Abstract
This is the first worldwide report of X. fastidiosa (Xf) subsp. pauca on Castanea sativa and the first characterization of Xf infection on this species. Plants located in three sites (in a long-term affected area in Apulia) were monitored for symptoms and bacterial concentrations in spring and summer, while microscopic analyses were performed to evaluate the pathogen distribution in the xylem vessels. All chestnut plants appeared asymptomatic but the Xf subsp. pauca strain “De Donno” was present at low concentrations (1.14 × 104 and 1.56 × 103 cfu mL−1 in April and June) and with a low incidence (respectively, 38% and 30%). The FISH-CLSM (Fluorescent In Situ Hybridization—Confocal Laser Scanning Microscope) analysis showed evident Xf occlusions but in a low percentage (9.2 ± 3.4%); these data can explain the lack of symptoms on the canopy. Furthermore, through a bibliographic analysis it emerged that Philaenus spumarius and Neophilaenus campestris, two Xf vectors present in Europe, are suckling feeding insects on chestnut trees and could be involved in the bacterial transmission to this species. Asymptomatic Xf host species can play a considerable role in new outbreak emergence or in the expansion of existing ones. So, it is essential to identify them to plan more effective monitoring activities.
1. Introduction
Xylella fastidiosa (Xf) is a quarantine pathogenic bacterium already found in different subspecies in various countries of the Mediterranean basin. It causes diseases to develop in plants such as olive trees, grapevines, almond trees, etc., and can compromise the entire production of entire crops [1]. Its diffusion is mainly due to the nutritional habits of the xylem vectors which transport the bacterium from one infected plant to a healthy one [2]. Trading and planting infected nursery material is also beneficial for the spread of the bacterium [3]. The first detection of the bacterium in Europe dates back to 2013 [4], when it was identified in the province of Lecce (Apulia, Italy) on olive trees strongly compromised by a disease now called “Olive Quick Decline Syndrome” (OQDS), and the genetic variant detected was X. fastidiosa subsp. pauca strain “De Donno” (ST53) [5]. After this first Italian detection, new outbreaks have been detected in other European countries [6].
Xylella fastidiosa is a polyphagous bacterium and it has been detected in 679 host species [7], but does not cause disease in most of these [8]. In 2013, it was already known that Xf (regardless of subspecies) had 132 host species belonging to 46 different families [9]. In 2016, the number of host species discovered had already risen to 329, belonging to 75 different families, and in 2018 it reached 563 species from 82 different botanical families [10]. It is evident that, after the detection of the bacterium in new world areas (e.g., Europe), even very far from the areas (Americas) of origin of the bacterium, the number of host species more than quintupled. This happened because the introduction of Xf in a new area usually generates the creation of new host–pathogen combinations [11] even without disease development. This is because, in a new area where the bacterium has been introduced and is spread by its vectors, it will inevitably be able to be injected into plant species that have never hosted the bacterium before. Furthermore, given the extreme polyphagy of Xf vectors and the ability of the bacterium to colonize hundreds of species, the number of host species is destined to continuously increase. Extending sampling to plants (even asymptomatic ones) on which the bacterium has never been reported, without limiting the monitoring to those already known, would help to find new host species and plan more effective monitoring. This is important because those species still unknown as hosts of Xf represent an inoculum source of epidemic expansion [11].
The asymptomatic and pauci-symptomatic host species, i.e., those plants which, when infected by a bacterium, develop belatedly mild symptoms or even show no symptoms at all, certainly play a role in the spread of Xf. For example, in Apulia, 36 plant species [12] were found to be infected and most of them are asymptomatic. The most important species which develop a slight desiccation when infected by X. fastidiosa subsp. pauca strain “De Donno” are Prunus dulcis and Prunus avium [13], while some olive cultivars [14] and various other plant species are asymptomatic (MIPAAF, Decree 24 January 2022). Therefore, these host species could act as intermediate plant hosts between other (highly) susceptible species, increasing the likelihood of new epidemic development. Moreover, if a plant species is not known to be a host, it may be not included in monitoring programs, reducing their effectiveness and increasing the risk of introducing Xf into uninfected regions by up to eight-fold [15].
Therefore, the identification of the Xf host species and the study of their distribution is particularly important if these hosts are asymptomatic, so as to make visual investigations ineffective.
Castanea sativa is a species distributed in Southern Europe (Spain, Italy, Balkan countries), in North Africa (especially Morocco), north-western Europe (Belgium, France) and Asia (Turkey, Armenia, Georgia, Azerbaijan, Syria) and thrives on territories with altitudes ranging from 200 to 1800 m above sea level, based on latitude, soil and climate characteristics [16]. In Europe, the chestnut represents an important plant species and covers an area of 2.5 million hectares, most of which are concentrated in Italy and France [16]. This relevance is mainly due to its use as a fruit production plant and in wood production but also for the numerous ecosystem services it performs [17]. In Italy, the chestnut is mainly found in six regions, namely Campania, Lazio, Tuscany, Emilia-Romagna, Piedmont and Veneto. Furthermore, in Italy this species is characterised by an enormous genetic variability due to the traditional method of propagation by seed [18]. In the Apulian territory, the chestnut tree has been introduced in some areas but is not widespread, probably due to the pedoclimatic characteristics. For example, the Salento is characterised above all by soils of calcareous origin [19] with basic or sub-basic pH [20], high summer temperatures and low annual rainfall, while chestnut prefers sub-acid soils and suffers from summer water stress [16].
In this work, a diagnostic investigation for Xf is carried out on several small groups of chestnut trees in Salento, coupled with the observation of any visible symptoms. In addition, the strain of the infected plants was characterised through MLST (Multi-Locus Sequence Typing) analysis and phylogenetic analysis, and some samples of infected branch were subjected to analysis with the FISH-CLSM (Fluorescence In Situ Hybridization coupled with Confocal Laser Scanning Microscopy) technique in order to observe the distribution of the bacterium in the xylem vessels.
