Abstract
Effectoromics has become integral to the identification of pathogen targets and/or host-resistant proteins for the genetic improvement of plants in agriculture and horticulture. Phytoplasmas are the causal agents of more than 100 plant diseases in economically important crops such as vegetables, spices, medicinal plants, ornamentals, palms, fruit trees, etc. To date, around 20 effectors in phytoplasmas have been experimentally validated but the list of putative effectors comprises hundreds of different proteins. Very few families (tribes) have been identified based on homology, such as the SAP05-like, SAP11-like, SAP54-like and TENGU-like families. The lack of conservation in amino acid sequences slows the progress of effectoromics in phytoplasmas since many effectors must be studied individually. Here, 717 phytoplasma effector candidates and 21 validated effectors were characterized in silico to identify common features. We identified functional domains in 153 effectors, while 585 had no known domains. The most frequently identified domain was the sequence-variable mosaic domain (SVM domain), widely distributed in 87 phytoplasma effectors. Searching for de novo amino acid motifs, 50 were found in the phytoplasma effector dataset; 696 amino acid sequences of effectors had at least 1 motif while 42 had no motif at all. These data allowed us to organize effectors into 15 tribes, uncovering, for the first time, evolutionary relationships largely masked by lack of sequence conservation among effectors. We also identified 42 eukaryotic linear motifs (ELMs) in phytoplasma effector sequences. Since the motifs are related to common functions, this novel organization of phytoplasma effectors may help further advance effectoromics research to combat phytoplasma infection in agriculture and horticulture.
1. Introduction
Phytoplasmas (Kingdom, Bacteria; Phylum, Mycoplasmatota; class, Mollicutes; genus, “Candidatus Phytoplasma” or “Ca. Phytoplasma”) are mycoplasma-like cell wall-less pathogenic microorganisms transmitted by phloem-feeding insect vectors [1,2,3]. These pathogens reside in plant phloem and cause severe damage to the agriculture and horticulture industries worldwide, with extensive yield losses in economically important crops such as vegetables, spices, medicinal plants, ornamentals, palms, fruit trees, among others [4]. In China, for example, more than 100 phytoplasma diseases have been reported [5].
Phytoplasmas secrete virulence proteins known as effectors, which interfere with host hormone signaling [6,7] and cause abnormal plant morphologies such as phyllody, virescence, chlorosis, and witches’ broom, among other symptoms [8,9].
Integrated management of the main phytoplasma diseases is expected to include the use of resistant plant material [10] and novel strategies for environmentally friendly insect vector control [11]. Because effectors are essential for the virulence of the pathogen, they are also susceptible targets for the control of phytoplasma-associated diseases [12,13]. They are also suitable tools for the identification of protein targets in the hosts, including the nucleotide-binding leucine-rich repeat (NLR) receptors (also called resistance proteins). Additionally, genetic engineering of NLR receptors can improve host recognition of pathogen effectors, enabling the production of resistant host lines [14,15].
To date, only around 20 phytoplasma effectors have been experimentally validated, which include SAP05 [16], SAP11 [17], SAP54/Phyl1 [18], TENGU [19,20], SPW1, SPW11, SWP12, SPW21 [21], Zaofeng3 and Zaofeng6 [22], and the membrane-bound proteins IdpA [23], Imp [24], VmpA [25] and Amp [26]. A number of novel effectors have been reported in the in silico characterization of phytoplasma genomes, for example, 257 hypothetical proteins in “Ca. Phytoplasma solani” [27] or 7 unique effectors in Flavescence dorée phytoplasma [28]. The list of phytoplasmas effectors keeps growing as more phytoplasma genomes are sequenced.
Different strategies have been used to classify effectors in other microbial kingdoms. In bacteria, effectors are classified according to the secretion system (type III, type IV) through which they are exported or translocated from the pathogen cell to the host [29,30]. In fungi, effectors that meet certain structural criteria such as small size, high cysteine content, presence of a signal peptide (SP) and absence of transmembrane domains (TMDs) are termed canonical or classical effectors, while those that do not meet some of these criteria are termed noncanonical [31,32,33]; this classification has helped to expand the size of fungal effectoromes, since the noncanonical effectors that were previously discarded contribute significantly to the overall effectorome [32]. Phytoplasma effectors are usually described as small, secreted proteins and like with fungi, they are secreted through the Sec-dependent type II secretion system [27,34,35], although recent developments have challenged this as a bona fide phytoplasma effector characteristic. Recently, six nonclassical effectors (ncSecPs) were identified in “Ca. Phytoplasma ziziphi”; these effectors lack signal peptides or translocation signals but are secreted through a Sec-independent secretion pathway. Agroinfiltration of these effectors in Nicotiana benthamiana Domin suppressed the hypersensitive response (HR) by enhancing the expression of the cell death suppressor genes PR-1 and PR-5 [36]. Similarly, a few known phytoplasma effectors are also transmembrane proteins, such as Imp (immunodominant membrane protein) [24] and Amp (antigenic membrane protein) [26], demonstrating that the phytoplasma effector definition must evolve from the current terminology, “secreted proteins”. The identification of larger and more complex effectoromes gives rise to further classifications of phytoplasma effector; thus, there is a need to identify effector families based on common characteristics that can further facilitate their organization and investigation. In other kingdoms, effector families, or “tribes”, have been defined based on sequence homology [37,38], common domains [39,40,41], or common motifs [42,43,44], among other criteria. The last two criteria are very useful, as many effectors do not share sequence homology among the organisms of a given microbial kingdom.
The inculturability of phytoplasma in artificial media greatly complicates research for specific cures. Effectoromics could become an effective approach to searching for targets in phytoplasmas. However, currently, only a few families/tribes of effector orthologs have been identified in a range of phylogenetically distant phytoplasmas: SAP11/SWP1 [45], SAP05, SAP54/PHYL1 and TENGU [19,46,47,48]. This lack of conservation slows the progress of functional characterization of phytoplasma effector candidates; most effector candidates must be studied individually and our knowledge about phytoplasma effectors is in its infancy, making functional characterization a challenge.
Using the UNIPROT database, a list of 738 amino acid sequences corresponding to nonredundant phytoplasma effectors was compiled (21 amino acid sequences corresponding to validated effectors and 717 amino acid sequences corresponding to effector candidates). This list was compiled using key words such as “Candidatus effectors”, “phytoplasma effectors”, “TENGU” and “SAP effectors”. Since programs that identify high levels of conservation have failed to find families in phytoplasma effectors beyond SAP05, SAP11, SAP54 and TENGU, we analyzed this dataset of 738 amino acid sequences through alternative methods, including Gene Ontology (GO), protein domains and motifs. Amino acid composition was also determined.
The search for de novo motifs enabled us to classify, for the first time, 696 phytoplasma effector candidates into 15 tribes. In other words, protein families that shared the same organization of multiple motifs were identified and grouped. The distribution of the tribes in the phytoplasmas revealed two evolutionary histories for their effectors. The tribes numbered 1, 6 and 7 were widespread in phytoplasmas, suggesting that their precursors occurred in common ancestors and were transmitted by vertical gene transfer, followed by duplication and rapid divergence in their evolution [49,50].
Other tribes (such as tribes 2, 3, 4, 5) showed discrete distribution, occurring only in phylogenetically related phytoplasmas; horizontal transmission to distant phytoplasmas was also observed in the members of these tribes.
Forty-two eukaryotic linear motifs, also known as short linear motifs (ELMs, SLiMs), were identified in the phytoplasma effectors. These SLiMs are novel tools that suggest the molecular mechanisms used by phytoplasma effectors during host infection. Phytoplasmas may hijack host targets by mimicry of key motifs contained in the regulators of critical host proteins [51].
The establishment of this new organization of phytoplasma effectors may help deepen our understanding of effectors in the race to combat phytoplasmas in agriculture and horticulture.
2. Materials and Methods
2.1. Protein Dataset
The phytoplasma effector dataset comprises 21 amino acid sequences corresponding to validated phytoplasma effectors taken from the scientific literature, and the list was further complemented by adding the results from a search of the UNIPROT database using the keywords SAP01, SAP02, SAP03, until SAP80, TENGU, phyllody, phyll, antigenic membrane protein (Amp), immunodominant membrane protein A (IdpA), immunodominant membrane protein (Imp), and variable membrane protein A (VmpA). All protein sequences were collected, and duplications were eliminated. The final dataset contained amino acid sequences corresponding to 21 true effectors and 717 effector candidates (total 738 amino acid sequences) and can be found in Table S1A and at https://github.com/Gisel-Carreon/Phytoplasma_effectors.
For a negative control set, 30 amino acid sequences corresponding to phytoplasma core proteins (proteins involved in essential metabolic activities) were randomly selected from validated genomes (draft genomes were excluded). The selected genomes for these core proteins were the onion yellows phytoplasma OY-M (T00154) (16SrI-B), the aster yellows witches’ broom phytoplasma AYWB (T00314) (16SrI-A), the “Ca. Phytoplasma mali” (T00729) (16SrX), the “Ca. Phytoplasma australiense” (T00752) (16SrXII), “Ca. Phytoplasma ziziphi” Jwb-nky (T05675) (16SrV-B), “Ca. Phytoplasma aurantifolia” (NCHU2014) (16SrII) and “Ca. Phytoplasma asteris” MBSP (CP015149) (16SrI); these validated genomes were found in the PUBMED database (https://pubmed.ncbi.nlm.nih.gov/; accessed on 14 July 2023); protein annotations were obtained from KEGG (https://www.genome.jp/kegg/genome/; accessed on 14 July 2023) (Table S1B).
2.2. In Silico Characterization of Phytoplasma Effectors
The phytoplasma effectors in the dataset were in silico characterized by amino acid length and amino acid composition (specifically of amino acids Cys, Ser, Leu, Lys, Asn and Trp), using a set of Perl scripts [32]. The presence of signal peptides and transmembrane domains were analyzed with SignalP v4.1 [52] and TMHMM v2.0 [53], respectively, both with default parameters, on mature proteins (without signal peptide).
For predicting nuclear localization signal (NLS), we used the NLStradamus program (http://www.moseslab.csb.utoronto.ca/NLStradamus/) [54]; the proteins were analyzed using the two-state static HMM method with a threshold score of 0.6.
2.3. Gene Ontology Distribution and Functional Annotation
Gene Ontology analysis for mapping biological process (BP), molecular function (MF) and cellular component (CC) were performed on the dataset of 738 amino acid sequences corresponding to phytoplasma effectors against the GO database integrated in the InterProScan v.86.0 tool under default parameters [55,56,57].
The GO functional classification histogram was plotted using the web server WEGO 2.0 (https://wego.genomics.cn/, accessed on 20 August 2023) [58]. The output file produced was the GO-native format without reference statement.
Functional domain identification was carried out using the InterproScan program v.86.0 in STANDALONE mode, which includes diverse source databases like the PFAM, CDD and SMART modules. We submitted the proteins in FASTA file as input and carried out the analysis under default parameters [57,59,60].
2.4. Classification of Effectors in Tribes Based on Motifs
The collection of 738 amino acid sequences corresponding to phytoplasma effectors was analyzed for motifs using the “Multiple Expectation-maximization for Motif Elicitation” (MEME) tool version 5.5.4, included in the “MEME suite” online platform (https://meme-suite.org/meme/) [61]. We used de novo motif discovery in classic mode, with 0 or 1 occurrence per sequence, and maximum 50 motifs as cut off.
The effector sequences that share various motifs were grouped together as a multigene family (tribe). To identify similarities in the patterns of amino acids, online multiple sequences alignments were conducted using the CLUSTALW (GenomeNet) software program (https://www.genome.jp/tools-bin/clustalw). Then, to create weighted sequence alignments, the WebLogo online server was utilized (https://weblogo.berkeley.edu/logo.cgi), using the CLUSTALW alignment file as input.
To determine whether these motifs were specific to phytoplasma effectors, phytoplasma non-effector core proteins were also screened for these motifs.
2.5. Search for Short Linear Motifs in True Phytoplasma Effectors
Short linear motifs (SLiMs, also often referred to as ELMs), were identified by submitting a pair of protein members of different classes of phytoplasma effectors (for example, two proteins for SAP11, two for Zaofeng effectors, two for ncSecPs, etc). The sequences, in a FASTA file, were submitted to the ELM database (http://elm.eu.org, accessed on 21 August 2023) [62]. ELM is a database of 3934 manually curated eukaryotic motifs. The analysis was run with default settings and a probability cutoff of 1 × 10−5.
ELMs found in the amino acid sequences of phytoplasma effectors were searched in the amino acid sequences of phytoplasma core proteins to determine whether they were specific to phytoplasma effector proteins.
3. Results
3.1. Protein Databases
Table 1 shows the list of UNIPROT database ID accessions for the amino acid sequences corresponding to the phytoplasma effectors used in this work.

Table 1.
List of phytoplasma effectors used in this work.
3.2. Characterization of Phytoplasma Effectors
To learn more about structural features of the known phytoplasma effectors, the dataset was analyzed for peptide length, amino acids composition, presence of signal peptide, presence of transmembrane domains (Table 2), among other characteristics. More than 90% were small peptides, with 400 amino acids or less. Consistent with the current belief, ~70% of the phytoplasma effectors are secreted and have a SP, but ~30% have TMDs, revealing that the non-classical effectors constitute an important subset in phytoplasma effectoromes.

