Selection and Validation of Reference Genes for qRT-PCR Analysis of Gene Expression in Tropaeolum majus (Nasturtium)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material, Candidate Reference Gene Isolation, and Primer Design
2.2. Total RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR Analysis
2.3. Statistical Analysis of Candidate Reference Genes
2.4. Validation of Selected Reference Genes
3. Results
3.1. Selection of Candidate Reference Genes and PCR Amplification
3.2. Expression Profiles of Candidate Reference Genes
3.3. Expression Stability of Candidate Reference Genes
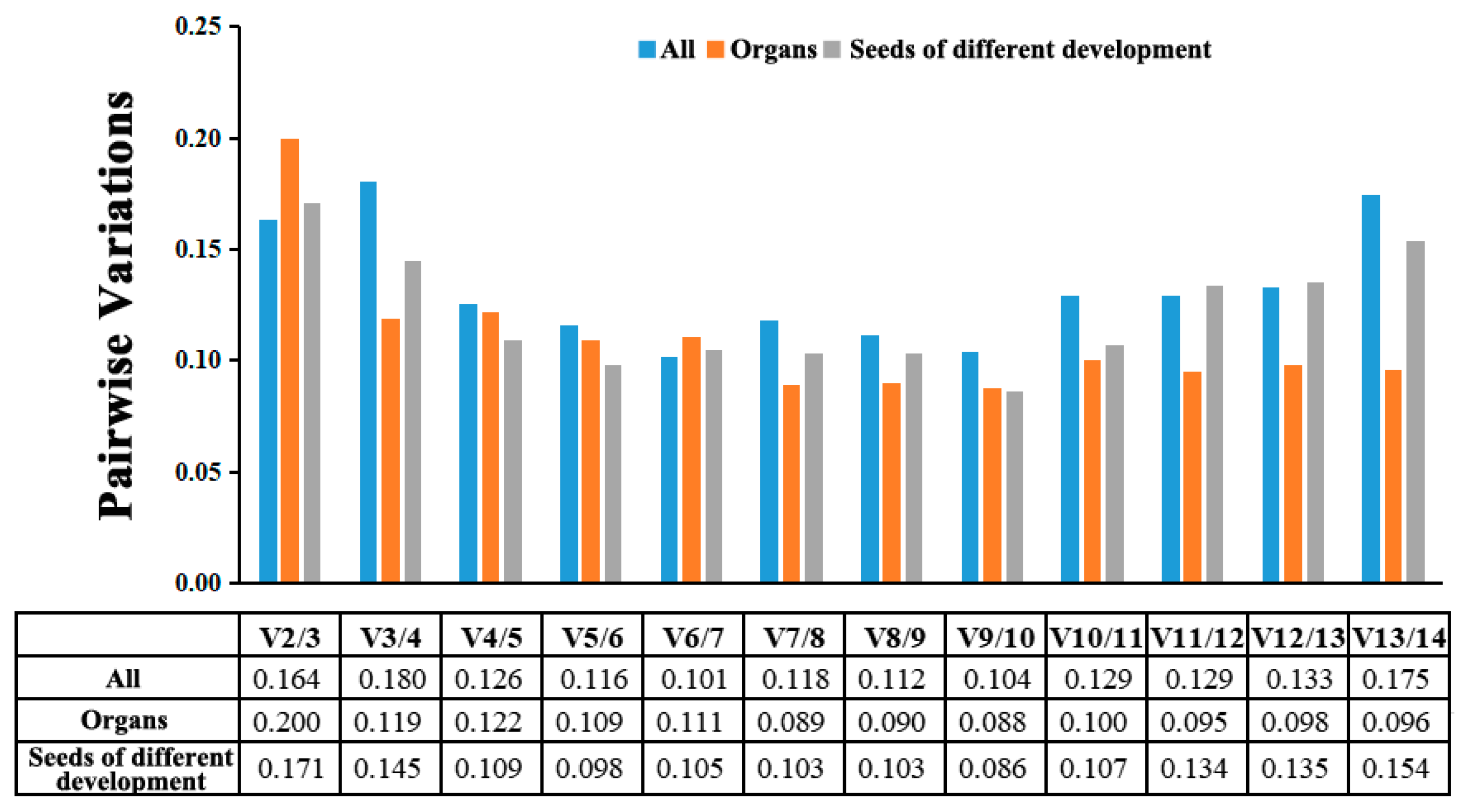
3.4. Validation of the Selected Reference Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bustin, S. Quantification of Mrna Using Real-Time Reverse Transcription PCR (RT-PCR): Trends and Problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef]
- Aleksandar, R.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to Reference Gene Selection for Quantitative Real-Time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar]
- Camila, C.; Scheible, W.-R.; Mueller-Roeber, B.; Ruzicic, S. A Quantitative RT-PCR Platform for High-Throughput Expression Profiling of 2500 Rice Transcription Factors. Plant Methods 2007, 3, 7. [Google Scholar]
- Vanguilder, H.D.; Vrana, K.E.; Freeman, W.M. Twenty-Five Years of Quantitative PCR for Gene Expression Analysis. Biotechniques 2008, 44, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Claire, G.; Mingam, A.; Charrier, B. Real-Time PCR: What Relevance to Plant Studies? J. Exp. Bot. 2004, 55, 1445–1454. [Google Scholar]
- Ma, R.; Sheng, X.; Yucheng, Z.; Bing, X.; Ren, W. Selection and Validation of Appropriate Reference Genes for Quantitative Real-Time PCR Analysis of Gene Expression in Lycoris aurea. Front. Plant Sci. 2016, 7, 536. [Google Scholar] [CrossRef]
- Liu, Y.P.; Zhang, Y.; Liu, F.; Liu, T.; Chen, J.Y.; Fu, G.; Zheng, C.Y.; Su, D.D.; Wang, Y.N.; Zhou, H.K.; et al. Establishment of Reference (Housekeeping) Genes via Quantitative Real-Time PCR for Investigation of the Genomic Basis of Abiotic Stress Resistance in Psammochloa villosa (Poaceae). J. Plant Physiol. 2022, 268, 153575. [Google Scholar] [CrossRef]
- Huggett, J.K.; Bustin, D.S.; Zumla, A. Real-Time RT-PCR Normalization; Strategies and Considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA Using Real-Time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef]