2. Materials and Methods
2.1. Plant Material, Symptom Observation and Diagnostic Analysis
The chestnut trees subjected to our monitoring are located in three municipalities (Martano, Corigliano d’ Otranto and Taviano) in the province of Lecce (Apulia, South Italy), an area demarcated as Xf-infected since 2015 [21]. Taviano is 30.9 km from Martano and 24.7 km from Corigliano as the crow flies, while between Martano and Corigliano there is a distance of 6.4 km. In Taviano and Corigliano, the cultivar present was “Nserta” (communication from orchard owners), while in Martano the cultivar was unknown. Then, in accordance with the “Guidelines for the survey of Xf in the EU territory” [22] indicating that, in open fields, plant surveys and sampling periods should be achieved during the vegetative season (not during the dormancy stage), in spring (April 2023) and summer (June 2023), leaf scorching symptoms were evaluated and diagnostic analyses were conducted to verify the presence of Xf. The presence of symptoms was evaluated using a pathometric scale proposed by Luvisi et al. [23].
A total of 15 plants were randomly sampled between all three sites (Figure 1). In the sites of Corigliano d’ Otranto and Taviano, the chestnut trees were close to olive trees heavily compromised by the bacterium, while around the Martano site almond and pistachio trees, as well as heavily compromised olive trees, were present. The pooled sample used for DNA extraction consisted of eight 1-year-old twigs randomly taken from each plant and representative of the analysed tree. The samples were processed for DNA extraction using the CTAB (Cetyltrimethyl Ammonium Bromide) method [24].
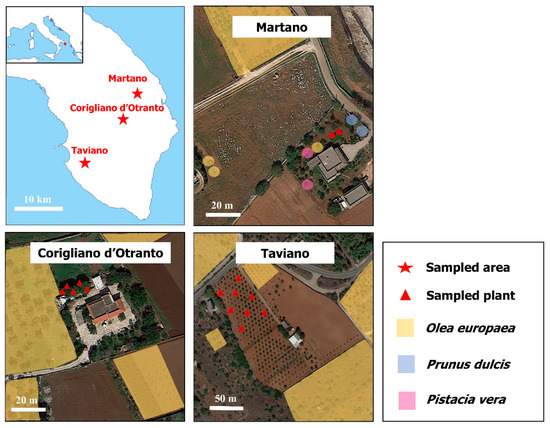
Figure 1.
Chestnut trees sampled for the survey. All sampled sites were located in areas where X. fastidiosa is considered endemic, and where infected olive, pistachio and almond trees were hosted.
The DNA was used as a template for Xf detection by TaqMan real-time PCR protocol with XF-F and XF-R primers, and with the XF-P probe proposed by Harper et al. [25]. The reactions were performed in a Real-Time thermal cycler (QuantStudio 3 Real-Time PCR System, Applied Biosystem, Foster City, CA, USA) in a final volume of 20 μL containing 10 µL of TaqMan Fast Advanced Master Mix (Applied Biosystem, Foster City, CA, USA), 300 nM of XF-F and XF-F primers, 100 nM of XF-P probe, ultrapure DNase/Rnase-free water (Carlo Erba Reagents S.r.l., Milan, Italy) and 3 µL of DNA (20 ng µL−1). The cycling conditions were as follows: initial UNG incubation at 50 °C for 2 min and polymerase activation at 95 °C for 20 s, followed by 40 cycles of 95 °C for 1 s and 60 °C for 20 s. Xf concentration, expressed as bacterial cfu mL−1, was inferred from Cq values using a standard curve with dilutions ranging from 102 to 107 cfu mL−1, as described by D’Attoma et al. [26]. Data related to the Xf detection in the analysed plants were used to evaluate the average incidence of infection (number of Xf-positive trees/total trees per site) and to estimate the average bacterial concentration in April and June (average value among the Xf concentrations detected in the Xf-positive plants for each site).
2.2. MLST and Phylogenetic Analysis
The genomic DNA extracted from one Xf-positive chestnut belonging to the group of plants located in Corigliano d’ Otranto were used as templates to amplify and sequence the seven MLST loci (leuA, petC, malF, cysG, holC, nuoL and gltT) and the non-MLST pilU gene following the procedure described by Yuan et al. [27]. Sequencing of the resulting PCR amplicons (in both directions) was performed by the Eurofins Genomics GmbH (Eurofins Genomics, Ebersberg, Germany). The MLST analysis was performed using Xylella fastidiosa PubMLST (https://pubmlst.org/organisms/xylella-fastidiosa, accessed on 29 May 2023). Sequence similarities were controlled with BLAST in NCBI (National Center for Biotechnology Information website (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (accessed on 29 May 2023). For phylogenetic analysis, the sequences of MLST loci obtained from our sample were aligned to reference sequences, each locus separately, with CLUSTALW [28]. The reference sequences were taken from Elbeaino et al. [29] and others uploaded to NCBI Genbank (Supplementary File S1). The alignments were then trimmed to span the same region and concatenated with MEGA 11 [30]. For phylogenetic analysis, trees supported by bootstrapping (1000 times) were generated by the maximum likelihood method implemented in MEGA 11 [30]. Branches with less than 50% bootstrap support were collapsed.