Table 2.
Characterization of phytoplasma effectors.
In terms of amino acid composition, almost 50% of the phytoplasma effectors are rich in lysine, ~20% are rich in leucine and ~15% in asparagine (>25 residues per sequence), while tryptophan and cysteine are rare amino acids; ~60% of the effectors have zero or one tryptophan and ~80% have zero or one cysteine residue.
3.3. Functional Categories of the Phytoplasma Effectors
For the 738 phytoplasma effectors, the Gene Ontology analysis identified annotations for 462 phytoplasma effectors, while 276 had no Gene Ontology annotations. Fourteen GO terms were identified: seven in the category “biological process” (phosphorelay signal transduction system, GO:0000160; regulation of DNA-templated transcription, GO:0006355; proteolysis, GO:0006508; protein targeting, GO:0006605; vesicle-mediated transport, GO:0016192; protein import, GO:0017038; protein refolding, GO:0042026); two in the category “cellular component” (plasma membrane, GO:0005886; membrane, GO:0016020); and five in the category “molecular function” (DNA binding, GO:0003677; ATP-dependent peptidase activity, GO:0004176; metalloendopeptidase activity, GO:0004222; ATP binding, GO:0005524; ATP-dependent protein-folding chaperone. GO:0140662). Figure 1 shows the distribution of GOs.
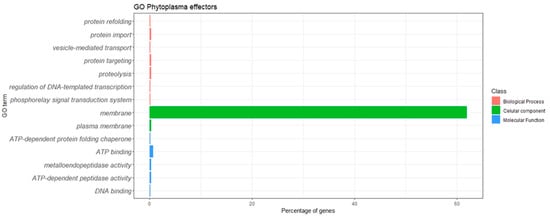
Figure 1.
Gene Ontology classification of the phytoplasma effectors.
The most represented GO term was GO:0016020, with 457 proteins assigned to the term “membrane”; as expected, this term was assigned to 298 known membrane-bound effectors (IdpA, Imp, VmpA and Amp from different phytoplasmas), but also to a few homologs to PHYL1, SAP01, SAP02/saP37/SAP76, SAP08, SAP09/SAP39, SAP34, SAP36, SAP40, SAP42, SAP48, SAP50, SAP53, SAP65, SAP66, SAP68, PME2ST, PME2 and 1 outer-surface lipoprotein. One hundred and twenty-two membrane effectors had no homology to known effectors; according to the UNIPROT database, six were assigned “putative effector”, four were assigned “effector protein” and one hundred and twelve were assigned “effector”. It was observed in this study that membrane-bound effectors are widely distributed in phytoplasmas, not to specific phytoplasma 16Sr groups.
Most effectors received multiple GO assignations; for example, the effectors A0A531Y2Y1 (SAP55-like protein of periwinkle leaf yellowing phytoplasma) and A0A0P7KH40 (SAP55-like of “Echinacea purpurea” witches’ broom phytoplasma) were classified with the GO terms GO:0005524 ATP-binding; GO:0004222 metalloendopeptidase activity; GO:0004176 ATP-dependent peptidase activity; and GO:0006508 proteolysis). Likewise, the effector A0A0G2SK05 (SecA translocation protein of Napier grass stunt phytoplasma) and A0A0G2SK62 (SecA translocation protein of Hyparrhenia grass white leaf phytoplasma) were assigned the terms GO:0005886 plasma membrane; GO:0017038 protein import; and GO:0006605 protein targeting. The effector B1Q3E7 (molecular chaperonin GroEL of “Ca. Phytoplasma japonicum”) was assigned GO:0140662 ATP-dependent protein-folding chaperone; GO:0005524 ATP binding; and GO:0042026 protein refolding.
The phytoplasma effectors retrieved a low number of GO terms, although some effectors had three to five GO terms assigned to them; the functions of the majority of these effectors could not be predicted through GO analysis.
3.4. Functional Domains in Phytoplasma Effectors
The use of effector-related domains is an emerging strategy for effector identification in other phytopathogens [61,62,63]. The phytoplasma effectors were analyzed with the program InterProScan version 5.39–77.0, which identified functional domains in 153 effectors, while 585 had no known domains (Figure 2).
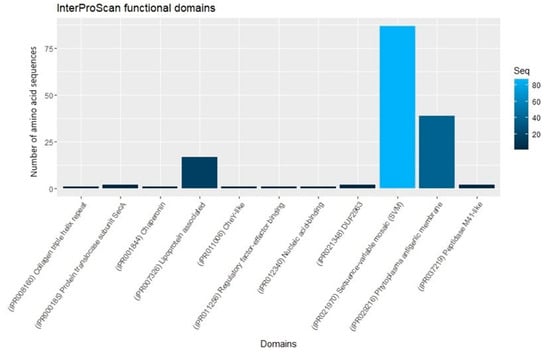
Figure 2.
InterProScan functional domains found in phytoplasma effectors.
The most frequent domain was the “SVM domain”, widely distributed in 87 phytoplasma effectors such as phyllody 1, Sap01, SAP04, SAP05, SAP11, SAP19, SAP21, SAP30, SAP40, SAP42, SAP43, SAP44, SAP45, SAP48, SAP49, SAP54, SAP56, SAP66, SAP67 and SAP68. The second most frequently found domain was “Phytoplasma antigenic membrane”, present in 39 phytoplasma antigenic membrane proteins (AMP). The third was “Lipoprotein associated”, present in 17 phytoplasma variable membrane proteins A (VmpA). Molecular chaperonin (GroEL) domain is present in the effector B1Q3E7 of “Ca. Phytoplasma japonicum”. The domain “response regulator transcription factor” was found in the effector A0A847N6X9 of “Ca. Phytoplasma sp” and the domain “collagen triple helix repeats” in the effector A0A421NUF2 of “Ca. Phytoplasma solani”. The domain DUF2963 was found in two effectors of Rapeseed phyllody phytoplasma, A0A859I9K5 (SAP02/SAP37/SAP76-like protein) and A0A859IA25 (SAP64-like protein). The domain “Nucleic acid-binding” in A0A0P7KDF6 of “Echinacea purpurea” witches’ broom phytoplasma and the domain peptidase M41-like was identified in A0A0P7KH40 and A0A531Y2Y1, two SAP55-like effectors of “Echinacea purpurea” witches’ broom phytoplasma and periwinkle leaf yellowing phytoplasma, respectively.
3.5. Identification of Effector Tribes in Phytoplasmas: Classification by Protein Motifs
In an effort to classify the phytoplasma effectors, set 1 (738 effectors) was analyzed using the MEME program to find de novo motif sequences. The search was conducted either on the full set #1 or in the set #1 but lacking the largest classes of effectors such as SAP11 (members) and VmpA (members). Both strategies rendered similar results. Figure 3 shows the 50 top amino acid motifs found in the phytoplasma effector dataset; 696 amino acid sequences of effectors have at least 1 motif while 42 have no motif at all.
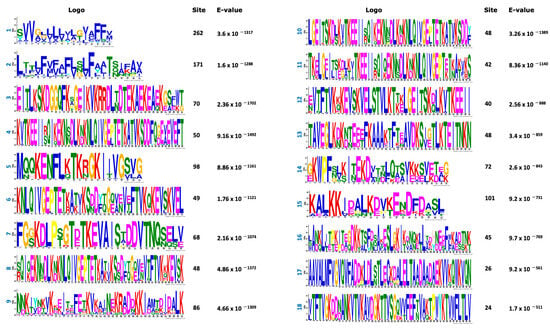
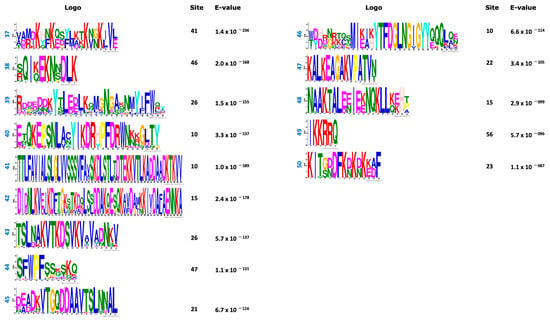
Figure 3.
Top 50 motifs found in the positive dataset (738 amino acid sequences) of phytoplasma effectors. The sites correspond to the number of effectors that contain that motif. E-value per each motif was included.
Based on the organization of these motifs, 15 effector tribes (families) were distinguished; each tribe consists of members that share various motifs in their sequences. Figure 4 shows the schematic representations of the organization of the motifs in the tribes. The WebLogo sequences of these 15 tribes are provided as Figures S1–S15. In some sequences, the gaps between the motifs were narrow and their WebLogo sequences appear as a single continuum sequence instead of various motifs (tribes 3, 5, 11 and 12), but all WebLogos have at least three motifs.
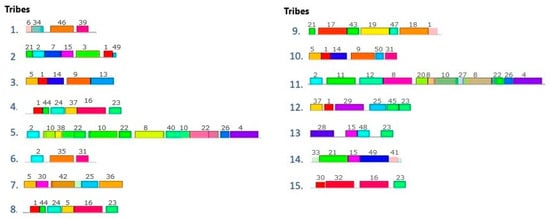
Figure 4.
Schematic representation of the organization of the motifs in the different effector tribes (effector families) in phytoplasmas. These configurations were found in the positive control dataset (738 amino acid sequences of phytoplasma effectors). Rectangles with the same color correspond to the same motif; the lines correspond to the gaps between the motifs in the amino acid sequences. The different sizes of the rectangles represent the different amino acid lengths of the motifs (different number of amino acids). The numbers above the rectangles correspond to the motifs enlisted in Figure 3.
The 15 phytoplasma protein tribes classified 696 effectors (Table 3) while 42 effectors were orphan sequences that do not belong to any tribe. Tribe 1 comprises the largest group of effectors (223 members from the set of 738 effectors). This effector tribe was found widely distributed in phytoplasmas with multiple effector members per genome (>6 members), and in some genomes with more than 18 members (Table 3). The effector tribes 6 and 7 also have a wide distribution among the phytoplasmas but have a smaller number of members per genome; for example, from family 7, only two or three members per genome were found.

Table 3.
Phytoplasma effector tribes and pattern distribution of effector members among phytoplasmas.
Other tribes were restricted to specific phytoplasmas and very few other phytoplasmas. For example, tribe 2 of “Ca. Phytoplasma solani”, tribe 3 of elm yellows phytoplasma, tribe 4 of apple proliferation phytoplasma and tribe 5 of alder yellows phytoplasma and Flavescence dorée phytoplasma.
To characterize the effector members of each tribe, their amino acid composition was analyzed. The amino acid asparagine was rich in effectors belonging to all tribes, except the effector members of tribes 2 and 14. The amino acid lysine was abundant in effector members of all tribes, especially tribes 5 and 11, with ~70 Lys per sequence. About 10 serine amino acids per sequence were observed, except in tribes 5 and 11, in which effector members contained >30 Ser per sequence, and in tribes 3 and 10, in which most effector members contained 4–5 Ser per sequence. Tryptophan and cysteine were poor in all effector members in all tribes; some patterns were observed with respect to these amino acids. Tribe 2 members have one Trp and zero Cys; in tribe 3, effector members have one Trp and one Cys, while in tribes 5 and 6, most effector members have neither Trp nor Cys (Table S2).
3.6. Short Linear Motifs in Phytoplasma Effectors
Short linear motifs (SLiMs), also known as ELMs (from eukaryotic linear motifs), are short linear peptides that have a specific sequence pattern (3–10 amino acids), which is recognized by interacting domains [64]. Usually, SLiMs are involved in transient key interactions with proteins, DNA or RNA, regulating cell processes such as cell signaling, cell cycle, protein degradation, etc. [51]. Many eukaryotic, bacterial and viral pathogens mimic SLiMs present in host cell proteins to hijack cellular processes as part of the infection cycle [65,66,67]. Therefore, we decided to explore SLiMs by analyzing a subset of 87 phytoplasma effectors in the ELM server (2 per each class in Table 1; each class contains at least 1 ID). Forty-two SLiMs were identified (Figure 5). In the top 10 most frequently identified motifs was the SLiM LIG_PDZ_Class_2, which has the pattern (VYF)X(VIL) and is present in diverse SAP effectors; this motif binds to a surface groove of PDZ domains of the target proteins. The PDZ is a ubiquitous motif of 80–90 amino acids found in the signaling proteins of bacteria, yeast, plants, viruses and animals [68]. Proteins containing PDZ motif anchor receptor proteins in the membrane to cytoskeleton, as well as help organize and hold together signaling complexes at the plasma membrane [69,70]. LIG_PDZ_Class_2-containing effectors may function close to the plasma membrane of the host to fulfill their functions.
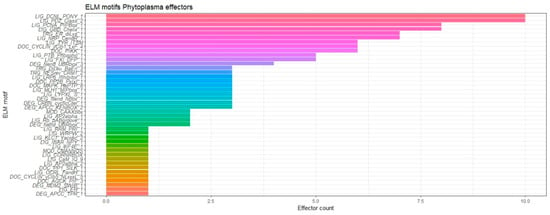
Figure 5.