- Kou, X.; Zhang, L.; Yang, S.; Li, G.; Ye, J. Selection and Validation of Reference Genes for Quantitative RT-PCR Analysis in Peach Fruit under Different Experimental Conditions. Sci. Hortic. 2017, 225, 195–203. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Nolan, T.; Pfaffl, M.W. Quantitative Real-Time RT-PCR—A Perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Thellin, O.; ElMoualij, B.; Heinen, E.; Zorzi, W. A Decade of Improvements in Quantification of Gene Expression and Internal Standard Selection. Biotechnol. Adv. 2009, 27, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Thellin, O.; Zorzi, W.; Lakaye, B.; De Borman, B.; Coumans, B.; Hennen, G.; Grisar, T.; Igout, A.; Heinen, E. Housekeeping Genes as Internal Standards: Use and Limits. J. Biotechnol. 1999, 75, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Kozera, B.; Rapacz, M. Reference Genes in Real-Time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef]
- Monteiro, F.; Sebastiana, M.; Pais, M.S.; Figueiredo, A. Reference Gene Selection and Validation for the Early Responses to Downy Mildew Infection in Susceptible and Resistant Vitis vinifera Cultivars. PLoS ONE 2013, 8, e72998. [Google Scholar] [CrossRef]
- Kiarash, J.G.; Wilde, H.D.; Amirmahani, F.; Moemeni, M.M.; Zaboli, M.; Nazari, M.; Moosavi, S.S.; Jamalvandi, M. Selection and Validation of Reference Genes for Normalization of QRT-PCR Gene Expression in Wheat (Triticum durum L.) under Drought and Salt Stresses. J. Genet. 2018, 97, 1433–1444. [Google Scholar] [CrossRef]
- Chapman, J.R.; Waldenström, J. With Reference to Reference Genes: A Systematic Review of Endogenous Controls in Gene Expression Studies. PLoS ONE 2015, 10, e0141853. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, Research0034. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The Miqe Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Hu, R.; Fan, C.; Li, H.; Zhang, Q.; Fu, Y.F. Evaluation of Putative Reference Genes for Gene Expression Normalization in Soybean by Quantitative Real-Time RT-PCR. BMC Mol. Biol. 2009, 10, 93. [Google Scholar] [CrossRef]
- Zhang, Y.; Han, X.; Chen, S.; Zheng, L.; He, X.; Liu, M.; Qiao, G.; Wang, Y.; Zhuo, R. Selection of Suitable Reference Genes for Quantitative Real-Time PCR Gene Expression Analysis in Salix matsudana under Different Abiotic Stresses. Sci. Rep. 2017, 7, 40290. [Google Scholar] [CrossRef]
- Suzuki, T.; Higgins, P.J.; Crawford, D.R. Control Selection for RNA Quantitation. Biotechniques 2000, 29, 332–337. [Google Scholar] [CrossRef]
- Ruan, W.; Lai, M. Actin, a Reliable Marker of Internal Control? Clin. Chim. Acta 2007, 385, 1–5. [Google Scholar] [CrossRef]