2.3. FISH-CLSM
To visualize the distribution of the bacterium Xf inside the xylem tissues, a FISH (Fluorescence In Situ Hybridization) together with a CLSM analysis (Confocal Laser Scanning Microscope LSM 700, Carl Zeiss, Jena, Germany) was conducted. One-year-old twigs of Xf-positive and Xf-negative plants (length 10 cm and diameter 0.5 cm) were first cut into small pieces (~1.5 cm). After initial sterilization with 70% ethanol for one minute, they were washed three times with sterile water, fixed in 4% paraformaldehyde in phosphate-buffered saline (1× PBS) overnight at room temperature and finally washed in buffer PBS for 10 min at room temperature. With the aid of the automatic tissue processor Leica TP 1020 (Leica Microsystems, Mannheim, Germany), the twig pieces were dehydrated through two successive 1 h incubations in 70, 80, 95 and 100% ethanol, followed by two consecutive 1 h incubations in xylene, and finally embedded in paraffin. The twig pieces were cut into 25 µm thick transverse sections with a HistoCore MULTICUT R semi-automatic rotary microtome (Leica Microsystems, Mannheim, Germany). Sections were then transferred to a 1:1 (v/v) PBS: 96% ethanol solution and stored at −20 °C until staining via FISH technique. To dissolve the paraffin, the sections were immersed in toluene for 3 min at 43 °C. FISH staining was performed using the Cy3-labeled X. fastidiosa-specific KO 210 probe and the protocol as proposed by Cardinale et al. [31]. The stained sections were mounted onto glass slides by using the Citifluor AF1 antifade reagent (Linaris Biologische Produkte GmbH, Dossenheim, Germany); the stained sections were stored for up to 4 days in the dark at 4 °C until the confocal laser-scanning microscope (Carl Zeiss LSM 700 laser scanning microscope, Jena, Germany) observations. Cy3 was excited with the 561 nm laser line; plant tissues were additionally excited with the 405 laser line to induce autofluorescence of the twig tissue. Emissions were detected in the range 570–613 nm for Cy3 and 420–480 nm for the plant autofluorescence.
2.4. Possible Vectors of X. fastidiosa for This Species
Xf is transmitted by insects that feed on xylem sap belonging to the order Hemiptera, superfamilies Cicadoidea, Cercopidea and Membracoidea, and in this work a bibliographic study on the potential vectors of Xf for this new host species was also performed. Firstly, a bibliographic search of the scientific literature was conducted in order to verify whether the association between C. sativa and known vectors of X. fastidiosa subsp. pauca strain “De Donno” had already been observed. Second, we searched for references regarding the association between other potential vectors belonging to the same superfamilies as the known vectors and C. sativa.
3. Results
3.1. Symptom Observation and Diagnostic Analysis
From the phytosanitary investigation conducted on chestnut plants hosted in the areas where Xylella is endemic, it emerged that no plant has ever developed leaf scorching symptoms attributable to Xf infection (Figure 2). Diagnostic analyses conducted in April and June showed that the average incidence of infection was 38% in April and 30% in June. The bacterial concentration detected on one-year-old branches also remained relatively low, with averages in April and June of 1.14 × 104 cfu mL−1 and 1.56 × 103 cfu mL−1, respectively (Table 1).

Figure 2.
Chestnut plants subjected to monitoring and sampling located at the site of Martano. The infected plants were asymptomatic, i.e., no leaf scorching symptoms attributable to X. fastidiosa were observed; (A) the plant on the left was positive for X. fastidiosa as determined by real-time PCR diagnostic analysis while the plant on the right was negative; (B) the leaves in the Xf-positive plant were healthy, as well as those in the (C) Xf-negative plant.

Table 1.
Incidence and average bacterial concentration detected on one-year-old branches of plants sampled in Martano, Corigliano d’Otranto and Taviano during the two sampling periods.
3.2. MLST and Phylogenetic Analysis
Identity searches in the pubMLST database (http://pubmlst.org/xfastidiosa/, accessed on 29 May 2023) indicated that the following alleles were present in our Xf found in Chestnut: 7 (leuA), 6 (petC), 24 (cysG), 10 (holC), 14 (gltT) and 16 (nuoL). It was not possible to assign the malF gene fragment since the sequence was incomplete, but through searching with a combination of loci on the pubMLST database, the closest genotypeturned out to be ST53. The sequence datasets obtained in this study are available in GenBank NCBI (www.ncbi.nlm.nih.gov, accessed on 29 June 2023) with accession numbers OR209769 to OR209776 and are collected in Supplementary File S2. MLST genes compared to BLAST in NCBI showed 98% to 100% similarity to sequences from isolates of X. fastidiosa subsp. pauca strain “De Donno” (cysG 99%, gltT 98%, holC 99%, leuA 100%, malF 100%, nuolL 98%, petC 99% and holC 99% with sequence accession number CP016610 relative to the complete genome of the isolate “Salento-2”), isolated from Olea europaea. Even the non-MLST pilU gene sequence was aligned with 99% nucleotide identity with the olive isolate “Salento-2”. The phylogenetic tree produced with the multi-locus combined analysis (MLST loci) confirmed the identity of the bacterium found on C. sativa. In fact, our bacteria clustered with several isolates of X. fastidiosa subsp. pauca strain “De Donno” detected on different species in Salento (Figure 3). The support bootstrap value in the intersection of the branches related to the bacterial strains UNISAL.CS and Salento-2 is low, since there is a very high value of similarity between sequences. In fact, if there is not a reasonable number of differences between all of sequences (they are almost the same), there will not be enough signal to establish phylogenetic relations and the two branches tend to collapse (considered the same lineage). In the Xf phylogenetic tree of the MLST analysis reported in Elbeaino et al. [29], branches with less than 75% bootstrap support were collapsed.
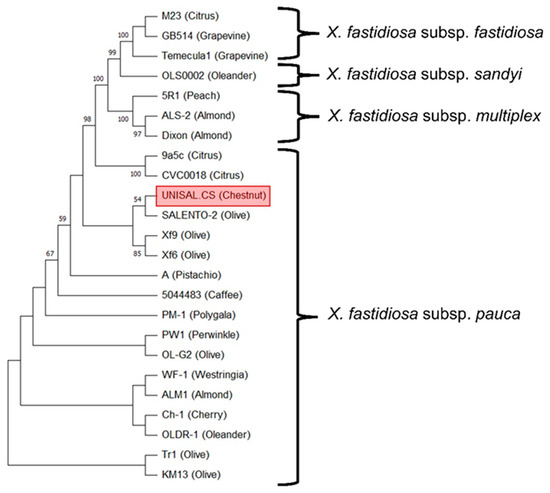
Figure 3.
