Wang, Y.; Ding, Z.T.; Zhao, L.; Xiao, J.; Wang, L.J.; Ding, S.B. Transcriptomic analysis of flower development in tea (Camellia sinensis (L.)). Gene 2017, 631, 39–51. [Google Scholar] [CrossRef]
- Wils, C.R.; Kaufmann, K. Gene-regulatory networks controlling inflorescence and flower development in Arabidopsis thaliana. BBA Gene Regul. Mech. 2017, 1860, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Boss, P.K.; Sensi, E.; Hua, C.; Davies, C.; Thomas, M.R. Cloning and characterisation of grapevine (Vitis vinifera L.) MADS-box genes expressed during inflorescence and berry development. Plant Sci. 2002, 162, 887–895. [Google Scholar] [CrossRef]
- Ding, L.; Kim, S.Y.; Michaels, S.D. FLOWERING LOCUS C EXPRESSOR family proteins regulate FLOWERING LOCUS C expression in both winter-annual and rapid-cycling Arabidopsis. Plant Physiol. 2013, 163, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Baudry, A.; Ito, S.; Song, Y.H.; Strait, A.A.; Kiba, T.; Lu, S.; Henriques, R.; Pruneda-Paz, J.L.; Chua, N.H.; Tobin, E.M.; et al. F-Box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 2010, 22, 606–622. [Google Scholar] [CrossRef] [PubMed]
- Sawa, M.; Kay, S.A. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 11698–11703. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.X.; Zhou, R.J.; Hu, Q.; Wei, W.L.; Liu, J. Conservation and divergence of the CONSTANS-Like (COL) genes related to flowering and circadian rhythm in Brassica napus. Front. Plant Sci. 2021, 12, 760379. [Google Scholar] [CrossRef] [PubMed]
- Wilkosz, R.; Schlappi, M. A gene expression screen identifies EARLI1 as a novel vernalization-responsive gene in Arabidopsis thaliana. Plant Mol. Biol. 2000, 44, 777–787. [Google Scholar] [CrossRef]
- Dai, Y.; Zhang, S.J.; Sun, X.; Li, G.L.; Yuan, L.Y.; Li, F.; Zhang, H.; Zhang, S.F.; Chen, G.H.; Wang, C.G.; et al. Comparative transcriptome analysis of gene expression and regulatory characteristics associated with different vernalization periods in Brassica rapa. Genes 2020, 11, 392. [Google Scholar] [CrossRef]
- Sun, I.H.; Kim, J.E.; Na, H. Flower bud differentiation in response to low temperature treatment and day-length extension treatment in radish (Raphanus sativus L.). Korean J. Breed. Sci. 2016, 48, 48–53. [Google Scholar] [CrossRef]
- Conti, L. Hormonal control of the floral transition: Can one catch them all? Dev. Biol. 2017, 430, 288–301. [Google Scholar] [CrossRef]
- Ma, Z.B.; Li, W.; Wang, H.P.; Yu, D.Q. WRKY transcription factors WRKY12 and WRKY13 interact with SPL10 to modulate age-mediated flowering. J. Integr. Plant Biol. 2020, 62, 1659–1673. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.Y.; Sun, Y.P.; Wang, B.T.; Yu, S.; Dai, H.Y.; Li, H.; Zhang, Z.H.; Zhang, J.X. Woodland strawberry WRKY71 acts as a promoter of flowering via a transcriptional regulatory cascade. Hortic. Res. 2020, 7, 137. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Li, L.; Ye, T.T.; Lu, Y.M.; Chen, X.; Wu, Y. The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J. Exp. Bot. 2013, 64, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; Dhandapani, V.; Rameneni, J.J.; Li, X.N.; Sivanandhan, G.; Choi, S.R.; Pang, W.X.; Im, S.; Lim, Y.P. Genome-wide analysis and characterization of Aux/IAA family genes in Brassica rapa. PLoS ONE 2016, 11, e0151522. [Google Scholar] [CrossRef] [PubMed]
- Renau-Morata, B.; Nebauer, S.G.; García-Carpintero, V.; Cañizares, J.; Minguet, E.G.; de los Mozos, M.; Molina, R.V. Flower induction and development in saffron: Timing and hormone signalling pathways. Ind. Crop. Prod. 2021, 164, 113370. [Google Scholar] [CrossRef]
- Blanc-Mathieu, R.; Dumas, R.; Turchi, L.; Lucas, J.; Parcy, F. Plant-TFClass: A structural classification for plant transcription factors. Trends Plant Sci. 2023, 21, S1360–S1385. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.G.; Zhang, L.P.; Li, D.B.; Wang, F.; Yu, D.Q. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 1963–1971. [Google Scholar] [CrossRef]
- Jiang, Y.J.; Liang, G.; Yang, S.Z.; Yu, D.Q. Arabidopsis WRKY57 functions as a node of convergence for Jasmonic acid- and Auxin-mediated signals in Jasmonic acid-induced leaf senescence. Plant Cell 2014, 26, 230–245. [Google Scholar] [CrossRef]