- Selvey, S.; Thompson, E.W.; Matthaei, K.; Lea, R.A.; Irving, M.G.; Griffiths, L.R. Beta-Actin--an Unsuitable Internal Control for RT-PCR. Mol. Cell. Probes 2001, 15, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Thorrez, L.; Van Deun, K.; Tranchevent, L.C.; Van Lommel, L.; Engelen, K.; Marchal, K.; Moreau, Y.; Van Mechelen, I.; Schuit, F. Using Ribosomal Protein Genes as Reference: A Tale of Caution. PLoS ONE 2008, 3, e1854. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.; Mauriat, M.; Pelloux, J.; Bellini, C.; Van Wuytswinkel, O. Towards a Systematic Validation of References in Real-Time RT-PCR. Plant Cell 2008, 20, 1734–1735. [Google Scholar] [CrossRef] [PubMed]
- Jakubczyk, K.; Janda, K.; Watychowicz, K.; Łukasiak, J.; Wolska, J. Garden Nasturtium (Tropaeolum majus L.)—A Source of Mineral Elements and Bioactive Compounds. Rocz. Panstw. Zakl. Hig. 2018, 69, 119–126. [Google Scholar] [PubMed]
- Pollard, M.R.; Stumpf, P.K. Long Chain (C(20) and C(22)) Fatty Acid Biosynthesis in Developing Seeds of Tropaeolum majus: An in Vivo Study. Plant Physiol. 1980, 66, 641–648. [Google Scholar] [CrossRef]
- Mietkiewska, E.; Giblin, E.M.; Wang, S.; Barton, D.L.; Dirpaul, J.; Brost, J.M.; Katavic, V.; Taylor, D.C. Seed-Specific Heterologous Expression of a Nasturtium Fae Gene in Arabidopsis Results in a Dramatic Increase in the Proportion of Erucic Acid. Plant Physiol. 2004, 136, 2665–2675. [Google Scholar] [CrossRef][Green Version]
- Sebastián, M.-S.; Favio, G.; Juan, F.A.; Natalia, P.-M. Molecular Framework Underlying Floral Bilateral Symmetry and Nectar Spur Development in Tropaeolum, an Atypical Member of the Brassicales. Am. J. Bot. 2021, 108, 1315–1330. [Google Scholar]
- Zhuang, H.; Fu, Y.; He, W.; Wang, L.; Wei, Y. Selection of Appropriate Reference Genes for Quantitative Real-Time PCR in Oxytropis ochrocephala Bunge Using Transcriptome Datasets under Abiotic Stress Treatments. Front. Plant Sci. 2015, 6, 475. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, L.; Zhang, Y.; Wang, G.; Song, D.; Zhang, Y. Selection and Validation of Appropriate Reference Genes for Quantitative Real-Time PCR Normalization in Staminate and Perfect Flowers of Andromonoecious Taihangia rupestris. Front. Plant Sci. 2017, 8, 729. [Google Scholar] [CrossRef]