Maximum likelihood (ML) method and general time reversible model were used for phylogenetic tree drawing, with 1000 bootstraps. Bootstrap values lower than 50 are not reported.
3.3. Xylella fastidiosa Distribution in C. sativa Xylem Vessels
The transverse sections of the 1-year-old infected branches showed clear Xf occlusions (Figure 4), while such occlusions were not observed in sections of twigs from plants with negative results in the diagnostic tests. On average, the infected plants had 9.2 ± 3.4% twig vessels colonised by Xf subsp. pauca, based on 20 representative confocal stacks (2500 visually checked vessels in total) from three infected plants and three independent FISH stains; no significant differences were found between the analysed trees (ANOVA, p = 0.49). The frequent colonization of adjacent vessels (Figure 4) suggested the well-known horizontal movement of the bacteria through pits connecting the vessels [31,32].
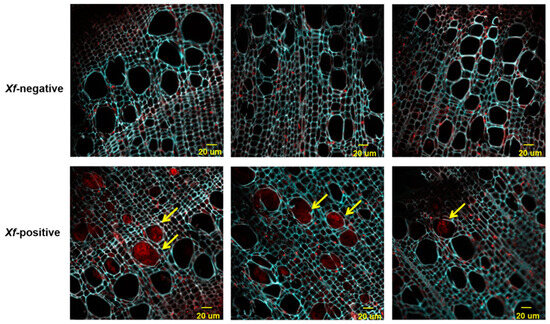
Figure 4.
Confocal microscopy images showing the colonization of C. sativa one-year-old twigs by fluorescence in situ hybridization (FISH)-stained X. fastidiosa. Red: signal of the X. fastidiosa-specific Cy5-labeled KO 210 FISH probe; cyan: autofluorescence of the plant tissue. Yellow arrows indicate the presence of Xf aggregates. These are representative images from several observations on twigs of different trees.
3.4. Possible Vectors of X. fastidiosa for This Species
Few studies have been published concerning the entomofauna of the Hemiptera (superfamilies Cycadoidea, Cercopidea and Membracoidea) on C. sativa. From the studies carried out, the vectors of the X. fastidiosa subsp. pauca strain “De Donno” detected on plants of C. sativa include Philaenus spumarius [33] and Neophilaenus campestris [34]. These two insects have been confirmed vectors for the epidemic that developed in Salento [35]. Philaenus signatus has been detected on C. sativa plants in Greece [33] and has already been considered as a potential vector of the X. fastidiosa subsp. pauca strain “De Donno” [36]. Insects belonging to the Cercopoidea and Membracoidea superfamilies but not yet recognised as Xf vectors were also found on the chestnut tree, including Cercopis sanguinolenta (Cercopoidea, Aphrophoridae); Aphrophora alni (Cercopoidea, Aphrophoridae); Alebra viridis and Alebra wahlbergi (Membracoidea, Cicadellidae); Arboridia spathulata (Membracoidea, Cicadellidae); and Empoasca vitis and Empoasca alsiosa (Membracoidea, Cicadellidae) [37,38,39].
4. Discussion
This work represents the first detection of X. fastidiosa subsp. pauca on chestnut trees (Castanea sativa) in plants located in Salento (Apulia, Italy). Another report of infected chestnut trees also occurred in Portugal, but the subspecies and related sequence type are still unknown and no other data regarding the infection and possible disease have been reported [7]. In the past, C. sativa does not seem to have been investigated as a host of Xf, not even in studies conducted on both European and American forest flora. Therefore, a characterization of Xf infection on this species has not yet been made, particularly through an evaluation of the symptoms, a quantification of the bacterial concentration and the frequency of the infection in the natural environment, and further microscopic analyses aiming to understand the ways in which the pathogen establishes in the xylem of this species.
With this work, through MLST and phylogenetic analysis we confirmed the first detection of X. fastidiosa subsp. pauca on C. sativa plants and also confirmed that it was the subsp. pauca ST53 genotype [7]. This result was expectable because the bacterium has been present for several years, and only the ST53 genotype was detected in the area where the chestnuts were sampled [21]. Furthermore, ST53 is the most reported genotype in natural infections, with about 50 host species detected between Italy, France and Costa Rica [40].
In the Fagaceae family, to which C. sativa belongs, there are numerous species recognised as Xf hosts, in particular in the genii Quercus and Fagus. The genus Quercus is included in the subfamily Quercoideae (like the Castanea genus), and in America, several species have been found naturally infected with Xf and symptomatic [41]. The symptomatic species found, and which develop a disease called bacterial leaf scorch [41] include Q. alba (subsp. unknown); Q. coccinea (subsp. multiplex); Q. falcata (subsp. multiplex); Q. imbricaria (subsp. sconosiuta); Q. incana (subsp. unknown); Q. laevis (subsp. multiplex); Q. laurifolia (subsp. unknown); Q. macrocarpa (subsp. multiplex); Q. nigra (subsp. multiplex); Q. palustris (subsp. multiplex); Q. phellos (subsp. multiplex); Q. rubra (subsp. multiplex); Q. shumardii (subsp. multiplex); Q. velutina (subsp. unknown); and Q. virginiana (subsp. unknown) [42]. As regards the genus Fagus (subfamily Fagoideae), in America the species Fagus crenata was reported as a symptomatic host of Xf (subsp. unknown), capable of developing leaf scorch [43].