Short linear motifs found in phytoplasma effectors. A set of 87 effectors was analyzed, selecting 2 effectors per each class in Table 1.
The SLiM LIG_DCNL_PONY_1 was as frequent as the LIG_PDZ_Class_2 motif, and it was also present in diverse SAP effectors. The LIG_DCNL_PONY_1 motif binds the ubiquitin-conjugating enzyme E2 M (UBE2M) and the ubiquitin-conjugating enzyme E2 F (UBE2F), which play diverse biological roles [51]. Phytoplasma effectors containing this motif may regulate their targets through proteolysis. Proteolytic activity has been described in the effectors SAP54 [18] and SAP05 [16], and may be the mechanism of action of other phytoplasma effectors as well. The LIG_PCNA_PIPBox_1 motif was identified in eight effectors and is a nuclear localization domain [51]. Several phytoplasma effectors, such as SAP11 and SPW1, target nuclear proteins in the host, including transcription factors, and the removal of the nuclear localization signal leads to loss of function [71,72]. The presence of the LIG_PCNA_PIPBox_1 domain may uncover other effectors that also function in the plant nuclei. In “Ca. Phytoplasma asteris”, ~7% of the effectorome is predicted to target the plant cell nuclei [34].
The SLiM LIG_GBD_Chelix_1 was found in eight effectors; this motif allows for the recruitment of the actin-regulatory proteins that initiates actin polymerization [73,74]. Polymerization of actin is a common molecular mechanism found in infections by pathogens in different kingdoms [74,75,76,77,78].
The SLiM LIG_NRP_CendR_1 was found in seven effectors that include several SAP-type effectors and the membrane antigenic IdpA. In humans, this domain binds to the neuropilin b1 domain binding site [51]. In addition to the very well-known interaction between the spike receptor protein of SARS-CoV-2 and the human angiotensin-converting enzyme 2 (ACE2), it was recently found that the spike receptor also targets neuropilin b1 [79], evidencing that the neuropilin b1 domain binding site is targetable by pathogens. This motif may be involved in infection by phytoplasmas, but the host target is largely unknown. The next motif was TRG_ER_diLys_1, and it was identified in seven effectors of the SAP-type and the membrane antigenic Amp and VmpA. This motif is an endoplasmic reticulum retrieving signal; it has been found in proteins from humans, rats and yeast. The proteins interact with the WD40 domain and G-beta repeat domain; the former is present in many transcription factors and E3ubiquitin ligase, and the latter in G proteins [51]. Although TRG_ER_diLys_1 has not been described in phytopathogens, it likely participates in phytoplasma pathogenicity.
The SLiM DOC_PIKK_1 was found in six effectors belonging to SAP05, SAP49, SAP54 and the membrane antigenic IdpA. This motif is a docking site for multiple phosphatidylinositol-3 kinase-related kinases (PIKKs) involved in cell cycle DNA damage checkpoints and oxidative stress, and response to DNA damage [51]. The DOC_PIKK_1 motif has been identified in the effector proteins CagA and Tir of the bacteria Legionella pneumophila and L. pasculli [80], but the precise role of this domain has not been established. The effectors with this domain may prevent DNA repair in the host caused by oxidative stress, or these effectors may inactivate PIKKs and suppress host signaling. The SLiM DOC_CYCLIN_yClb1_LxF_4, present in SAP09, SAP20, SAP49 and SAP61, is a docking site present in substrates and inhibitors of the M-phase cyclins Clb1/2 [81]. These effectors possibly interfere in the cell cycle of the host cell. The SLiM LIG_TYR_ITSM occurred in SAP43, SAP49 and SAP50. This motif binds to and is regulated by SH2 adaptor molecules, and is critical for the activation and termination of signal transduction pathways [51]. The motif LIG_TYR_ITSM was identified in effector proteins of Corynebacterium diphtheria [82], but its role is unknown. The SLiMs LIG_FXI_DFP_1 and LIG_PTB_Phospho_1 are in the 10th position, each one identified in five effectors. LIG_FXI_DFP_1 was found in SAP39, SAP43 and PME2; this motif is a disulfide-linked dimer, each subunit containing four apple domains (A1-4) and a C-terminal trypsin-like catalytic domain. The DFP binds to the second apple domain of coagulation factor XI and plasma kallikrein heavy chain [83]; nanobodies against factor XI apple 3 domain inhibit its protein–protein interaction, evidencing the importance of these domains [84]. These proteins are typically present in mammals and absent in plants, but coagulation factor XI and plasma kallikrein are serine proteases [85]; plant serine proteases play key roles in plant defense [86,87]. The phytoplasma effectors containing LIG_FXI_DFP_1 may target some of those proteases. The last motif in the top 10 motifs identified here was LIG_PTB_Phospho_1, present in SAP08, SAP67 and PHYL1. The LIG_PTB_Phospho_1 motif binds short peptides with a core Asn-X-X-Tyr motif, phosphorylated on the Tyr residue. To the best of our knowledge, the motif Asn-X-X-Tyr motif has not been described in plants, making it difficult to predict the plant targets for effectors harboring the LIG_PTB_Phospho_1 motif, but tyrosine kinases involved in defense responses are possible targets of effectors with this motif [88,89,90,91]. Tyrosine phosphorylation plays an important role in plant cell signaling [90], and plant and animal tyrosine kinases share ancestral origin [91]. Table 4 corresponds to a summary of the information about the top 10 SLiMs.

Table 4.
The top 10 SLiMs (ELMs) found in phytoplasma effectors.
4. Discussion
Phytoplasmas are diverse pathogens that cause severe problems in agriculture and horticulture. Integrated control management of the main phytoplasma diseases is expected to include early detection [92], the use of nonantibiotic antimicrobials [93], the use of resistant plant material [10] and novel strategies for environmentally friendly insect vector control [11]. In line with these strategies, effectoromics may play a key role in crop protection since effectors are targetable genes [12,13], or they may be used as “hunter genes” to detect genes encoding resistance proteins [94,95]. These strategies are already strengthening certain genetic improvement programs [96,97].
Effectoromics is currently a rapidly growing and evolving area. Until recently, the description of effectors in bacteria, fungi and phytoplasmas was “small, TMD-lacking secreted proteins” [27,32,34]. It is now clear that noncanonical (or non-classical) effectors that do not meet these criteria also exist [98]. In fungi and oomycetes, the description of effectors has been rapidly changing, since validated novel effectors with sizes larger than 400 amino acids or with TMDs, or without a signal peptide, among other non-classical characteristics, have been discovered [98]. Most reports have identified effectors in phytoplasma genomes while looking for proteins with signal peptides, and the absence of TMDs. Contrarily, Debonneville et al. (2022) [28] recently drew attention to effectors with TMD in Flavescence dorée phytoplasma, and Gao et al. (2023) [36] demonstrated the existence of secreted effectors that lack a signal peptide (SP) but are secreted through Sec 2-independent secretion pathway in “Ca. Phytoplasma ziziphi”, evidencing that non-classical effectors also exist in phytoplasmas. Here, the analysis of 738 phytoplasma effectors revealed that non-classical effectors are more common in phytoplasmas than previously believed. Approximately 30% of the known effectors lack SP, ~30% have TMDs and almost 10% are greater than 400 amino acids in length (Table 2). This list of phytoplasma effectors comprises membrane-bound proteins such as IdpA, Imp, VmpA and Amp. Based on more traditional descriptions of effectors as “no TMD, secreted proteins”, these proteins should be excluded, but it is known that the Amp protein interacts with the actin of insect vectors [99], and recently, Wang et al. (2023) [26] showed that the expression of this protein in Nicotiana tabacum inhibits plant host defense and promotes infection by the rice orange leaf phytoplasma, validating this protein as a true effector.
Currently, the list of experimentally validated phytoplasma effectors is less than 25, and functional effectoromics is tedious and only slowly advancing. In phytoplasma effectors, homology has been described only for the effector families/tribes SAP05, SAP11, SAP54 and TENGU; other homology-based families/tribes such as IdpA, Imp, VmpA and Amp add to the list, but the number of effector families/tribes still remains very limited. This lack of conservation makes it difficult to progress in the functional characterization of phytoplasma effectors, since each effector must be studied individually. Homology is also usually low or nonexistent among effectors from other taxonomic kingdoms [100,101].
The Gene Ontology database is the world’s largest source of information on gene function. Unfortunately, the result of the GO analysis for the phytoplasma effectors was not very informative. Fourteen GO terms were retrieved but eight terms were assigned to the same effectors, leaving most effectors unassigned; the Gene Ontology result reflects our lack of knowledge about the functions of most phytoplasma effectors. In total, 462 phytoplasma effectors received a GO assignation; the GO term “membrane” was the largest one, assigned to 457 effectors. Of these 457 effectors, 298 correspond to Amp, Stamp, VmpA, IdpA and Imp; 37 correspond to SAP-like proteins; and 122 correspond to novel phytoplasma effectors. As Debonneville et al. (2022) [28] evidenced, most unknown phytoplasma effectors may be transmembrane proteins. It is expected that in coming times, the phytoplasma effectoromes routinely include transmembrane proteins.
The second strategy to explore possible functions involved the identification of protein domains. The most frequently identified domain was the SVM domain, widely distributed in 87 PHYL1 and SAP phytoplasma effectors. The SVM domain is a signal sequence about 30 amino acids in length, which has been reported as a unique feature of phytoplasmal genome architecture [102]. The genomes of ancestral relatives of phytoplasmas have no SVM-like structures and it is supposed that this domain arose from ancient phage attacks to phytoplasmas [103]. The second and third most frequent domains in phytoplasma effectors were restricted to AMP and VmpA, respectively, and are related with their location on the membrane surface. Some domains suggest the functions of the effectors that contain them, such as “chaperonin (GroEL) domain”, “response regulator transcription factor”, “Nucleic acid-binding”, and “Peptidase M41-like domain”. This highlights new opportunities for future research on these effectors.
The classification of effectors in tribes (protein families that share motifs) is useful to accelerate effectoromics [98,104,105,106], since shared motifs are related to common functions [107,108], and it was the third strategy that was followed here. Fifteen families or tribes were distinguished based on the different combinations found of protein (de novo) motifs. The occurrence of these effector tribes in the phytoplasma genomes revealed two evolutionary histories: the tribes 1, 6 and 7 are widely distributed, suggesting they come from the common ancestor of the phytoplasmas and were inherited by vertical transfer while other groups of phytoplasmas do not have effector members in these tribes, suggesting gene loss, which is common in the evolutionary histories of effectors [109,110].
Other tribes (2, 3, 4 and 5) were restricted to particular phytoplasmas; these effector tribes probably arose through the interaction and coevolution with the hosts. For example, the tribe 3 is characteristic of phytoplasmas in group 16SrV: elm yellows phytoplasma genome (16SrV), alder yellows phytoplasma (16SrV-A), “Ca. Phytoplasma vitis” (Flavescence dorée phytoplasma) (16SrV-C); in the alder yellows phytoplasma genome, this tribe has been amplified. Tribe 5 was also found principally in phytoplasmas belonging to the 16SrV phytoplasma group: the alder yellows phytoplasma (16SrV-A), Ziziphus jujuba witches’ broom phytoplasma (16SrV-B) and Flavescence dorée phytoplasma (16SrV-C), with 17, 2 and 15 effectors per genome, respectively, suggesting that this tribe comes from a common ancestor of phytoplasmas belonging to the phytoplasma 16SrV group and became amplified in certain genomes. One or two members of tribe 5 are present in the genome of phytoplasmas of other 16Sr phytoplasma groups. This tribe probably arose in phytoplasmas of the phytoplasma group 16SrV and arrived at phytoplasmas of other 16Sr phytoplasma groups by horizontal gene transfer (gain of genes), which is also a common mechanism of genome evolution in other organisms for the genomic content of effectors [109,111,112,113,114]. Gain and loss of effectors are common in all microbial kingdoms, and drive the patchy/discontinuous distribution typical of an effector’s phylogenetic distribution [115,116].
De novo motifs were useful in the organization of nonhomologous effectors, and this classification may help accelerate the discovery of effector functions by studying only a few members per tribe as representatives of that family. In our analysis, de novo motifs did not reveal much information about the effector functions. Known motifs (SLiM or ELM) were identified in the repository of the ELM server, which is a comprehensive database of known experimentally validated motifs [51]. Although the ELM acronym means “eukaryotic linear motif”, the search for ELMs is also useful for studies on pathogenic prokaryotes, since pathogens mimic features of critical host proteins to hijack their cell machinery, promoting infection of the host [66,117,118,119]. Effectors that do not share high overall sequence identity, but share motifs, domains or similar tridimensional structures, may share similar functions and are termed “functional orthologs” [120].