- Demidenko, N.V.; Logacheva, M.D.; Penin, A.A. Selection and Validation of Reference Genes for Quantitative Real-Time PCR in Buckwheat (Fagopyrum esculentum) Based on Transcriptome Sequence Data. PLoS ONE 2011, 6, e19434. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De Novo Transcript Sequence Reconstruction from RNA-Seq Using the Trinity Platform for Reference Generation and Analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Ginzinger, D.G. Gene Quantification Using Real-Time Quantitative PCR: An Emerging Technology Hits the Mainstream. Exp. Hematol. 2002, 30, 503–512. [Google Scholar] [CrossRef]
- Jiang, Q.; Wang, F.; Li, M.Y.; Ma, J.; Tan, G.F.; Xiong, A.S. Selection of Suitable Reference Genes for QPCR Normalization under Abiotic Stresses in Oenanthe javanica (Bi.) Dc. PLoS ONE 2014, 9, e92262. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of Stable Housekeeping Genes, Differentially Regulated Target Genes and Sample Integrity: Bestkeeper—Excel-Based Tool Using Pair-Wise Correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. Mirdeepfinder: A miRNA Analysis Tool for Deep Sequencing of Plant Small RNAs. Plant. Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Chang, E.; Shi, S.; Liu, J.; Cheng, T.; Xue, L.; Yang, X.; Yang, W.; Lan, Q.; Jiang, Z. Selection of Reference Genes for Quantitative Gene Expression Studies in Platycladus orientalis (Cupressaceae) Using Real-Time PCR. PLoS ONE 2012, 7, e33278. [Google Scholar] [CrossRef]
- Xiao, X.; Ma, J.; Wang, J.; Wu, X.; Li, P.; Yao, Y. Validation of Suitable Reference Genes for Gene Expression Analysis in the Halophyte salicornia Europaea by Real-Time Quantitative PCR. Front. Plant Sci. 2014, 5, 788. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods A Companion Methods Enzymol. 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Qi, J.; Zhang, G.; Xu, J.; Tao, A.; Fang, P.; Su, J. Selection of Reliable Reference Genes for Quantitative Real-Time PCR Gene Expression Analysis in Jute (Corchorus capsularis) under Stress Treatments. Front. Plant Sci. 2015, 6, 848. [Google Scholar] [CrossRef]
- Qi, S.; Yang, L.; Wen, X.; Hong, Y.; Song, X.; Zhang, M.; Dai, S. Reference Gene Selection for RT-PCR Analysis of Flower Development in Chrysanthemum morifolium and Chrysanthemum lavandulifolium. Front. Plant Sci. 2016, 7, 287. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, L.; Li, W.; Han, S.; Yang, W.; Qi, L. Reference Gene Selection for Quantitative Real-Time PCR Normalization in Caragana intermedia under Different Abiotic Stress Conditions. PLoS ONE 2013, 8, e53196. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Yan, H.; Jiang, X.; Zhang, Y.; Zhang, X.; Ji, Y.; Zeng, B.; Xu, B.; Yin, G.; Lee, S.; et al. Reference Gene Selection for Quantitative Real-Time Reverse-Transcriptase PCR in Orchard grass Subjected to Various Abiotic Stresses. Gene 2014, 553, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Long, J.; Liu, S.W.; Yang, Z.W.; Zhu, Q.J.; Zhao, X.L.; Peng, C. Selection and Validation of Reference Genes for Mrna Expression by Quantitative Real-Time PCR Analysis in Neolamarckia cadamba. Sci. Rep. 2018, 8, 9311. [Google Scholar] [CrossRef]