A Q. ilex plant was found to be infected with X. fastidiosa subsp. pauca ST53 in Corsica (France), but the data need to be reconfirmed because Q. ilex may not be a systemic host of the bacterium [44,45], while there are no data related to the detection of X. fastidiosa subsp. pauca ST53 (“De Donno” strain) in Q. ilex in Salento. The ST53 genotype present in Apulia has a different behaviour compared to other genotypes of the subsp. pauca; in fact, it does not infect citrus plants, as happens, for example, in Brazil with other genotypes (ST11, ST12, ST13) that cause Citrus Variegated Chlorosis [46,47]. This genotype, being recently described as apparently very polyphagous, could be detected in new host species different from those already known for the subsp. pauca. In the case of the chestnut trees found infected in Salento, however, no symptoms of leaf scorching were ever observed, and the bacterial concentration detected was low compared to that found in susceptible olive trees and grapevine (6.77 × 106 and 6.0 × 105 cfu mL−1, respectively) [13,48]. Therefore, given the low bacterial concentration detected and the absence of symptoms, C. sativa could be considered a tolerant species. This feature would make this species potentially useful for reforestation projects in environments where X. fastidiosa subsp. pauca ST53 is considered endemic, but at the same time it could represent a very dangerous species in environments where there is a risk of introduction of Xf, because it could represent a source of bacterial inoculation and, therefore, favour the birth and expansion of an outbreak in a hidden way. Furthermore, given the low concentration detected and the typical erratic distribution of the pathogen [49], the incidence of false negatives could be high. Unfortunately, it is not known whether different genotypes of Xf can cause disease in chestnut, and it is possible that the potential resistance that plants are showing against the ST53 genotype is not being shown for other genotypes, which could cause disease. In fact, the different response of various plant species to the infection of different Xf genotypes is known [13,50,51]. Almond trees, for example, when infected by Xf subsp. fastidiosa, develop more severe symptoms compared with infection caused by subsp. multiplex. Furthermore, in almond trees, the Xf subsp. fastidiosa is capable of causing the disease with a lower bacterial concentration than that necessary for the subsp. multiplex [52].
In many species, indeed, Xf appears to behave like a commensal endophyte. In this case, the bacterium is not pathogenic, and coexists with the host without causing damage [53,54]. It is not yet clear why the bacterium becomes pathogenic in some of the host species, but it has been hypothesised that the compatibility between the wall-degrading enzymes produced by Xf and the carbohydrate composition of the pit membranes may be decisive in the onset of the disease [55]. It has also been hypothesised that the composition of the lipopolysaccharide O antigen could play a role in the recognition of the pathogen by determining whether the type of association with the plant will be commensal or parasitic [53]. In our case, we can hypothesize an example of commensalism between the bacterium and the plant, but it would be premature because further investigations to confirm this are necessary.
The first microscopy investigation conducted on twigs using the KO210 probe (specific for Xylella fastidiosa) [31] allowed the detection of occlusions in adjacent vessels, typical of the horizontal movement of Xf. However, the xylem vessels occluded by the bacterium represented a low percentage. These data can explain the lack of symptom development on the canopy; further investigations are necessary to understand the mechanisms involved in the resistance of this species against the bacterium.
Regarding the bibliographic research on the entomofauna potentially involved in the transmission of Xf to the chestnut tree, further studies are needed. However, it is known that the vectors of Xf discovered in Salento can feed on numerous hosts. Philaenus spumarius is known to be the main vector of Xf in Puglia and the most widespread potential vector in Europe [56]. Furthermore, this insect is highly polyphagous: the young stages can feed on many herbaceous species and the adults, with their movement, increase the number of host species also reaching trees and shrubs [57]. Neophilaenus campestris is also a very polyphagous species but much less abundant than P. spumarius [58]. Philaenus italosignus is present but not widespread in Southern Italy [59] and has never been found on chestnut plants. Overall, the polyphagy of the vectors confirms the fact that C. sativa, characterised by generally softer leaves and branches than the olive tree, may also represent a Trojan horse in the Xylella spread process.
5. Conclusions
This first worldwide detection of X. fastidiosa subsp. pauca in C. sativa must be a signal both for the countries where the bacterium is present and for the countries which, having predisposing conditions (favourable climate, host plants and vectors), are considered at risk of pathogen introduction. It is unknown whether the chestnut remains asymptomatic when infected by all Xf subspecies and what the level of susceptibility among the different chestnut cultivars is. Therefore, it is necessary to start monitoring activities on this species especially in the areas where the bacterium has been detected. In fact, as reported in this work, although this is the first known survey for Xf infection on chestnut plants, infected plants were detected in all three sampled sites, despite the relatively large distance between them. Furthermore, with the aim of characterising the infection by Xf in chestnuts in more detail, it would be very useful to plan and implement experiments in a controlled environment, artificially infecting different chestnut cultivars with the most widespread genotypes in Europe, then evaluating the immediate and long-term effects of infection, symptom severity, anatomical–morphological, metabolic and genetic plant responses. This will allow an evaluation of which genotypes cause a disease in chestnut trees, and at what bacterial concentration the plant activates the hypersensitivity that triggers the disease. That is indeed important for all the main fruit tree species. It will be also necessary to deepen the studies concerning the entomofauna and, in particular, the Xf vectors capable of feeding on the chestnut tree. This work shows how knowledge of all host species, including the pauci-symptomatic and asymptomatic ones, and of the vectors present in a given area, is essential to make containment measures truly effective.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae9121315/s1, File S1: Accession numbers of the Xylella fastidiosa multilocus sequence typing (MLST) sequences used to build the phylogenetic tree; File S2: Xylella fastidiosa multilocus sequence typing (MLST) and non-MLST sequences obtained in this study from the sequencing of the amplified DNA regions: cysG, gltT, holC, leuA, malF, nuoL, petC and pilU.