ELM analysis can only be performed on a single protein sequence, which is a tedious task when high-throughput analysis is required. As such, the complete dataset of 738 phytoplasma effector proteins could not be analyzed; instead, we analyzed a subset of 87 amino acid sequences, which represented all classes of phytoplasma effector proteins known to date, and we were able to identify 42 SLiMs. Each of these protein signatures occurs in different classes of effectors, for example, SAPs and Vamp share the SLiM TRG_ER_diLys_1. We revised, in detail, the top 10 SLiMs (those most frequently found in phytoplasma effectors) and found that some SAP effectors may regulate their targets through proteolysis, either binding ubiquitin-conjugating enzyme or docking serine proteases. Proteolysis plays a central role in plant–pathogen interaction; both actors, pathogen and host, use proteases for defense and attack [121,122,123]. Some phytoplasmas effectors with protease activity are SAP05 and SAP54 [16], but the identification of the SLiMs LIG_DCNL_PONY_1 and LIG_FXI_DFP_1 suggests that proteolysis plays a more important role than evidenced in the pathogenicity of phytoplasmas [51]. Other known mechanisms of pathogens of other kingdoms were also revealed as probable infection mechanisms in phytoplasmas, for example, actin polymerization by effectors containing the motif LIG_GBD_Chelix_1 [73,74], or targeting of the host nuclei and transcription factors by effectors with the motif LIG_PCNA_PIPBox_1 [51]. Other host targets are more difficult to predict, such as the target for the SLiM LIG_NRP_CendR_1 that binds to the human neuropilin b1 domain binding site [51], or the motif LIG_PTB_Phospho_1 that binds short peptides with a core Asn-X-X-Tyr motif [51], which has not been found in plant proteins. Other effectors seem to be localized part of the time in the cytosolic and part of the time bound to membrane proteins, approaching the plasma membrane to interact with their targets [124]. Other effectors appear to regulate cell cycle or signaling pathways [125,126]. Interestingly, some of the top 10 SLiMs have been identified in bacterial effectors such as DOC_PIKK_1 identified in the effector proteins CagA and Tir of the bacteria L. pneumophila and L. pasculli [80] and the motif LIG_TYR_ITSM identified in effector proteins of C. diphtheria [82], reinforcing the potential role of these motifs in phytoplasmas for the successful infection of the host.
Pathogens and hosts are constantly coevolving, and SLiMs/ELMs are becoming increasingly studied in pathogens, as these mimic critical host motifs, allowing mimicry peptides (mimitopes) to sabotage key host processes [117,127,128,129,130,131]. Since these SLiMs are abundant and critically important in pathogenesis, they are now being studied as novel drug targets [65,103], either to screen for novel molecules [118], or by docking-based peptide design that disrupts mimitope–host target interaction [132,133,134]. To the best of our knowledge, this is the first report of motifs (known motifs and de novo motifs) in phytoplasma effectors. Some effectors that share common characteristics may target the same protein in the host, named hub proteins, which play key roles in the host cell [135]. For example, the effectors IPI-01 and IPI-04 from the oomycete Phytophthora infestans are able to interact and interfere with functionality of the RB protein, an NLR immune receptor of Solanum bulbocastanum Dunal [136]. In other example, the type III Effectors NopT and NopP of rhizobial bacteria target the hub protein GmPBS1, which is key for nodulation in soybean [137]. The identification of common targets for different members of phytoplasma effector families/tribes may enable the identification of key host proteins for further research. Since all these analyses are in silico, further experimental validation is necessary. Interaction between effectors and host targets may be confirmed through in vitro double-hybrid tests and in vivo through the bimolecular fluorescence complementation (BiFC) assay [72]. The host’s hub proteins may be used as molecular markers to assist genetic improvement programs or for direct genetic manipulation through CRISPR-Cas9 technology for the development of phytoplasma-resistant plants [138]. Likewise, the identification of targets in insect vectors may enable us to control phytoplasmas through the modulation of gene expression in insects with gene silencing [139,140]. In addition to their practical applications, these experimental evaluations also uncover the mechanisms used by phytoplasma effectors and effector roles during host–pathogen interactions. Findings here in the amino acid sequences of phytoplasma effectors open exciting lines of research to unravel effector roles and explore novel strategies for phytoplasma control.
In addition to the novel insights from the identification of protein domains, de novo motifs and known motifs, the analysis of amino acid compositions revealed interesting results about phytoplasma effectors. They were found to be rich in the amino acids Lys, Leu and Asn, and poor in Trp and Cys. A similar composition of amino acids was reported by Singh and Lakhanpaul (2020) [141] in ortholog proteins of SAP54, which were rich in the amino acids asparagine (19.7%), leucine (16%) and lysine (10.9%), while cysteine and tryptophan were absent.
The proteomes of the unicellular eukaryotes Plasmodium falciparum, P. berghei, P. chabaudi and P. yoelii have been dominated by Lys-rich, Asn-rich and Ile-rich proteins [142]. The genomes of these parasites are AT-rich, as phytoplasma genomes usually are [35,103,143], which leads to an abundance of the trinucleotide AAT, coding for Asn [142,144]. In P. falciparum, the causal agent of the malaria disease, the Asn-rich proteins are fibrillar [145], and their abnormal extracellular accumulation leads to the formation of the amyloid, irreversible insoluble protein aggregates that deposit in organs and tissues and affect the heart, kidneys, liver, nerves and digestive system of the human host [146]. For this biochemical behavior, these proteins have been named “prion-like” [147]. Prions have been linked with diseases in eukaryotes, but a prion-like protein was first described in Clostridium botulinum that behaved like a prion when it was expressed in Escherichia coli bacteria and yeast [148]. To date, it is known that prion-like proteins are part of bacterial proteomes, and the largest fractions are found in the Mycoplasma genus [149], a relative of the Phytoplasma genus; both belong to the bacterial class Mollicutes but the former are bacterial parasites of animal pathogens [150]. Prions may be ancient in the history of life [151,152] since they have been found in phages of various groups of bacteria and archaea [153], and the spike protein of the SARS-CoV-2 virus [154]; therefore, their presence in phytoplasmas would not be rare. Prionic signatures should be identified in phytoplasma proteomes and may help to unravel the function of some members in the effectoromes. The prion-like aggregated structure probably confers protection to the phytoplasma effectors against proteolytic degradation [155]. Richness in Asn in the proteins is typically associated with regions of low complexity, probably forming bulging domains [142]; in P. falciparum, these low-complexity regions in the proteins play diverse roles throughout the parasite’s lifecycle, from mediating protein–protein interactions to enabling the parasite to evade the host immune system [156]. In the phytoplasma effectors named phyllogens, Iwabuchi et al. [157] demonstrated the importance of asparagine and asparagine–lysine amino acids in the induction of changes in flower morphology.
In the case of Lys-rich proteins, Lys-rich secreted effectors were recently reported in the fungus Fusarium oxysporum f. sp. lycopersici and were found regulated by acetylation [158]. Acetylation is a dynamic and reversible posttranslational modification that regulates protein functions through modifying enzymatic activity, interactions with DNA, protein stability, protein localization and the interaction with other proteins [159]. Most of the genes of F. oxysporum f. sp. lycopersici for Lys-rich proteins were upregulated during initial infection of the plant host [158], revealing that they are indeed involved in pathogenesis. In prokaryotes, lysine acetylation regulates proteins involved in transcription, translation, pathways associated with central metabolism and stress responses [160]. These regulatory functions are consistent with the roles of a number of phytoplasma effectors that bind other proteins and regulate gene transcription and the metabolisms of phytoregulators [18,71,161]; however, whether acetylation occurs in phytoplasma proteins remains to be determined.
The amino acid leucine contributes to the aliphatic index in proteins. Leu is common to both plant receptors that recognize pathogen effectors [162,163] and bacterial effectors, such as the SlrP effector from Salmonella enterica that targets the human chaperone ERdj3 [164,165]; the SspH1, SspH2 and Slrp effectors of S. typhimurium required for normal pathogenesis in animal models [166]; the XC1553 effector from Xanthomonas campestris pv. campestris that is recognized in vascular tissue of Arabidopsis thaliana [167]; the YopM effector from Yersinia pestis, which has a C-terminal E3 ligase domain (NEL domain) [168]; and the GALA5 effector from Ralstonia syzygii [169], among others. Leu-rich proteins are widely distributed in bacteria, playing important roles in various protein–protein interaction processes [170,171,172]; this suggests that this effector feature existed before the phytoplasmas arose from their bacterial ancestor [141,173]. However, all these bacterial effectors are secreted through the type III secretion system [167,174], while most of the effectors of phytoplasmas are secreted through the Sec-dependent pathway [34]. Leucine also plays a role in pathogenicity in other microorganisms, for example, in fungi and oomycetes, the motif RXLR at the C-terminus of some effectors often suppresses plant immunity [44,175].
Regarding the amino acid cysteine, it forms inter- or intramolecular disulfide bonds, which confer thermal stability to the protein structure under oxidizing conditions [176]. In fungi, many effectors are Cys-rich proteins; this characteristic confers stability in the acidic environment of the host apoplast [177,178]. Conversely, in phytoplasma effectors, the presence of the amino acid Cys was found to be limited or absent. Miseta and Csutora [179] analyzed the content of Cys in the proteins of phylogenetically distant organisms and found an evolutionary trend favoring the incorporation of more cysteine residues into proteins of more complex organisms, as an “order-promoting” condition [179,180]. Stability in phytoplasma effectors is probably driven by other means, for example, through low-complexity regions or prionic signatures in the proteins. In the case of Trp, this amino acid is usually rare in proteins, since it is encoded by only one codon. Further research is needed to establish the functional significance of these amino acid patterns.
All these novel findings shed light on the world of effectoromics in phytoplasmas. It will be interesting to address some of the hypotheses that are raised from these computational results.
5. Conclusions
Conventional functional analysis using Gene Ontology and the search for protein domains resulted in poorly informative data.
The computational characterization uncovered novel features of phytoplasma effectors:
The search for de novo motifs enabled the classification of phytoplasma effectors in 15 tribes or motif-based families; some of them are widespread in phytoplasmas, while other tribes are only associated with particular phytoplasmas, evidencing at least two different evolutionary histories in phytoplasma effectors.
The presence of SliMs or ELMs suggested that phytoplasmas also employ common strategies used by pathogens from other kingdoms. They may mimic host protein interactors that bind to other host proteins with critical roles.
The identification of de novo motifs proved essential for the organization of the phytoplasma effectors. The classification of the phytoplasma effectors in tribes, together with the identification of SLiMs/ELMs, are promissory first steps in the discovery of probable functional orthologs in phytoplasmas.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae9111228/s1, Figure S1: WebLogo plot of phytoplasma effectors belonging to tribe 1; Figure S2: WebLogo plot of phytoplasma effectors belonging to tribe 2; Figure S3: WebLogo plot of phytoplasma effectors belonging to tribe 3; Figure S4: WebLogo plot of phytoplasma effectors belonging to tribe 4; Figure S5: WebLogo plot of phytoplasma effectors belonging to tribe 5; Figure S6: WebLogo plot of phytoplasma effectors belonging to tribe 6; Figure S7: WebLogo plot of phytoplasma effectors belonging to tribe 7; Figure S8: WebLogo plot of phytoplasma effectors belonging to tribe 8; Figure S9: WebLogo plot of phytoplasma effectors belonging to tribe 9; Figure S10: WebLogo plot of phytoplasma effectors belonging to tribe 10; Figure S11: WebLogo plot of phytoplasma effectors belonging to tribe 11; Figure S12: WebLogo plot of phytoplasma effectors belonging to tribe 12; Figure S13: WebLogo plot of phytoplasma effectors belonging to tribe 13; Figure S14: WebLogo plot of phytoplasma effectors belonging to tribe 14; Figure S15: WebLogo plot of phytoplasma effectors belonging to tribe 15; Table S1A, phytoplasma effector dataset; Table S1B, phytoplasma non-effectors, essential (core) proteins; Table S2. Phytoplasma effector tribes and amino acid composition of effector members.
Author Contributions
Conceptualization: B.C.-C.; data curation and formal analysis: K.G.C.-A. and S.E.V.-L.; original draft preparation: B.C.-C.; writing—review and editing: B.C.-C., K.G.C.-A., S.E.V.-L. and L.S.-C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Consejo Nacional de Humanidades, Ciencias y Tecnologías (CONAHCyT), México, grant number FOP16-2021-01 No. 320993.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Data are provided in the manuscript and Supplementary Materials. Amino acid sequences of phytoplasma effectors are available at https://github.com/Gisel-Carreon/Phytoplasma_effectors.