- Galli, V.; Borowski, J.M.; Perin, E.C.; Rda, S.M.; Labonde, J.; Idos, S.P.; Silva, S.D.; Rombaldi, C.V. Validation of Reference Genes for Accurate Normalization of Gene Expression for Real Time-Quantitative PCR in Strawberry Fruits Using Different Cultivars and Osmotic Stresses. Gene 2015, 554, 205–214. [Google Scholar] [CrossRef]
- Duan, M.; Wang, J.; Zhang, X.; Yang, H.; Wang, H.; Qiu, Y.; Song, J.; Guo, Y.; Li, X. Identification of Optimal Reference Genes for Expression Analysis in Radish (Raphanus sativus L.) and Its Relatives Based on Expression Stability. Front. Plant Sci. 2017, 8, 1605. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, N.; Si, H.; Calderón-Urrea, A. Selection and Validation of Reference Genes for RT-QPCR Analysis in Potato under Abiotic Stress. Plant Methods 2017, 13, 85. [Google Scholar] [CrossRef]
- Ho-Youn, K.; Saha, P.; Farcuh, M.; Li, B.; Sadka, A.; Blumwald, E. RNA-Seq Analysis of Spatiotemporal Gene Expression Patterns During Fruit Development Revealed Reference Genes for Transcript Normalization in Plums. Plant Mol. Biol. Report. 2015, 33, 1634–1649. [Google Scholar]
- Li, J.; Han, J.; Hu, Y.; Yang, J. Selection of Reference Genes for Quantitative Real-Time PCR during Flower Development in Tree Peony (Paeonia suffruticosa Andr.). Front. Plant Sci. 2016, 7, 516. [Google Scholar] [CrossRef] [PubMed]
- Niu, K.; Shi, Y.; Ma, H. Selection of Candidate Reference Genes for Gene Expression Analysis in Kentucky Bluegrass (Poa pratensis L.) under Abiotic Stress. Front. Plant Sci. 2017, 8, 193. [Google Scholar] [CrossRef] [PubMed]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-Wide Identification and Testing of Superior Reference Genes for Transcript Normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef]
- Nicot, N.; Hausman, J.F.; Hoffmann, L.; Evers, D. Housekeeping Gene Selection for Real-Time RT-PCR Normalization in Potato during Biotic and Abiotic Stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef]
- Jian, B.; Liu, B.; Bi, Y.; Hou, W.; Wu, C.; Han, T. Validation of Internal Control for Gene Expression Study in Soybean by Quantitative Real-Time PCR. BMC Mol. Biol. 2008, 9, 59. [Google Scholar] [CrossRef]