Author Contributions
Conceptualization, L.D.B. and A.L.; investigation, D.G., E.S. and G.C.; data curation, D.G. and G.C.; writing—original draft preparation, D.G, E.S. and G.C.; writing—review and editing, E.S., D.G., A.L. and A.G.D.D.; supervision, L.D.B., A.L. and A.G.D.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Acknowledgments
We would like to thank the farmers for their support in field observations.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Saponari, M.; Giampetruzzi, A.; Loconsole, G.; Boscia, D.; Saldarelli, P. Xylella fastidiosa in Olive in Apulia: Where We Stand. Phytopathology 2019, 109, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Picciotti, U.; Araujo Dalbon, V.; Ciancio, A.; Colagiero, M.; Cozzi, G.; De Bellis, L.; Finetti-Sialer, M.M.; Greco, D.; Ippolito, A.; Lahbib, N.; et al. “Ectomosphere”: Insects and Microorganism Interactions. Microorganisms 2023, 9, 440. [Google Scholar] [CrossRef] [PubMed]
- Loconsole, G.; Zicca, S.; Manco, L.; El Hatib, O.; Altamura, G.; Potere, O.; Elicio, V.; Valentini, F.; Boscia, D.; Saponari, M. Diagnostic Procedures to Detect Xylella fastidiosa in Nursery Stocks and Consignments of Plants for Planting. Agriculture 2021, 11, 922. [Google Scholar] [CrossRef]
- Saponari, M.; Boscia, D.; Nigro, F.; Martelli, G.P. Identification of DNA sequences related to Xylella fastidiosa in oleander, almond and olive trees exhibiting leaf scorch symptoms in Apulia (Southern Italy). J. Plant Pathol. 2013, 95, 3. [Google Scholar] [CrossRef]
- Martelli, G.P.; Boscia, D.; Porcelli, F.; Saponari, M. The olive quick decline syndrome in South-east Italy: A threatening phytosanitary emergency. Eur. J. Plant Pathol. 2016, 144, 235–243. [Google Scholar] [CrossRef]
- Greco, D.; Aprile, A.; De Bellis, L.; Luvisi, A. Diseases Caused by Xylella fastidiosa in Prunus Genus: An Overview of the Research on an Increasingly Widespread Pathogen. Front. Plant Sci. 2021, 13, 712452. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority); Gibin, D.; Pasinato, L.; Delbianco, A. Scientific Report on the update of the Xylella spp. host plant database—Systematic literature search up to 31 December 2022. EFSA J. 2023, 21, 8061. [Google Scholar] [CrossRef]
- EFSA Panel on Plant Health (PLH); Bragard, C.; Dehnen-Schmutz, K.; Di Serio, F.; Gonthier, P.; Jacques, M.A.; Jaques Miret, J.A.; Justesen, A.F.; MacLeod, A.; Magnusson, C.S.; et al. Update of the Scientific Opinion on the risks to plant health posed by Xylella fastidiosa in the EU territory. EFSA J. 2019, 17, 5665. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority). Statement of EFSA on host plants, entry and spread pathways and risk reduction options for Xylella fastidiosa Wells et al. EFSA J. 2013, 11, 3468. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority). Scientific report on the update of the Xylella spp. host plant database. EFSA J. 2018, 16, 5408. [Google Scholar] [CrossRef]
- Hopkins, D.L.; Purcell, A.H. Xylella fastidiosa: Cause of Pierce’s Disease of Grapevine and Other Emergent Diseases. Plant Dis. 2002, 86, 1056–1066. [Google Scholar] [CrossRef] [PubMed]
- Regione Puglia, Piante Specificate Sensibili Alla Xylella fastidiosa Sottospecie pauca Riscontrate in Puglia. 2023. Available online: www.emergenzaxylella.it (accessed on 21 October 2023).
- Saponari, M.; Boscia, D.; Altamura, G.; Loconsole, G.; Zicca, S.; D’Attoma, G.; Morelli, M.; Palmisano, M.; Saponari, A.; Tavano, D.; et al. Isolation and pathogenicity of Xylella fastidiosa associated to the olive quick decline syndrome in Southern Italy. Sci. Rep. 2017, 7, 17723. [Google Scholar] [CrossRef] [PubMed]
- Vergine, M.; Pavan, S.; Negro, C.; Nicolì, F.; Greco, D.; Sabella, E.; Aprile, A.; Ricciardi, L.; De Bellis, L.; Luvisi, A. Phenolic characterization of olive genotypes potentially resistant to Xylella. J. Plant Interact. 2022, 17, 462–474. [Google Scholar] [CrossRef]
- White, S.M.; Bullock, J.M.; Hooftman, D.A.P.; Chapman, D.S. Modelling the spread and control of Xylella fastidiosa in the early stages of invasion in Apulia, Italy. Biol. Invasions 2017, 19, 1825–1837. [Google Scholar] [CrossRef] [PubMed]
- Conedera, M.; Tinner, W.; Krebs, P.; de Rigo, D.; Caudullo, G. Castanea sativa in Europe: Distribution, habitat, usage and threats. In European Atlas of Forest Tree Species; San-Miguel-Ayanz, J., de Rigo, D., Caudullo, G., Houston Durrant, T., Mauri, A., Eds.; European Commission: Luxembourg, 2016; p. e0125e0+. [Google Scholar]
- Beccaro, G.L.; Donno, D.; Lione, G.G.; De Biaggi, M.; Gamba, G.; Rapalino, S.; Riondato, I.; Gonthier, P.; Mellano, M.G. Castanea spp. Agrobiodiversity Conservation: Genotype Influence on Chemical and Sensorial Traits of Cultivars Grown on the Same Clonal Rootstock. Foods 2020, 9, 1062. [Google Scholar] [CrossRef] [PubMed]
- Alessandri, S.; Krznar, M.; Ajolfi, D.; Ramos Cabrer, A.M.; Pereira-Lorenzo, S.; Dondini, L. Genetic Diversity of Castanea sativa Mill. Accessions from the Tuscan-Emilian Apennines and Emilia Romagna Region (Italy). Agronomy 2020, 10, 1319. [Google Scholar] [CrossRef]
- Costantini, E.A.C.; Dazzi, C. The Soils of Italy, World Soils Book Series. Geol. Geomorphol. 2013, 3, 39–56. [Google Scholar] [CrossRef]
- Del Coco, L.; Migoni, D.; Girelli, C.R.; Angilè, F.; Scortichini, M.; Fanizzi, F.P. Soil and Leaf Ionome Heterogeneity in Xylella fastidiosa subsp. pauca-Infected, Non-Infected and Treated Olive Groves in Apulia, Italy. Plants 2020, 9, 760. [Google Scholar] [CrossRef]
- Official Bulletin of the Apulia Region (BURP) n. 15 of 29-01-2015. Available online: https://burp.regione.puglia.it/documents/20135/899904/DELIBERAZIONE+DEL+CONSIGLIO+REGIONALE+20+gennaio+2015%2C+n.293+%28id+4729422%29.pdf/aff953bd-db88-ffcc-c79c-c2fc9fa69f86?t=1622798897598 (accessed on 7 July 2023).