Acknowledgments
The authors thank CONAHCyT. for the scholarships granted to Sara Elena Vila Luna (550897) for her doctoral studies, and Karla Gisel Carreón Anguiano (application 4120040) for her postdoctoral research.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bertaccini, A. Phytoplasmas: Diversity, Taxonomy, and Epidemiology. Front. Biosci. 2007, 12, 673. [Google Scholar] [CrossRef] [PubMed]
- Hogenhout, S.A.; Oshima, K.; Ammar, E.; Kakizawa, S.; Kingdom, H.N.; Namba, S. Phytoplasmas: Bacteria That Manipulate Plants and Insects. Mol. Plant Pathol. 2008, 9, 403–423. [Google Scholar] [CrossRef] [PubMed]
- Oshima, K.; Maejima, K.; Namba, S. Genomic and Evolutionary Aspects of Phytoplasmas. Front. Microbiol. 2013, 4, 230. [Google Scholar] [CrossRef] [PubMed]
- Kirdat, K.; Tiwarekar, B.; Sathe, S.; Yadav, A. From Sequences to Species: Charting the Phytoplasma Classification and Taxonomy in the Era of Taxogenomics. Front. Microbiol. 2023, 14, 1123783. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-Y.; Zhang, R.-Y.; Li, J.; Li, Y.-H.; Shan, H.-L.; Li, W.-F.; Huang, Y.-K. The Diversity, Distribution and Status of Phytoplasma Diseases in China. Front. Sustain. Food Syst. 2022, 6, 943080. [Google Scholar] [CrossRef]
- Sugio, A.; MacLean, A.M.; Kingdom, H.N.; Grieve, V.M.; Manimekalai, R.; Hogenhout, S.A. Diverse Targets of Phytoplasma Effectors: From Plant Development to Defense Against Insects. Annu. Rev. Phytopathol. 2011, 49, 175–195. [Google Scholar] [CrossRef] [PubMed]
- Rashid, U.; Bilal, S.; Bhat, K.A.; Shah, T.A.; Wani, T.A.; Bhat, F.A.; Mughal, M.N.; Nazir, N. Phytoplasma Effectors and Their Role in Plant-Insect Interaction. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 1136–1148. [Google Scholar] [CrossRef]
- Bertaccini, A. Plants and Phytoplasmas: When Bacteria Modify Plants. Plants 2022, 11, 1425. [Google Scholar] [CrossRef]
- Hemmati, C.; Nikooei, M.; Al-Subhi, A.M.; Al-Sadi, A.M. History and Current Status of Phytoplasma Diseases in the Middle East. Biology 2021, 10, 226. [Google Scholar] [CrossRef]
- Oliveira, M.J.R.A.; Roriz, M.; Vasconcelos, M.W.; Bertaccini, A.; Carvalho, S.M.P. Conventional and Novel Approaches for Managing “Flavescence Dorée” in Grapevine: Knowledge Gaps and Future Prospects. Plant Pathol. 2019, 68, 3–17. [Google Scholar] [CrossRef]
- Tzec-Simá, M.; Félix, J.W.; Granados-Alegría, M.; Aparicio Ortiz, M.; Juárez-Monroy, D.; Mayo-Ruiz, D.; Vivas-López, S.; Gómez-Tah, R.; Canto-Canché, B.; Berezovski, M.V. Potential of Omics to Control Diseases and Pests in the Coconut Tree. Agronomy 2022, 12, 3164. [Google Scholar] [CrossRef]
- Galetto, L.; Abbà, S.; Rossi, M.; Ripamonti, M.; Palmano, S.; Bosco, D.; Marzachì, C. Silencing of ATP Synthase β Reduces Phytoplasma Multiplication in a Leafhopper Vector. J. Insect Physiol. 2021, 128, 104176. [Google Scholar] [CrossRef] [PubMed]
- Tomkins, M.; Kliot, A.; Marée, A.F.; Hogenhout, S.A. A Multi-Layered Mechanistic Modelling Approach to Understand How Effector Genes Extend beyond Phytoplasma to Modulate Plant Hosts, Insect Vectors and the Environment. Curr. Opin. Plant Biol. 2018, 44, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Cadiou, L.; Brunisholz, F.; Cesari, S.; Kroj, T. Molecular Engineering of Plant Immune Receptors for Tailored Crop Disease Resistance. Curr. Opin. Plant Biol. 2023, 74, 102381. [Google Scholar] [CrossRef] [PubMed]
- Zdrzałek, R.; Stone, C.; De La Concepcion, J.C.; Banfield, M.J.; Bentham, A.R. Pathways to Engineering Plant Intracellular NLR Immune Receptors. Curr. Opin. Plant Biol. 2023, 74, 102380. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; MacLean, A.M.; Sugio, A.; Maqbool, A.; Busscher, M.; Cho, S.-T.; Kamoun, S.; Kuo, C.-H.; Immink, R.G.H.; Hogenhout, S.A. Parasitic Modulation of Host Development by Ubiquitin-Independent Protein Degradation. Cell 2021, 184, 5201–5214.e12. [Google Scholar] [CrossRef] [PubMed]
- Sugio, A.; MacLean, A.M.; Hogenhout, S.A. The Small Phytoplasma Virulence Effector SAP 11 Contains Distinct Domains Required for Nuclear Targeting and CIN—TCP Binding and Destabilization. New Phytol. 2014, 202, 838–848. [Google Scholar] [CrossRef]
- MacLean, A.M.; Orlovskis, Z.; Kowitwanich, K.; Zdziarska, A.M.; Angenent, G.C.; Immink, R.G.H.; Hogenhout, S.A. Phytoplasma Effector SAP54 Hijacks Plant Reproduction by Degrading MADS-Box Proteins and Promotes Insect Colonization in a RAD23-Dependent Manner. PLoS Biol. 2014, 12, e1001835. [Google Scholar] [CrossRef]
- Sugawara, K.; Honma, Y.; Komatsu, K.; Himeno, M.; Oshima, K.; Namba, S. The Alteration of Plant Morphology by Small Peptides Released from the Proteolytic Processing of the Bacterial Peptide TENGU. Plant Physiol. 2013, 162, 2005–2014. [Google Scholar] [CrossRef]
- Hoshi, A.; Oshima, K.; Kakizawa, S.; Ishii, Y.; Ozeki, J.; Hashimoto, M.; Komatsu, K.; Kagiwada, S.; Yamaji, Y.; Namba, S. A Unique Virulence Factor for Proliferation and Dwarfism in Plants Identified from a Phytopathogenic Bacterium. Proc. Natl. Acad. Sci. USA 2009, 106, 6416–6421. [Google Scholar] [CrossRef]
- Wang, N.; Yang, H.; Yin, Z.; Liu, W.; Sun, L.; Wu, Y. Phytoplasma Effector SWP1 Induces Witches’ Broom Symptom by Destabilizing the TCP Transcription Factor BRANCHED1: Phytoplasma Effector SWP1 Destabilizes BRC1. Mol. Plant Pathol. 2018, 19, 2623–2634. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Chen, L.; Ye, X.; Tan, B.; Zheng, X.; Cheng, J.; Wang, W.; Yang, Q.; Zhang, Y.; Li, J.; et al. Phytoplasma Effector Zaofeng6 Induces Shoot Proliferation by Decreasing the Expression of ZjTCP7 in Ziziphus Jujuba. Hortic. Res. 2022, 9, uhab032. [Google Scholar] [CrossRef] [PubMed]
- Kakizawa, S.; Oshima, K.; Ishii, Y.; Hoshi, A.; Maejima, K.; Jung, H.-Y.; Yamaji, Y.; Namba, S. Cloning of Immunodominant Membrane Protein Genes of Phytoplasmas and Their in Planta Expression. FEMS Microbiol. Lett. 2009, 293, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Boonrod, K.; Munteanu, B.; Jarausch, B.; Jarausch, W.; Krczal, G. An Immunodominant Membrane Protein (Imp) of ‘ Candidatus Phytoplasma Mali’ Binds to Plant Actin. Mol. Plant-Microbe Interact. 2012, 25, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Arricau-Bouvery, N.; Duret, S.; Dubrana, M.-P.; Desqué, D.; Eveillard, S.; Brocard, L.; Malembic-Maher, S.; Foissac, X. Interactions between the Flavescence Dorée Phytoplasma and Its Insect Vector Indicate Lectin-Type Adhesion Mediated by the Adhesin VmpA. Sci. Rep. 2021, 11, 11222. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, X.; Zhou, S.; Zhang, X.; Zhu, Y.; Chen, B.; Huang, X.; Yang, X.; Zhou, G.; Zhang, T. The Antigenic Membrane Protein (Amp) of Rice Orange Leaf Phytoplasma Suppresses Host Defenses and Is Involved in Pathogenicity. Int. J. Mol. Sci. 2023, 24, 4494. [Google Scholar] [CrossRef]
- Music, M.S.; Samarzija, I.; Hogenhout, S.A.; Haryono, M.; Cho, S.-T.; Kuo, C.-H. The Genome of ‘Candidatus Phytoplasma Solani’ Strain SA-1 Is Highly Dynamic and Prone to Adopting Foreign Sequences. Syst. Appl. Microbiol. 2019, 42, 117–127. [Google Scholar] [CrossRef]
- Debonneville, C.; Mandelli, L.; Brodard, J.; Groux, R.; Roquis, D.; Schumpp, O. The Complete Genome of the “Flavescence Dorée” Phytoplasma Reveals Characteristics of Low Genome Plasticity. Biology 2022, 11, 953. [Google Scholar] [CrossRef]
- Wagner, N.; Teper, D.; Pupko, T. Predicting Type III Effector Proteins Using the Effectidor Web Server. In Bacterial Virulence; Gal-Mor, O., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2022; Volume 2427, pp. 25–36. ISBN 978-1-07-161970-4. [Google Scholar]
- Noroy, C.; Lefrançois, T.; Meyer, D.F. Searching Algorithm for Type IV Effector Proteins (S4TE) 2.0: Improved Tools for Type IV Effector Prediction, Analysis and Comparison in Proteobacteria. PLoS Comput. Biol. 2019, 15, e1006847. [Google Scholar] [CrossRef]
- Sperschneider, J.; Dodds, P.N.; Gardiner, D.M.; Singh, K.B.; Taylor, J.M. Improved Prediction of Fungal Effector Proteins from Secretomes with EffectorP 2.0. Mol. Plant Pathol. 2018, 19, 2094–2110. [Google Scholar] [CrossRef]
- Carreón-Anguiano, K.G.; Islas-Flores, I.; Vega-Arreguín, J.; Sáenz-Carbonell, L.; Canto-Canché, B. EffHunter: A Tool for Prediction of Effector Protein Candidates in Fungal Proteomic Databases. Biomolecules 2020, 10, 712. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.A.; McDowell, J.M. Recent Advances in Understanding of Fungal and Oomycete Effectors. Curr. Opin. Plant Biol. 2022, 68, 102228. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Correa, V.R.; Toruño, T.Y.; Ammar, E.-D.; Kamoun, S.; Hogenhout, S.A. AY-WB Phytoplasma Secretes a Protein That Targets Plant Cell Nuclei. Mol. Plant-Microbe Interact. 2009, 22, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.M.; Lin, Y.-C.; Li, J.-R.; Chien, Y.-Y.; Wang, C.-J.; Chou, L.; Wang, C.-W.; Chiu, Y.-C.; Kuo, C.-H.; Yang, J.-Y. Accelerating Complete Phytoplasma Genome Assembly by Immunoprecipitation-Based Enrichment and MinION-Based DNA Sequencing for Comparative Analyses. Front. Microbiol. 2021, 12, 766221. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Ren, Z.; Zhao, W.; Li, W. Candidatus Phytoplasma ziziphi Encodes Non-Classically Secreted Proteins That Suppress Hypersensitive Cell Death Response in Nicotiana Benthamiana. Phytopathol. Res. 2023, 5, 11. [Google Scholar] [CrossRef]
- Stern, D.L.; Han, C. Gene Structure-Based Homology Search Identifies Highly Divergent Putative Effector Gene Family. Genome Biol. Evol. 2022, 14, evac069. [Google Scholar] [CrossRef]
- Jones, D.A.B.; Moolhuijzen, P.M.; Hane, J.K. Remote Homology Clustering Identifies Lowly Conserved Families of Effector Proteins in Plant-Pathogenic Fungi. Microb. Genom. 2021, 7, 000637. [Google Scholar] [CrossRef]
- Dean, P. Functional Domains and Motifs of Bacterial Type III Effector Proteins and Their Roles in Infection. FEMS Microbiol. Rev. 2011, 35, 1100–1125. [Google Scholar] [CrossRef]
- Seong, K.; Krasileva, K.V. Computational Structural Genomics Unravels Common Folds and Novel Families in the Secretome of Fungal Phytopathogen Magnaporthe Oryzae. Mol. Plant-Microbe Interact. 2021, 34, 1267–1280. [Google Scholar] [CrossRef]
- Rocafort, M.; Bowen, J.K.; Hassing, B.; Cox, M.P.; McGreal, B.; De La Rosa, S.; Plummer, K.M.; Bradshaw, R.E.; Mesarich, C.H. The Venturia Inaequalis Effector Repertoire Is Dominated by Expanded Families with Predicted Structural Similarity, but Unrelated Sequence, to Avirulence Proteins from Other Plant-Pathogenic Fungi. BMC Biol. 2022, 20, 246. [Google Scholar] [CrossRef]