- Die, J.V.; Román, B.; Nadal, S.; González-Verdejo, C.I. Evaluation of Candidate Reference Genes for Expression Studies in Pisum sativum under Different Experimental Conditions. Planta 2010, 232, 145–153. [Google Scholar] [CrossRef]
- Yang, Y.; Hou, S.; Cui, G.; Chen, S.; Wei, J.; Huang, L. Characterization of Reference Genes for Quantitative Real-Time PCR Analysis in Various Tissues of Salvia miltiorrhiza. Mol. Biol. Rep. 2010, 37, 507–513. [Google Scholar] [CrossRef]
- Lilly, S.T.; Drummond, R.S.; Pearson, M.N.; MacDiarmid, R.M. Identification and Validation of Reference Genes for Normalization of Transcripts from Virus-Infected Arabidopsis thaliana. Mol. Plant Microbe Interact. 2011, 24, 294–304. [Google Scholar] [CrossRef]
- Li, X.S.; Yang, H.L.; Zhang, D.Y.; Zhang, Y.M.; Wood, A.J. Reference Gene Selection in the Desert Plant Eremosparton Songoricum. Int. J. Mol. Sci. 2012, 13, 6944–6963. [Google Scholar] [CrossRef]
- Expósito-Rodríguez, M.; Borges, A.A.; Borges-Pérez, A.; Pérez, J.A. Selection of Internal Control Genes for Quantitative Real-Time RT-PCR Studies during Tomato Development Process. BMC Plant Biol. 2008, 8, 131. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Nijhawan, A.; Tyagi, A.K.; Khurana, J.P. Validation of Housekeeping Genes as Internal Control for Studying Gene Expression in Rice by Quantitative Real-Time PCR. Biochem. Biophys. Res. Commun. 2006, 345, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Narsai, R.; Ivanova, A.; Ng, S.; Whelan, J. Defining Reference Genes in Oryza sativa Using Organ, Development, Biotic and Abiotic Transcriptome Datasets. BMC Plant Biol. 2010, 10, 56. [Google Scholar] [CrossRef] [PubMed]
- González-Agüero, M.; García-Rojas, M.; Di Genova, A.; Correa, J.; Maass, A.; Orellana, A.; Hinrichsen, P. Identification of Two Putative Reference Genes from Grapevine Suitable for Gene Expression Analysis in Berry and Related Tissues Derived from RNA-Seq Data. BMC Genom. 2013, 14, 878. [Google Scholar] [CrossRef]
- Libault, M.; Thibivilliers, S.; Bilgin, D.D.; Radwan, O.; Benitez, M.; Clough, S.J.; Stacey, G. Identification of Four Soybean Reference Genes for Gene Expression Normalization. Plant Genome 2008, 1, 44–54. [Google Scholar] [CrossRef]
- Løvdal, T.; Lillo, C. Reference Gene Selection for Quantitative Real-Time PCR Normalization in Tomato Subjected to Nitrogen, Cold, and Light Stress. Anal. Biochem. 2009, 387, 238–242. [Google Scholar] [CrossRef]
- Ji, Y.; Tu, P.; Wang, K.; Gao, F.; Yang, W.; Zhu, Y.; Li, S. Defining Reference Genes for Quantitative Real-Time PCR Analysis of Anther Development in Rice. Acta Biochim. Biophys. Sin. 2014, 46, 305–312. [Google Scholar] [CrossRef]