- Chen, W.; Hao, W.J.; Xu, Y.X.; Zheng, C.; Ni, D.J.; Yao, M.Z.; Chen, L. Isolation and characterization of CsWRKY7, a subgroup IId WRKY transcription factor from Camellia sinensis, linked to development in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 2815. [Google Scholar] [CrossRef]
- Sheldon, C.C.; Hills, M.J.; Lister, C.; Dean, C.; Dennis, E.S.; Peacock, W.J. Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc. Natl. Acad. Sci. USA 2008, 105, 2214–2219. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Suh, S.S.; Park, E.; Cho, E.; Ahn, J.H.; Kim, S.G.; Lee, J.S.; Kwon, Y.M.; Lee, I. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 2000, 14, 2366–2376. [Google Scholar] [CrossRef] [PubMed]
- Scortecci, K.C.; Michaels, S.D.; Amasino, R.M. Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 2001, 26, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Theißen, G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001, 4, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, I. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 2010, 61, 2247–2254. [Google Scholar] [CrossRef]
- Michaels, S.D.; Ditta, G.; Gustafson-Brown, C.; Pelaz, S.; Yanofsky, M.; Amasino, R.M. AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J. 2003, 33, 867–874. [Google Scholar] [CrossRef]
- Zhang, Y.; Schwarz, S.; Saedler, H.; Huijser, P. SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol. Biol. 2007, 63, 429–439. [Google Scholar] [CrossRef]
- Jung, J.H.; Ju, Y.; Seo, P.J.; Lee, J.H.; Park, C.M. The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 2012, 69, 577–588. [Google Scholar] [CrossRef]
- Yu, S.; Galvão, V.C.; Zhang, Y.C.; Horrer, D.; Zhang, T.Q.; Hao, Y.H.; Feng, Y.Q.; Wang, S.; Schmid, M.; Wang, J.W. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA PROMOTER BINDING-LIKE transcription factors. Plant Cell 2012, 24, 3320–3332. [Google Scholar] [CrossRef]
- Schmid, M.; Uhlenhaut, N.H.; Godard, F.; Demar, M.; Bressan, R.; Weigel, D.; Lohmann, J.U. Dissection of floral induction pathways using global expression analysis. Development 2003, 130, 6001–6012. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, Z.L.; Yang, Y.X.; Chen, X.Q.; Chen, G.P. Function annotation of an SBP-box gene in Arabidopsis based on analysis of co-expression networks and promoters. Int. J. Mol. Sci. 2009, 10, 116–132. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Hyun, Y.B.; Richter, R.; Vincent, C.; Martinez-Gallegos, R.; Porri, A.; Coupland, G. Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Dev. Cell 2016, 37, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.N.; Liu, R.X.; Chen, Y.; Cui, J.T.; Ge, W.; Zhang, K.Z. Molecular identification and functional verification of SPL9 and SPL15 of lilium. Mol. Genet. Genom. 2022, 297, 63–74. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chai, W.; He, X.; Wen, B.; Jiang, Y.; Zhang, Z.; Bai, R.; Zhang, X.; Xu, J.; Hou, L.; Li, M.; et al. The Molecular Mechanism of Relatively Low-Temperature-Induced Broccoli Flower Bud Differentiation Revealed by Transcriptomic Profiling. Horticulturae 2023, 9, 1353. https://doi.org/10.3390/horticulturae9121353
Chai W, He X, Wen B, Jiang Y, Zhang Z, Bai R, Zhang X, Xu J, Hou L, Li M, et al. The Molecular Mechanism of Relatively Low-Temperature-Induced Broccoli Flower Bud Differentiation Revealed by Transcriptomic Profiling. Horticulturae. 2023; 9(12):1353. https://doi.org/10.3390/horticulturae9121353
Chicago/Turabian StyleChai, Wenchen, Xia He, Boyue Wen, Yajie Jiang, Zixuan Zhang, Rui Bai, Xinling Zhang, Jin Xu, Leiping Hou, Meilan Li, and et al. 2023. "The Molecular Mechanism of Relatively Low-Temperature-Induced Broccoli Flower Bud Differentiation Revealed by Transcriptomic Profiling" Horticulturae 9, no. 12: 1353. https://doi.org/10.3390/horticulturae9121353
APA StyleChai, W., He, X., Wen, B., Jiang, Y., Zhang, Z., Bai, R., Zhang, X., Xu, J., Hou, L., Li, M., & Zhang, J. (2023). The Molecular Mechanism of Relatively Low-Temperature-Induced Broccoli Flower Bud Differentiation Revealed by Transcriptomic Profiling. Horticulturae, 9(12), 1353. https://doi.org/10.3390/horticulturae9121353