- Jiang, R.H.Y.; Tripathy, S.; Govers, F.; Tyler, B.M. RXLR Effector Reservoir in Two Phytophthora Species Is Dominated by a Single Rapidly Evolving Superfamily with More than 700 Members. Proc. Natl. Acad. Sci. USA 2008, 105, 4874–4879. [Google Scholar] [CrossRef] [PubMed]
- Solé, M.; Popa, C.; Mith, O.; Sohn, K.H.; Jones, J.D.G.; Deslandes, L.; Valls, M. The Awr Gene Family Encodes a Novel Class of Ralstonia Solanacearum Type III Effectors Displaying Virulence and Avirulence Activities. Mol. Plant-Microbe Interact. 2012, 25, 941–953. [Google Scholar] [CrossRef]
- Liu, L.; Xu, L.; Jia, Q.; Pan, R.; Oelmüller, R.; Zhang, W.; Wu, C. Arms Race: Diverse Effector Proteins with Conserved Motifs. Plant Signal. Behav. 2019, 14, 1557008. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.H.; Tan, C.M.; Wu, C.-T.; Lin, T.-H.; Jiang, S.-Y.; Liu, R.-C.; Tsai, M.-C.; Su, L.-W.; Yang, J.-Y. Alterations of Plant Architecture and Phase Transition by the Phytoplasma Virulence Factor SAP11. J. Exp. Bot. 2018, 69, 5389–5401. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.-T.; Kung, H.-J.; Huang, W.; Hogenhout, S.A.; Kuo, C.-H. Species Boundaries and Molecular Markers for the Classification of 16SrI Phytoplasmas Inferred by Genome Analysis. Front. Microbiol. 2020, 11, 1531. [Google Scholar] [CrossRef]
- Maejima, K.; Iwai, R.; Himeno, M.; Komatsu, K.; Kitazawa, Y.; Fujita, N.; Ishikawa, K.; Fukuoka, M.; Minato, N.; Yamaji, Y.; et al. Recognition of Floral Homeotic MADS Domain Transcription Factors by a Phytoplasmal Effector, Phyllogen, Induces Phyllody. Plant J. 2014, 78, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Li, Y.; Chen, W.; Yang, H.Z.; Zhang, P.H.; Wu, Y.F. Identification of Wheat Blue Dwarf Phytoplasma Effectors Targeting Plant Proliferation and Defence Responses. Plant Pathol. 2017, 67, 603–609. [Google Scholar] [CrossRef]
- Lipsitch, M.; Siller, S.; Nowak, M.A. The evolution of virulence in pathogens with vertical and horizontal transmission. Evolution 1996, 50, 1729–1741. [Google Scholar] [CrossRef]
- Russell, S.L. Transmission Mode Is Associated with Environment Type and Taxa across Bacteria-Eukaryote Symbioses: A Systematic Review and Meta-Analysis. FEMS Microbiol. Lett. 2019, 366, fnz013. [Google Scholar] [CrossRef]
- Kumar, M.; Michael, S.; Alvarado-Valverde, J.; Mészáros, B.; Sámano-Sánchez, H.; Zeke, A.; Dobson, L.; Lazar, T.; Örd, M.; Nagpal, A.; et al. The Eukaryotic Linear Motif Resource: 2022 Release. Nucleic Acids Res. 2022, 50, D497–D508. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating Signal Peptides from Transmembrane Regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete genomes11Edited by F. Cohen. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Nguyen Ba, A.N.; Pogoutse, A.; Provart, N.; Moses, A.M. NLStradamus: A Simple Hidden Markov Model for Nuclear Localization Signal Prediction. BMC Bioinform. 2009, 10, 202. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Carbon, S.; Douglass, E.; Good, B.M.; Unni, D.R.; Harris, N.L.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; Hartline, E.; et al. The Gene Ontology Resource: Enriching a GOld Mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A Web Tool for Plotting GO Annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro Protein Families and Domains Database: 20 Years On. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Kumar, M.; Gouw, M.; Michael, S.; Sámano-Sánchez, H.; Pancsa, R.; Glavina, J.; Diakogianni, A.; Valverde, J.A.; Bukirova, D.; Čalyševa, J.; et al. ELM—The Eukaryotic Linear Motif Resource in 2020. Nucleic Acids Res. 2020, 48, D296–D306. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Wei, W.; Wu, Y.; Zhou, Y.; Peng, F.; Zhang, S.; Chen, P.; Xu, X. BcCFEM1, a CFEM Domain-Containing Protein with Putative GPI-Anchored Site, Is Involved in Pathogenicity, Conidial Production, and Stress Tolerance in Botrytis Cinerea. Front. Microbiol. 2017, 8, 1807. [Google Scholar] [CrossRef] [PubMed]
- Wood, K.J.; Nur, M.; Gil, J.; Fletcher, K.; Lakeman, K.; Gann, D.; Gothberg, A.; Khuu, T.; Kopetzky, J.; Naqvi, S.; et al. Effector Prediction and Characterization in the Oomycete Pathogen Bremia Lactucae Reveal Host-Recognized WY Domain Proteins That Lack the Canonical RXLR Motif. PLoS Pathog. 2020, 16, e1009012. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Zheng, X.; Wang, X.; Yang, J.; Zheng, X.; Zeng, Q.; Li, J.; Zhuge, Q.; Xiong, Q. Systematic Identification and Functional Characterization of the CFEM Proteins in Poplar Fungus Marssonina Brunnea. Front. Cell. Infect. Microbiol. 2022, 12, 1045615. [Google Scholar] [CrossRef] [PubMed]
- Van Roey, K.; Uyar, B.; Weatheritt, R.J.; Dinkel, H.; Seiler, M.; Budd, A.; Gibson, T.J.; Davey, N.E. Short Linear Motifs: Ubiquitous and Functionally Diverse Protein Interaction Modules Directing Cell Regulation. Chem. Rev. 2014, 114, 6733–6778. [Google Scholar] [CrossRef] [PubMed]
- Davey, N.E.; Travé, G.; Gibson, T.J. How Viruses Hijack Cell Regulation. Trends Biochem. Sci. 2011, 36, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Sámano-Sánchez, H.; Gibson, T.J. Mimicry of Short Linear Motifs by Bacterial Pathogens: A Drugging Opportunity. Trends Biochem. Sci. 2020, 45, 526–544. [Google Scholar] [CrossRef] [PubMed]
- Wadie, B.; Kleshchevnikov, V.; Sandaltzopoulou, E.; Benz, C.; Petsalaki, E. Use of Viral Motif Mimicry Improves the Proteome-Wide Discovery of Human Linear Motifs. Cell Rep. 2022, 39, 110764. [Google Scholar] [CrossRef]
- Ponting, C.P. Evidence for PDZ Domains in Bacteria, Yeast, and Plants: Novel PDZ Domains. Protein Sci. 1997, 6, 464–468. [Google Scholar] [CrossRef]
- Fanning, A.S.; Anderson, J.M. PDZ Domains: Fundamental Building Blocks in the Organization of Protein Complexes at the Plasma Membrane. J. Clin. Investig. 1999, 103, 767–772. [Google Scholar] [CrossRef]
- Christensen, N.R.; Čalyševa, J.; Fernandes, E.F.A.; Lüchow, S.; Clemmensen, L.S.; Haugaard-Kedström, L.M.; Strømgaard, K. PDZ Domains as Drug Targets. Adv. Ther. 2019, 2, 1800143. [Google Scholar] [CrossRef] [PubMed]
- Bai, B.; Zhang, G.; Pei, B.; Song, Q.; Hao, X.; Zhao, L.; Wu, Y. The Function of the Phytoplasma Effector SWP12 Depends on the Properties of Two Key Amino Acids. J. Biol. Chem. 2023, 299, 103052. [Google Scholar] [CrossRef] [PubMed]
- Strohmayer, A.; Schwarz, T.; Braun, M.; Krczal, G.; Boonrod, K. The Effect of the Anticipated Nuclear Localization Sequence of ‘Candidatus Phytoplasma Mali’ SAP11-like Protein on Localization of the Protein and Destabilization of TCP Transcription Factor. Microorganisms 2021, 9, 1756. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.-C.; Skehan, B.M.; Campellone, K.G.; Leong, J.M.; Rosen, M.K. Structural Mechanism of WASP Activation by the Enterohaemorrhagic E. Coli Effector EspFU. Nature 2008, 454, 1009–1013. [Google Scholar] [CrossRef]
- Mészáros, B.; Sámano-Sánchez, H.; Alvarado-Valverde, J.; Čalyševa, J.; Martínez-Pérez, E.; Alves, R.; Shields, D.C.; Kumar, M.; Rippmann, F.; Chemes, L.B.; et al. Short Linear Motif Candidates in the Cell Entry System Used by SARS-CoV-2 and Their Potential Therapeutic Implications. Sci. Signal. 2021, 14, eabd0334. [Google Scholar] [CrossRef] [PubMed]
- Choe, J.E.; Welch, M.D. Actin-Based Motility of Bacterial Pathogens: Mechanistic Diversity and Its Impact on Virulence. Pathog. Dis. 2016, 74, ftw099. [Google Scholar] [CrossRef] [PubMed]
- Guiney, D.G.; Lesnick, M. Targeting of the Actin Cytoskeleton during Infection by Salmonella Strains. Clin. Immunol. 2005, 114, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.; Shaw, B.D. The Role of Actin, Fimbrin and Endocytosis in Growth of Hyphae in Aspergillus Nidulans. Mol. Microbiol. 2008, 68, 690–705. [Google Scholar] [CrossRef]
- Wang, J.; Lian, N.; Zhang, Y.; Man, Y.; Chen, L.; Yang, H.; Lin, J.; Jing, Y. The Cytoskeleton in Plant Immunity: Dynamics, Regulation, and Function. Int. J. Mol. Sci. 2022, 23, 15553. [Google Scholar] [CrossRef]
- Alnomasy, S.F. Virus-Receptor Interactions of SARS-CoV-2 Spikereceptor-Binding Domain and Human Neuropilin-1 B1 Domain. Saudi J. Biol. Sci. 2021, 28, 3926–3928. [Google Scholar] [CrossRef]
- Sampietro, D.; Sámano-Sánchez, H.; Davey, N.E.; Sharan, M.; Mészáros, B.; Gibson, T.J.; Kumar, M. Conserved SQ and QS Motifs in Bacterial Effectors Suggest Pathogen Interplay with the ATM Kinase Family during Infection. bioRxiv 2018, 364117. [Google Scholar] [CrossRef]
- Örd, M.; Venta, R.; Möll, K.; Valk, E.; Loog, M. Cyclin-Specific Docking Mechanisms Reveal the Complexity of M-CDK Function in the Cell Cycle. Mol. Cell 2019, 75, 76–89.e3. [Google Scholar] [CrossRef]
- Weerasekera, D.; Stengel, F.; Sticht, H.; De Mattos Guaraldi, A.L.; Burkovski, A.; Azevedo Antunes, C. The C-Terminal Coiled-Coil Domain of Corynebacterium Diphtheriae DIP0733 Is Crucial for Interaction with Epithelial Cells and Pathogenicity in Invertebrate Animal Model Systems. BMC Microbiol. 2018, 18, 106. [Google Scholar] [CrossRef] [PubMed]
- Bar Barroeta, A.; Marquart, J.A.; Bakhtiari, K.; Meijer, A.B.; Urbanus, R.T.; Meijers, J.C.M. Nanobodies against Factor XI Apple 3 Domain Inhibit Binding of Factor IX and Reveal a Novel Binding Site for High Molecular Weight Kininogen. J. Thromb. Haemost. 2022, 20, 2538–2549. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Barroeta, A.B.; Wong, S.S.; Kim, H.J.; Pathak, M.; Dreveny, I.; Meijers, J.C.M.; Emsley, J. Structures of Factor XI and Prekallikrein Bound to Domain 6 of High–Molecular Weight Kininogen Reveal Alternate Domain 6 Conformations and Exosites. J. Thromb. Haemost. 2023, 21, 2378–2389. [Google Scholar] [CrossRef]
- Balakireva, A.; Zamyatnin, A. Indispensable Role of Proteases in Plant Innate Immunity. Int. J. Mol. Sci. 2018, 19, 629. [Google Scholar] [CrossRef]
- Ekchaweng, K.; Evangelisti, E.; Schornack, S.; Tian, M.; Churngchow, N. The Plant Defense and Pathogen Counterdefense Mediated by Hevea Brasiliensis Serine Protease HbSPA and Phytophthora Palmivora Extracellular Protease Inhibitor PpEPI10. PLoS ONE 2017, 12, e0175795. [Google Scholar] [CrossRef]
- Bender, K.W.; Zipfel, C. Paradigms of Receptor Kinase Signaling in Plants. Biochem. J. 2023, 480, 835–854. [Google Scholar] [CrossRef]
- Cock, J.M.; Vanoosthuyse, V.; Gaude, T. Receptor Kinase Signalling in Plants and Animals: Distinct Molecular Systems with Mechanistic Similarities. Curr. Opin. Cell Biol. 2002, 14, 230–236. [Google Scholar] [CrossRef]