- Olejnik, P.; Mądrzak, C.J.; Nuc, K. Cyclophilins and Their Functions in Abiotic Stress and Plant-Microbe Interactions. Biomolecules 2021, 11, 1390. [Google Scholar] [CrossRef]
- Qu, R.; Miao, Y.; Cui, Y.; Cao, Y.; Zhou, Y.; Tang, X. Selection of Reference Genes for the Quantitative Real-Time PCR Normalization of Gene Expression in Isatis indigotica Fortune. BMC Mol. Biol. 2019, 20, 9. [Google Scholar] [CrossRef]
- Nguyen, D.Q.; Eamens, A.L.; Grof, C.P.L. Reference Gene Identification for Reliable Normalisation of Quantitative RT-PCR Data in Setaria viridis. Plant Methods 2018, 14, 24. [Google Scholar] [CrossRef]
- Hossain, M.S.; Ahmed, R.; Haque, M.S.; Alam, M.M.; Islam, M.S. Identification and Validation of Reference Genes for Real-Time Quantitative RT-PCR Analysis in Jute. BMC Mol. Biol. 2019, 20, 13. [Google Scholar] [CrossRef] [PubMed]
- Morgante, C.V.; Guimarães, P.M.; Martins, A.C.; Araújo, A.C.; Leal-Bertioli, S.C.; Bertioli, D.J.; Brasileiro, A.C. Reference Genes for Quantitative Reverse Transcription-Polymerase Chain Reaction Expression Studies in Wild and Cultivated Peanut. BMC Res. Notes 2011, 4, 339. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Yang, Z.; Samma, M.K.; Wang, R.; Shen, W. Systematic Validation of Candidate Reference Genes for QRT-PCR Normalization under Iron Deficiency in Arabidopsis. Biometals 2013, 26, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Štajner, N.; Cregeen, S.; Javornik, B. Evaluation of Reference Genes for RT-QPCR Expression Studies in Hop (Humulus lupulus L.) During Infection with Vascular Pathogen Verticillium Albo-Atrum. PLoS ONE 2013, 8, e68228. [Google Scholar] [CrossRef]
- Zhao, Y.; Luo, J.; Xu, S.; Wang, W.; Liu, T.; Han, C.; Chen, Y.; Kong, L. Selection of Reference Genes for Gene Expression Normalization in Peucedanum praeruptorum Dunn under Abiotic Stresses, Hormone Treatments and Different Tissues. PLoS ONE 2016, 11, e0152356. [Google Scholar] [CrossRef]
- Delporte, M.; Legrand, G.; Hilbert, J.L.; Gagneul, D. Selection and Validation of Reference Genes for Quantitative Real-Time PCR Analysis of Gene Expression in Cichorium intybus. Front. Plant Sci. 2015, 6, 651. [Google Scholar] [CrossRef]
- Fan, C.; Ma, J.; Guo, Q.; Li, X.; Wang, H.; Lu, M. Selection of Reference Genes for Quantitative Real-Time PCR in Bamboo (Phyllostachys edulis). PLoS ONE 2013, 8, e56573. [Google Scholar] [CrossRef]