- European Commission Directorate-General for Health and Food Safety—Safety of the Food Chain Plant health. In Guidelines for the Survey of Xylella fastidiosa (Wells et al.) in the Union Territory; European Commission: Brussels, Belgium, 2015.
- Luvisi, A.; Aprile, A.; Sabella, E.; Vergine, M.; Nutricati, E.; Miceli, A.; Negro, C.; De Bellis, L. Xylella fastidiosa subsp. pauca (CoDiRO strain) infection in four olive (Olea europaea L.) cultivars: Profile of phenolic compounds in leaves and progression of leaf scorch symptoms. Phytopathol. Mediterr. 2017, 56, 259–273. [Google Scholar] [CrossRef]
- Loconsole, G.; Potere, O.; Boscia, D.; Altamura, G.; Djelouah, K.; Elbeaino, T.; Frascheri, D.; Lorusso, D.; Palmisano, F.; Pollastro, P.; et al. Detection of Xylella fastidiosa in olive trees by molecular and serological methods. J. Plant Pathol. 2014, 96, 7–14. [Google Scholar] [CrossRef]
- Harper, S.J.; Ward, L.I.; Clover, G.R.G. Development of LAMP and Real-Time PCR Methods for the Rapid Detection of Xylella fastidiosa for Quarantine and Field Applications. Phytopathology 2010, 100, 1282–1288. [Google Scholar] [CrossRef] [PubMed]
- D’Attoma, G.; Morelli, M.; Saldarelli, P.; Saponari, M.; Giampetruzzi, A.; Boscia, D.; Savino, V.N.; De La Fuente, L.; Cobine, P.A. Ionomic Differences between Susceptible and Resistant Olive Cultivars Infected by Xylella fastidiosa in the Outbreak Area of Salento, Italy. Pathogens 2019, 8, 272. [Google Scholar] [CrossRef]
- Yuan, X.; Morano, L.; Bromley, R.; Spring-Pearson, S.; Stouthamer, R.; Nunney, L. Multilocus sequence typing of Xylella fastidiosa causing Pierce’s disease and oleander leaf scorch in the United States. Phytopathology 2010, 100, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Elbeaino, T.; Valentini, F.; Abou Kubaa, R.; Moubarak, P.; Yaseen, T.; Digiaro, M. Multilocus sequence typing of Xylella fastidiosa isolated from olive affected by ‘olive quick decline syndrome’ in Italy. Phytopathol. Mediterr. 2014, 53, 533–542. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, M.; Luvisi, A.; Meyer, J.B.; Sabella, E.; De Bellis, L.; Cruz, A.C.; Ampatzidis, Y.; Cherubini, P. Specific fluorescence in situ hybridization (FISH) test to highlight colonization of xylem vessels by Xylella fastidiosa in naturally infected olive trees (Olea europaea L.). Front. Plant Sci. 2018, 9, 431. [Google Scholar] [CrossRef]
- Sabella, E.; Luvisi, A.; Aprile, A.; Negro, C.; Vergine, M.; Nicolì, F.; Miceli, A.; De Bellis, L. Xylella fastidiosa induces differential expression of lignification related-genes and lignin accumulation in tolerant olive trees cv. Leccino. J. Plant. Phys. 2018, 220, 60–68. [Google Scholar] [CrossRef]
- Drosopoulos, S. New data on the nature and origin of colour polymorphism in the spittlebug genus Philaenus (Hemiptera: Aphorophoridae). Ann. Soc. Entomol. 2003, 39, 31–42. [Google Scholar] [CrossRef]
- Kaplan, C.; Turanlı, T.; Çeliker, N.M. Pest species in chestnut growing areas of Izmir and Manisa provinces and their economic importance. In Proceedings of the VIII International Scientific Agriculture Symposium, Jahorina, Bosnia and Herzegovina, 5–8 October 2017; pp. 756–762. [Google Scholar]
- Elbeaino, T.; Yaseen, T.; Valentini, F.; Ben Moussa, I.E.; Mazzoni, V.; D’onghia, A.M. Identification of three potential insect vectors of Xylella fastidiosa in Southern Italy. Phytopathol. Mediterr. 2014, 53, 328–332. [Google Scholar] [CrossRef]
- Lahbib, N.; Picciotti, U.; Bouhachem, S.; Garganese, F.; Porcelli, F. Morphs of Philaenus species, candidate Xylella fastidiosa vectors. Bull. Insectology 2022, 75, 197–209. [Google Scholar]
- Mazzoni, V. Contribution to the knowledge of the Auchenorrhyncha (Hemiptera Fulgoromorpha and Cicadomorpha) of Tuscany (Italy). Redia 2005, 88, 85–102. [Google Scholar]
- EFSA PLH Panel (EFSA Panel on Plant Health). Scientific Opinion on the risks to plant health posed by Xylella fastidiosa in the EU territory, with the identification and evaluation of risk reduction options. EFSA J. 2015, 13, 3989. [Google Scholar] [CrossRef]
- Demichelis, S.; Bosco, D. Host-plant relationship and life history of some Alebra species in Italy (Auchenorrhyncha: Cicadellide). Eur. J. Entomol. 1995, 92, 683–690. [Google Scholar]
- EFSA (European Food Safety Authority); Delbianco, A.; Gibin, D.; Pasinato, L.; Boscia, D.; Morelli, M. Update of the Xylella spp. host plant database—systematic literature search up to 30 June 2022. EFSA J. 2023, 21, 7726. [Google Scholar] [CrossRef]