- Luan, S. Tyrosine Phosphorylation in Plant Cell Signaling. Proc. Natl. Acad. Sci. USA 2002, 99, 11567–11569. [Google Scholar] [CrossRef]
- Shiu, S.-H.; Bleecker, A.B. Receptor-like Kinases from Arabidopsis Form a Monophyletic Gene Family Related to Animal Receptor Kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 10763–10768. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; Manimekalai, R. Phytoplasma Diseases of Plants: Molecular Diagnostics and Way Forward. World J. Microbiol. Biotechnol. 2021, 37, 102. [Google Scholar] [CrossRef] [PubMed]
- Bertaccini, A. Containment of Phytoplasma-Associated Plant Diseases by Antibiotics and Other Antimicrobial Molecules. Antibiotics 2021, 10, 1398. [Google Scholar] [CrossRef] [PubMed]
- Domazakis, E.; Lin, X.; Aguilera-Galvez, C.; Wouters, D.; Bijsterbosch, G.; Wolters, P.J.; Vleeshouwers, V.G.A.A. Effectoromics-Based Identification of Cell Surface Receptors in Potato. In Plant Pattern Recognition Receptors; Shan, L., He, P., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; Volume 1578, pp. 337–353. ISBN 978-1-4939-6858-9. [Google Scholar]
- Vleeshouwers, V.G.A.A.; Rietman, H.; Krenek, P.; Champouret, N.; Young, C.; Oh, S.-K.; Wang, M.; Bouwmeester, K.; Vosman, B.; Visser, R.G.F.; et al. Effector Genomics Accelerates Discovery and Functional Profiling of Potato Disease Resistance and Phytophthora Infestans Avirulence Genes. PLoS ONE 2008, 3, e2875. [Google Scholar] [CrossRef] [PubMed]
- Cesari, S.; Xi, Y.; Declerck, N.; Chalvon, V.; Mammri, L.; Pugnière, M.; Henriquet, C.; De Guillen, K.; Chochois, V.; Padilla, A.; et al. New Recognition Specificity in a Plant Immune Receptor by Molecular Engineering of Its Integrated Domain. Nat. Commun. 2022, 13, 1524. [Google Scholar] [CrossRef]
- Maidment, J.H.; Shimizu, M.; Bentham, A.R.; Vera, S.; Franceschetti, M.; Longya, A.; Stevenson, C.E.; De La Concepcion, J.C.; Białas, A.; Kamoun, S.; et al. Effector Target-Guided Engineering of an Integrated Domain Expands the Disease Resistance Profile of a Rice NLR Immune Receptor. eLife 2023, 12, e81123. [Google Scholar] [CrossRef]
- Carreón-Anguiano, K.G.; Todd, J.N.A.; Chi-Manzanero, B.H.; Couoh-Dzul, O.J.; Islas-Flores, I.; Canto-Canché, B. WideEffHunter: An Algorithm to Predict Canonical and Non-Canonical Effectors in Fungi and Oomycetes. Int. J. Mol. Sci. 2022, 23, 13567. [Google Scholar] [CrossRef]
- Rashidi, M.; Galetto, L.; Bosco, D.; Bulgarelli, A.; Vallino, M.; Veratti, F.; Marzachì, C. Role of the Major Antigenic Membrane Protein in Phytoplasma Transmission by Two Insect Vector Species. BMC Microbiol. 2015, 15, 193. [Google Scholar] [CrossRef]
- Mak, H.; Thurston, T.L.M. Interesting Biochemistries in the Structure and Function of Bacterial Effectors. Front. Cell. Infect. Microbiol. 2021, 11, 608860. [Google Scholar] [CrossRef]
- Rozano, L.; Jones, D.A.B.; Hane, J.K.; Mancera, R.L. Template-Based Modelling of the Structure of Fungal Effector Proteins. Mol. Biotechnol. 2023, 1–30. [Google Scholar] [CrossRef]
- Jomantiene, R.; Zhao, Y.; Davis, R.E. Sequence-Variable Mosaics: Composites of Recurrent Transposition Characterizing the Genomes of Phylogenetically Diverse Phytoplasmas. DNA Cell Biol. 2007, 26, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Davis, R.E.; Jomantiene, R.; Zhao, Y. Ancient, Recurrent Phage Attacks and Recombination Shaped Dynamic Sequence-Variable Mosaics at the Root of Phytoplasma Genome Evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 11827–11832. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; McLellan, H.; Boevink, P.C.; Birch, P.R.J. All Roads Lead to Susceptibility: The Many Modes of Action of Fungal and Oomycete Intracellular Effectors. Plant Commun. 2020, 1, 100050. [Google Scholar] [CrossRef]
- Lorrain, C.; Hecker, A.; Duplessis, S. Effector-Mining in the Poplar Rust Fungus Melampsora Larici-Populina Secretome. Front. Plant Sci. 2015, 6, 1051. [Google Scholar] [CrossRef] [PubMed]
- Saunders, D.G.O.; Win, J.; Cano, L.M.; Szabo, L.J.; Kamoun, S.; Raffaele, S. Using Hierarchical Clustering of Secreted Protein Families to Classify and Rank Candidate Effectors of Rust Fungi. PLoS ONE 2012, 7, e29847. [Google Scholar] [CrossRef]
- Espadaler, J.; Querol, E.; Aviles, F.X.; Oliva, B. Identification of Function-Associated Loop Motifs and Application to Protein Function Prediction. Bioinformatics 2006, 22, 2237–2243. [Google Scholar] [CrossRef]
- Polacco, B.J.; Babbitt, P.C. Automated Discovery of 3D Motifs for Protein Function Annotation. Bioinformatics 2006, 22, 723–730. [Google Scholar] [CrossRef]
- Fouché, S.; Plissonneau, C.; Croll, D. The Birth and Death of Effectors in Rapidly Evolving Filamentous Pathogen Genomes. Curr. Opin. Microbiol. 2018, 46, 34–42. [Google Scholar] [CrossRef]
- Latorre, S.M.; Reyes-Avila, C.S.; Malmgren, A.; Win, J.; Kamoun, S.; Burbano, H.A. Differential Loss of Effector Genes in Three Recently Expanded Pandemic Clonal Lineages of the Rice Blast Fungus. BMC Biol. 2020, 18, 88. [Google Scholar] [CrossRef]
- Huang, J.; Gogarten, J. Ancient Horizontal Gene Transfer Can Benefit Phylogenetic Reconstruction. Trends Genet. 2006, 22, 361–366. [Google Scholar] [CrossRef]
- Omelchenko, M.V.; Makarova, K.S.; Wolf, Y.I.; Rogozin, I.B.; Koonin, E.V. Evolution of Mosaic Operons by Horizontal Gene Transfer and Gene Displacement in Situ. Genome Biology. Genome Biol. 2003, 4, R55. [Google Scholar] [CrossRef]
- Schweizer, G.; Münch, K.; Mannhaupt, G.; Schirawski, J.; Kahmann, R.; Dutheil, J.Y. Positively Selected Effector Genes and Their Contribution to Virulence in the Smut Fungus Sporisorium Reilianum. Genome Biol. Evol. 2018, 10, 629–645. [Google Scholar] [CrossRef]
- Tiwari, P.; Bae, H. Horizontal Gene Transfer and Endophytes: An Implication for the Acquisition of Novel Traits. Plants 2020, 9, 305. [Google Scholar] [CrossRef]
- Dillon, M.M.; Almeida, R.N.D.; Laflamme, B.; Martel, A.; Weir, B.S.; Desveaux, D.; Guttman, D.S. Molecular Evolution of Pseudomonas Syringae Type III Secreted Effector Proteins. Front. Plant Sci. 2019, 10, 418. [Google Scholar] [CrossRef]
- Morgado, S.; Vicente, A.C. Diversity and Distribution of Type VI Secretion System Gene Clusters in Bacterial Plasmids. Sci. Rep. 2022, 12, 8249. [Google Scholar] [CrossRef] [PubMed]
- Elkhaligy, H.; Balbin, C.A.; Siltberg-Liberles, J. Comparative Analysis of Structural Features in SLiMs from Eukaryotes, Bacteria, and Viruses with Importance for Host-Pathogen Interactions. Pathogens 2022, 11, 583. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.; Singhal, N.; Kumar, M. Investigating the Eukaryotic Host-like SLiMs in Microbial Mimitopes and Their Potential as Novel Drug Targets for Treating Autoimmune Diseases. Front. Microbiol. 2022, 13, 1039188. [Google Scholar] [CrossRef] [PubMed]
- Tayal, S.; Bhatia, V.; Mehrotra, T.; Bhatnagar, S. ImitateDB: A Database for Domain and Motif Mimicry Incorporating Host and Pathogen Protein Interactions. Amino Acids 2022, 54, 923–934. [Google Scholar] [CrossRef] [PubMed]
- Stergiopoulos, I.; Van Den Burg, H.A.; Ökmen, B.; Beenen, H.G.; Van Liere, S.; Kema, G.H.J.; De Wit, P.J.G.M. Tomato Cf Resistance Proteins Mediate Recognition of Cognate Homologous Effectors from Fungi Pathogenic on Dicots and Monocots. Proc. Natl. Acad. Sci. USA 2010, 107, 7610–7615. [Google Scholar] [CrossRef] [PubMed]
- Figaj, D.; Ambroziak, P.; Przepiora, T.; Skorko-Glonek, J. The Role of Proteases in the Virulence of Plant Pathogenic Bacteria. Int. J. Mol. Sci. 2019, 20, 672. [Google Scholar] [CrossRef]
- Pearson, J.S.; Giogha, C.; Mühlen, S.; Nachbur, U.; Pham, C.L.L.; Zhang, Y.; Hildebrand, J.M.; Oates, C.V.; Lung, T.W.F.; Ingle, D.; et al. EspL Is a Bacterial Cysteine Protease Effector That Cleaves RHIM Proteins to Block Necroptosis and Inflammation. Nat. Microbiol. 2017, 2, 16258. [Google Scholar] [CrossRef]
- Thomas, E.; Van Der Hoorn, R. Ten Prominent Host Proteases in Plant-Pathogen Interactions. Int. J. Mol. Sci. 2018, 19, 639. [Google Scholar] [CrossRef]
- Zhang, S.; Li, C.; Si, J.; Han, Z.; Chen, D. Action Mechanisms of Effectors in Plant-Pathogen Interaction. Int. J. Mol. Sci. 2022, 23, 6758. [Google Scholar] [CrossRef] [PubMed]
- Mukhtar, M.S.; McCormack, M.E.; Argueso, C.T.; Pajerowska-Mukhtar, K.M. Pathogen Tactics to Manipulate Plant Cell Death. Curr. Biol. 2016, 26, R608–R619. [Google Scholar] [CrossRef]
- Alto, N.M.; Orth, K. Subversion of Cell Signaling by Pathogens. Cold Spring Harb. Perspect. Biol. 2012, 4, a006114. [Google Scholar] [CrossRef]
- Davey, N.E.; Simonetti, L.; Ivarsson, Y. The next Wave of Interactomics: Mapping the SLiM-Based Interactions of the Intrinsically Disordered Proteome. Curr. Opin. Struct. Biol. 2023, 80, 102593. [Google Scholar] [CrossRef] [PubMed]
- Goswami, S.; Samanta, D.; Duraivelan, K. Molecular Mimicry of Host Short Linear Motif-Mediated Interactions Utilised by Viruses for Entry. Mol. Biol. Rep. 2023, 50, 4665–4673. [Google Scholar] [CrossRef]
- Irwin, N.A.T.; Pittis, A.A.; Richards, T.A.; Keeling, P.J. Systematic Evaluation of Horizontal Gene Transfer between Eukaryotes and Viruses. Nat. Microbiol. 2021, 7, 327–336. [Google Scholar] [CrossRef]
- Martins, Y.C.; Jurberg, A.D.; Daniel-Ribeiro, C.T. Visiting Molecular Mimicry Once More: Pathogenicity, Virulence, and Autoimmunity. Microorganisms 2023, 11, 1472. [Google Scholar] [CrossRef]
- Mondino, S.; Schmidt, S.; Buchrieser, C. Molecular Mimicry: A Paradigm of Host-Microbe Coevolution Illustrated by Legionella. mBio 2020, 11, e01201-20. [Google Scholar] [CrossRef]
- Chang, L.; Mondal, A.; Perez, A. Towards Rational Computational Peptide Design. Front. Bioinforma. 2022, 2, 1046493. [Google Scholar] [CrossRef] [PubMed]
- Delaunay, M.; Ha-Duong, T. Computational Design of Cyclic Peptides to Inhibit Protein-Peptide Interactions. Biophys. Chem. 2023, 296, 106987. [Google Scholar] [CrossRef] [PubMed]
- Kazmirchuk, T.D.D.; Bradbury-Jost, C.; Withey, T.A.; Gessese, T.; Azad, T.; Samanfar, B.; Dehne, F.; Golshani, A. Peptides of a Feather: How Computation Is Taking Peptide Therapeutics under Its Wing. Genes 2023, 14, 1194. [Google Scholar] [CrossRef] [PubMed]
- Todd, J.N.A.; Carreón-Anguiano, K.G.; Islas-Flores, I.; Canto-Canché, B. Fungal Effectoromics: A World in Constant Evolution. Int. J. Mol. Sci. 2022, 23, 13433. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Song, J. NLR Immune Receptor RB Is Differentially Targeted by Two Homologous but Functionally Distinct Effector Proteins. Plant Commun. 2021, 2, 100236. [Google Scholar] [CrossRef]
- Li, D.; Zhu, Z.; Deng, X.; Zou, J.; Ma, C.; Li, C.; Yin, T.; Liu, C.; Wang, J.; Chen, Q.; et al. GmPBS1, a Hub Gene Interacting with Rhizobial Type-III Effectors NopT and NopP, Regulates Soybean Nodulation. Agronomy 2023, 13, 1242. [Google Scholar] [CrossRef]
- Ijaz, M.; Khan, F.; Zaki, H.E.M.; Khan, M.M.; Radwan, K.S.A.; Jiang, Y.; Qian, J.; Ahmed, T.; Shahid, M.S.; Luo, J.; et al. Recent Trends and Advancements in CRISPR-Based Tools for Enhancing Resistance against Plant Pathogens. Plants 2023, 12, 1911. [Google Scholar] [CrossRef]
- Pacheco, I.D.S.; Galdeano, D.M.; Maluta, N.K.P.; Lopes, J.R.S.; Machado, M.A. Gene Silencing of Diaphorina Citri Candidate Effectors Promotes Changes in Feeding Behaviors. Sci. Rep. 2020, 10, 5992. [Google Scholar] [CrossRef]
- Wang, H.; Shi, S.; Hua, W. Advances of Herbivore-Secreted Elicitors and Effectors in Plant-Insect Interactions. Front. Plant Sci. 2023, 14, 1176048. [Google Scholar] [CrossRef]
- Singh, A.; Lakhanpaul, S. Detection, Characterization and Evolutionary Aspects of S54LP of SP (SAP54 Like Protein of Sesame Phyllody): A Phytoplasma Effector Molecule Associated with Phyllody Development in Sesame (Sesamum indicum L.). Physiol. Mol. Biol. Plants 2020, 26, 445–458. [Google Scholar] [CrossRef]
- Filisetti, D.; Théobald-Dietrich, A.; Mahmoudi, N.; Rudinger-Thirion, J.; Candolfi, E.; Frugier, M. Aminoacylation of Plasmodium Falciparum tRNAAsn and Insights in the Synthesis of Asparagine Repeats. J. Biol. Chem. 2013, 288, 36361–36371. [Google Scholar] [CrossRef]
- Liefting, L.W.; Kirkpatrick, B.C. Cosmid Cloning and Sample Sequencing of the Genome of the Uncultivable Mollicute, Western X-Disease Phytoplasma, Using DNA Purified by Pulsed-Field Gel Electrophoresis. FEMS Microbiol. Lett. 2003, 221, 203–211. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Muralidharan, V.; Goldberg, D.E. Asparagine Repeats in Plasmodium Falciparum Proteins: Good for Nothing? PLoS Pathog. 2013, 9, e1003488. [Google Scholar] [CrossRef] [PubMed]
- Rajapandi, T. Chaperoning of Asparagine Repeat-Containing Proteins in Plasmodium Falciparum. J. Parasit. Dis. 2020, 44, 687–693. [Google Scholar] [CrossRef]
- Moles, E.; Valle-Delgado, J.J.; Urbán, P.; Azcárate, I.G.; Bautista, J.M.; Selva, J.; Egea, G.; Ventura, S.; Fernàndez-Busquets, X. Possible Roles of Amyloids in Malaria Pathophysiology. Future Sci. OA 2015, 1, fso.15.43. [Google Scholar] [CrossRef] [PubMed]
- Pallarès, I.; De Groot, N.S.; Iglesias, V.; Sant’Anna, R.; Biosca, A.; Fernàndez-Busquets, X.; Ventura, S. Discovering Putative Prion-Like Proteins in Plasmodium Falciparum: A Computational and Experimental Analysis. Front. Microbiol. 2018, 9, 1737. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.Q. Prion-like Protein Spotted in Bacteria for the First Time. Nature 2017. [Google Scholar] [CrossRef]
- Harrison, P.M. Evolutionary Behaviour of Bacterial Prion-like Proteins. PLoS ONE 2019, 14, e0213030. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L.; Chung, W.-C.; Lin, C.-P.; Kuo, C.-H. Comparative Analysis of Gene Content Evolution in Phytoplasmas and Mycoplasmas. PLoS ONE 2012, 7, e34407. [Google Scholar] [CrossRef]
- Jheeta, S.; Chatzitheodoridis, E.; Devine, K.; Block, J. The Way Forward for the Origin of Life: Prions and Prion-Like Molecules First Hypothesis. Life 2021, 11, 872. [Google Scholar] [CrossRef]
- Navarro, S.; Marinelli, P.; Diaz-Caballero, M.; Ventura, S. The Prion-like RNA-Processing Protein HNRPDL Forms Inherently Toxic Amyloid-like Inclusion Bodies in Bacteria. Microb. Cell Factories 2015, 14, 102. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tetz, G.; Tetz, V. Prion-Like Domains in Phagobiota. Front. Microbiol. 2017, 8, 2239. [Google Scholar] [CrossRef] [PubMed]
- Tetz, G.; Tetz, V. Prion-like Domains in Spike Protein of SARS-CoV-2 Differ across Its Variants and Enable Changes in Affinity to ACE2. Microorganisms 2022, 10, 280. [Google Scholar] [CrossRef] [PubMed]
- Shahnawaz, M.; Park, K.-W.; Mukherjee, A.; Diaz-Espinoza, R.; Soto, C. Prion-like Characteristics of the Bacterial Protein Microcin E492. Sci. Rep. 2017, 7, 45720. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.M.; Nofal, S.D.; McLaughlin, E.J.; Osborne, A.R. Repetitive Sequences in Malaria Parasite Proteins. FEMS Microbiol. Rev. 2017, 41, 923–940. [Google Scholar] [CrossRef]
- Iwabuchi, N.; Kitazawa, Y.; Maejima, K.; Koinuma, H.; Miyazaki, A.; Matsumoto, O.; Suzuki, T.; Nijo, T.; Oshima, K.; Namba, S.; et al. Functional Variation in Phyllogen, a Phyllody-inducing Phytoplasma Effector Family, Attributable to a Single Amino Acid Polymorphism. Mol. Plant Pathol. 2020, 21, 1322–1336. [Google Scholar] [CrossRef]
- Li, J.; Gao, M.; Gabriel, D.W.; Liang, W.; Song, L. Secretome-Wide Analysis of Lysine Acetylation in Fusarium Oxysporum f. Sp. Lycopersici Provides Novel Insights Into Infection-Related Proteins. Front. Microbiol. 2020, 11, 559440. [Google Scholar] [CrossRef]
- Narita, T.; Weinert, B.T.; Choudhary, C. Functions and Mechanisms of Non-Histone Protein Acetylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 156–174. [Google Scholar] [CrossRef]
- Jones, J.D.; O’Connor, C.D. Protein Acetylation in Prokaryotes. Proteomics 2011, 11, 3012–3022. [Google Scholar] [CrossRef]
- Aurin, M.-B.; Haupt, M.; Görlach, M.; Rümpler, F.; Theißen, G. Structural Requirements of the Phytoplasma Effector Protein SAP54 for Causing Homeotic Transformation of Floral Organs. Mol. Plant-Microbe Interact. 2020, 33, 1129–1141. [Google Scholar] [CrossRef]
- Bauer, S.; Yu, D.; Lawson, A.W.; Saur, I.M.L.; Frantzeskakis, L.; Kracher, B.; Logemann, E.; Chai, J.; Maekawa, T.; Schulze-Lefert, P. The Leucine-Rich Repeats in Allelic Barley MLA Immune Receptors Define Specificity towards Sequence-Unrelated Powdery Mildew Avirulence Effectors with a Predicted Common RNase-like Fold. PLoS Pathog. 2021, 17, e1009223. [Google Scholar] [CrossRef] [PubMed]
- Qi, D.; DeYoung, B.J.; Innes, R.W. Structure-Function Analysis of the Coiled-Coil and Leucine-Rich Repeat Domains of the RPS5 Disease Resistance Protein. Plant Physiol. 2012, 158, 1819–1832. [Google Scholar] [CrossRef] [PubMed]
- Bernal-Bayard, J.; Cardenal-Muñoz, E.; Ramos-Morales, F. The Salmonella Type III Secretion Effector, Salmonella Leucine-Rich Repeat Protein (SlrP), Targets the Human Chaperone ERdj3. J. Biol. Chem. 2010, 285, 16360–16368. [Google Scholar] [CrossRef] [PubMed]
- Pillay, T.D.; Hettiarachchi, S.U.; Gan, J.; Diaz-Del-Olmo, I.; Yu, X.-J.; Muench, J.H.; Thurston, T.L.M.; Pearson, J.S. Speaking the Host Language: How Salmonella Effector Proteins Manipulate the Host: This Article Is Part of the Bacterial Cell Envelopes Collection. Microbiology 2023, 169, 001342. [Google Scholar] [CrossRef] [PubMed]
- Miao, E.A.; Scherer, C.A.; Tsolis, R.M.; Kingsley, R.A.; Adams, L.G.; Baumler, A.J.; Miller, S.I. Salmonella Typhimurium Leucine-Rich Repeat Proteins Are Targeted to the SPI1 and SPI2 Type III Secretion Systems. Mol. Microbiol. 1999, 34, 850–864. [Google Scholar] [CrossRef]
- Xu, R.-Q.; Blanvillain, S.; Feng, J.-X.; Jiang, B.-L.; Li, X.-Z.; Wei, H.-Y.; Kroj, T.; Lauber, E.; Roby, D.; Chen, B.; et al. AvrAC Xcc8004, a Type III Effector with a Leucine-Rich Repeat Domain from Xanthomonas campestris Pathovar campestris Confers Avirulence in Vascular Tissues of Arabidopsis Thaliana Ecotype Col-0. J. Bacteriol. 2008, 190, 343–355. [Google Scholar] [CrossRef]
- Hu, Y.; Huang, H.; Hui, X.; Cheng, X.; White, A.P.; Zhao, Z.; Wang, Y. Distribution and Evolution of Yersinia Leucine-Rich Repeat Proteins. Infect. Immun. 2016, 84, 2243–2254. [Google Scholar] [CrossRef]
- Remigi, P.; Anisimova, M.; Guidot, A.; Genin, S.; Peeters, N. Functional Diversification of the GALA Type III Effector Family Contributes to Ralstonia Solanacearum Adaptation on Different Plant Hosts. New Phytol. 2011, 192, 976–987. [Google Scholar] [CrossRef]
- Loimaranta, V.; Hytönen, J.; Pulliainen, A.T.; Sharma, A.; Tenovuo, J.; Strömberg, N.; Finne, J. Leucine-Rich Repeats of Bacterial Surface Proteins Serve as Common Pattern Recognition Motifs of Human Scavenger Receptor Gp340. J. Biol. Chem. 2009, 284, 18614–18623. [Google Scholar] [CrossRef]
- Eshghi, A.; Gaultney, R.A.; England, P.; Brûlé, S.; Miras, I.; Sato, H.; Coburn, J.; Bellalou, J.; Moriarty, T.J.; Haouz, A.; et al. An Extracellular Leptospira interrogans Leucine-rich Repeat Protein Binds Human E- and VE-cadherins. Cell. Microbiol. 2019, 21, e12949. [Google Scholar] [CrossRef]
- Kibby, E.M.; Conte, A.N.; Burroughs, A.M.; Nagy, T.A.; Vargas, J.A.; Whalen, L.A.; Aravind, L.; Whiteley, A.T. Bacterial NLR-Related Proteins Protect against Phage. Cell 2023, 186, 2410–2424.e18. [Google Scholar] [CrossRef] [PubMed]
- Galetto, L.; Fletcher, J.; Bosco, D.; Turina, M.; Wayadande, A.; Marzachì, C. Characterization of Putative Membrane Protein Genes of the ‘Candidatus Phytoplasma Asteris’, Chrysanthemum Yellows Isolate. Can. J. Microbiol. 2008, 54, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Padilla-Ramos, R.; Salas-Muñoz, S.; Velásquez-Valle, R.; Reveles-Torres, L.R. Un Nuevo Enfoque Molecular En El Estudio de La Interacción Parásito-Hospedero. Rev. Mex. Fitopatol. Mex. J. Phytopathol. 2019, 37, 95–114. [Google Scholar] [CrossRef]
- Bhopatkar, A.A.; Uversky, V.N.; Rangachari, V. Disorder and Cysteines in Proteins: A Design for Orchestration of Conformational See-Saw and Modulatory Functions. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2020; Volume 174, pp. 331–373. ISBN 978-0-12-822615-5. [Google Scholar]
- Singh, Y.; Nair, A.M.; Verma, P.K. Surviving the Odds: From Perception to Survival of Fungal Phytopathogens under Host-Generated Oxidative Burst. Plant Commun. 2021, 2, 100142. [Google Scholar] [CrossRef]
- Zhang, Z.-H.; Jin, J.-H.; Sheng, G.-L.; Xing, Y.-P.; Liu, W.; Zhou, X.; Liu, Y.-Q.; Chen, X.-R. A Small Cysteine-Rich Phytotoxic Protein of Phytophthora capsici Functions as Both Plant Defense Elicitor and Virulence Factor. Mol. Plant-Microbe Interact. 2021, 34, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Miseta, A.; Csutora, P. Relationship Between the Occurrence of Cysteine in Proteins and the Complexity of Organisms. Mol. Biol. Evol. 2000, 17, 1232–1239. [Google Scholar] [CrossRef]
- Gérard, C.; Carrière, F.; Receveur-Bréchot, V.; Launay, H.; Gontero, B. A Trajectory of Discovery: Metabolic Regulation by the Conditionally Disordered Chloroplast Protein, CP12. Biomolecules 2022, 12, 1047. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).