- Gould, A.B.; Lashomb, J.H. Bacterial Leaf Scorch (BLS) of Shade Trees. Plant Health Instr. 2007. [Google Scholar] [CrossRef]
- Tkaczyk, M. Worldwide review of bacterial diseases of oaks (Quercus sp.) and their potential threat to trees in Central Europe. For. Int. J. For. Res. 2022, 96, 425–433. [Google Scholar] [CrossRef]
- Huang, Q.; Wenbin, L.; Hartung, J.S. Association of Xylella fastidiosa with leaf scorch in Japanese beech bonsai. Can. J. Plant Pathol. 2003, 25, 401–405. [Google Scholar] [CrossRef]
- EFSA Panel on Plant Health (PLH); Jeger, M.; Bragard, C.; Caffier, D.; Chatzivassiliou, E.; Dehnen-Schmutz, K.; Gilioli, G.; Grégoire, J.-C.; Jaques Miret, J.A.; MacLeod, A.; et al. Statement on susceptibility of Citrus spp., Quercus ilex and Vitis spp. to Xylella fastidiosa strain CoDiRO. EFSA J. 2016, 14, 4601. [Google Scholar] [CrossRef]
- Denancé, N.; Legendre, B.; Briand, M.; Olivier, V.; de Boisseson, C.; Poliakoff, F.; Jacques, M.A. Several subspecies and sequence types are associated with the emergence of Xylella fastidiosa in natural settings in France. Plant Pathol. 2017, 66, 1054–1064. [Google Scholar] [CrossRef]
- Nunney, L.; Yuan, X.; Bromley, R.E.; Stouthamer, R. Detecting genetic introgression: High levels of intersubspecific recombination found in Xylella fastidiosa in Brazil. Appl. Environ. Microbiol. 2012, 78, 4702–4714. [Google Scholar] [CrossRef] [PubMed]
- Nunney, L.; Ortiz, B.; Russell, S.A.; Ruiz Sànchez, R.; Stouthamer, R. The Complex Biogeography of the Plant Pathogen Xylella fastidiosa: Genetic Evidence of Introductions and Subspecific Introgression in Central America. PLoS ONE 2014, 9, e112463. [Google Scholar] [CrossRef] [PubMed]
- Krivanek, A.F.; Walker, M.A. Vitis resistance to Pierce’s disease is characterized by differential Xylella fastidiosa populations in stems and leaves. Phytopathology 2005, 95, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Nicolì, F.; Negro, C.; Nutricati, E.; Vergine, M.; Aprile, A.; Sabella, E.; Damiano, G.; De Bellis, L.; Luvisi, A. Accumulation of Azelaic Acid in Xylella fastidiosa-Infected Olive Trees: A Mobile Metabolite for Health Screening. Phytopathology 2019, 109, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Janse, J.; Obradovic, A. Xylella fastidiosa: Its biology, diagnosis, control and risks. J. Plant Pathol. 2010, 92, 35–148. [Google Scholar]
- Nunne, L.; Azad, H.; Stouthamer, R. An Experimental test of the Host-Plant Range of Nonrecombinant Strains of North American Xylella fastidiosa subsp. multiplex. Phytopathology 2019, 109, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Groves, R.L.; Chen, J.; Civerolo, E.L.; Freeman, M.W.; Viveros, M.A. Spatial Analysis of Almond Leaf Scorch Disease in the San Joaquin Valley of California: Factors Affecting Pathogen Distribution and Spread. Plant Dis. 2005, 89, 581–589. [Google Scholar] [CrossRef]
- Rapicavoli, J.N.; Blanco-Ulate, B.; Muszyński, A.; Figueroa-Balderas, R.; Morales-Cruz, A.; Azadi, P.; Dobruchowska, J.M.; Castro, C.; Cantu, D.; Roper, M.C. Lipopolysaccharide O-antigen delays plant innate immune recognition of Xylella fastidiosa. Nat. Commun. 2018, 9, 390. [Google Scholar] [CrossRef]
- Chatterjee, S.; Almeida, R.P.P.; Lindow, S. Living in two Worlds: The Plant and Insect Lifestyles of Xylella fastidiosa. Annu. Rev. Phytopathol. 2008, 46, 243–271. [Google Scholar] [CrossRef]
- Sun, Q.; Greve, L.C.; Labavitch, J.M. Polysaccharide compositions of intervessel pit membranes contribute to Pierce’s disease resistance of grapevines. Plant Physiol. 2011, 155, 1976–1987. [Google Scholar] [CrossRef][Green Version]
- Cornara, D.; Cavalieri, V.; Dongiovanni, C.; Altamura, G.; Palmisano, F.; Bosco, D.; Porcelli, F.; Almeida, R.P.P.; Saponari, M. Transmission of Xylella fastidiosa by naturally infected Philaenus spumarius (Hemiptera, Aphrophoridae) to different host plants. J. Appl. Entomol. 2017, 141, 80–87. [Google Scholar] [CrossRef]
- Gonella, E.; Picciau, L.; Pippinato, L.; Cavagna, B.; Alma, A. Host plant identification in the generalist xylem feeder Philaenus spumarius through gut content analysis. Entomol. Exp. Appl. 2020, 168, 890–899. [Google Scholar] [CrossRef]
- Martelli, G.P. The current status of the quick decline syndrome of olive in Southern Italy. Phytoparasitica 2016, 44, 1–10. [Google Scholar] [CrossRef]
- Cavalieri, V.; Altamura, G.; Fumarola, G.; di Carolo, M.; Saponari, M.; Cornara, D.; Bosco, D.; Dongiovanni, C. Transmission of Xylella fastidiosa subspecies pauca Sequence Type 53 by Different Insect Species. Insects 2019, 10, 324. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).