- Tjasa, R.; Stajner, N.; Bandelj, D.; Javornik, B.; Jakse, J. Validation of Candidate Reference Genes in RT-QPCR Studies of Developing Olive Fruit and Expression Analysis of Four Genes Involved in Fatty Acids Metabolism. Mol. Breed. 2013, 32, 211–222. [Google Scholar]
- Chandna, R.; Augustine, R.; Bisht, N.C. Evaluation of Candidate Reference Genes for Gene Expression Normalization in Brassica juncea Using Real Time Quantitative RT-PCR. PLoS ONE 2012, 7, e36918. [Google Scholar] [CrossRef]
- de Carvalho, K.; Filho, J.C.B.; Santos, T.B.D.; de Souza, S.G.; Vieira, L.G.; Pereira, L.F.; Domingues, D.S. Nitrogen Starvation, Salt and Heat Stress in Coffee (Coffea arabica L.): Identification and Validation of New Genes for QPCR Normalization. Mol. Biotechnol. 2013, 53, 315–325. [Google Scholar] [CrossRef]
- Reid, K.E.; Olsson, N.; Schlosser, J.; Peng, F.; Lund, S.T. An Optimized Grapevine RNA Isolation Procedure and Statistical Determination of Reference Genes for Real-Time RT-PCR during Berry Development. BMC Plant Biol. 2006, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.K.; Schultink, A.; Keegstra, K.; Wilkerson, C.G.; Pauly, M. RNA-Seq Analysis of Developing Nasturtium Seeds (Tropaeolum majus): Identification and Characterization of an Additional Galactosyltransferase Involved in Xyloglucan Biosynthesis. Mol. Plant 2012, 5, 984–992. [Google Scholar] [CrossRef] [PubMed]
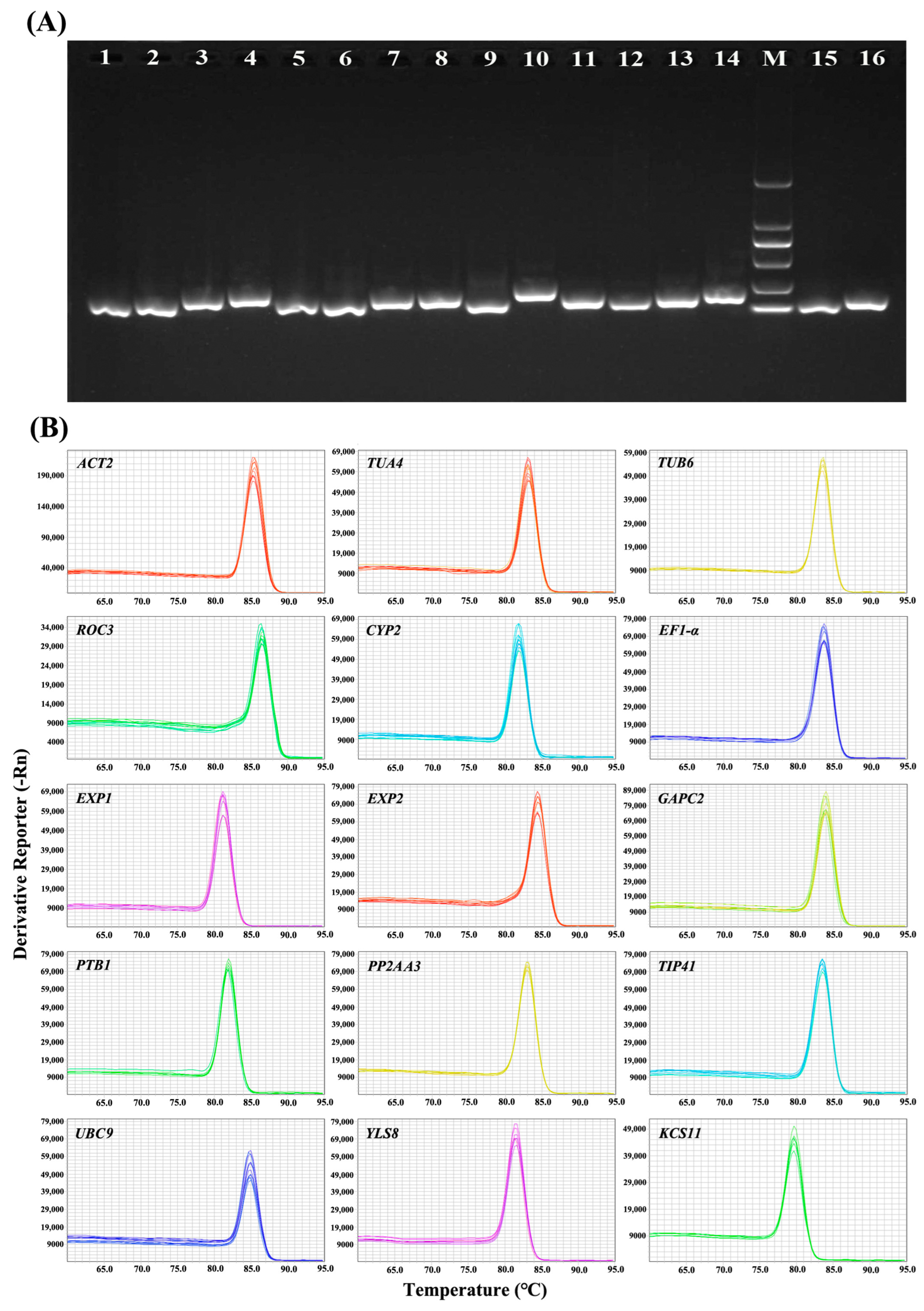



| Gene Name | Gene Symbol | Arabidopsis Homologue | Primer Sequence (Forward/Reverse) | Size (bp) | Efficiency (%) | R2 |
|---|---|---|---|---|---|---|
| Actin-2 | ACT2 | AT3G18780 | CAGACCGTATGAGCAAGGAGA | 178 | 98.07 | 0.9979 |
| ATTGATGGACCAGACTCGTCG | ||||||
| Glyceraldehyde-3-phosphate dehydrogenase GAPC2 | GAPC2 | AT1G13440 | GGCTGCTATCAAGGAGGAGTCTG | 203 | 115.20 | 0.9913 |
| GATCAACCACCCGGGTACTGT | ||||||
| Tubulin alpha-4 chain | TUA4 | AT1G04820 | GCATTGGTATGTTGGTGAGGGT | 134 | 95.79 | 0.9942 |
| ACTCATCCCCATCATCATCTTCC | ||||||
| Tubulin beta-6 chain | TUB6 | AT5G12250 | ATGTTCAGGAGGGTGAGTGA | 168 | 104.13 | 0.9996 |
| CTCATCAGCGGTAGCATCTTG | ||||||
| Peptidyl-prolyl cis-trans isomerase CYP19-1 ROC3 | ROC3 | AT2G16600 | CTCAGTTCTTCGTCTGTACCGAG | 213 | 98.84 | 0.9985 |
| AGAAGCGCAGATCCTCCACT | ||||||
| Cyclophilin-like peptidyl-prolyl cis-trans isomerase 2 | CYP2 | AT4G33060 | CAGCTCAGTTGAAGTTTGTGCC | 235 | 99.89 | 0.9964 |
| CAGCCTCATCCAGGTTATAGAACTC | ||||||
| GTP binding Elongation factor EFTu/EF1A | EF1-α | AT1G07920 | ATCTCCAAGGATGGGCAGAC | 168 | 108.64 | 1.0000 |
| TCCAACCTTCTTCAGGTAGGAAG | ||||||
| Expressed protein 1 (Dimethylallyl, adenosine tRNA methylthiotransferase) | EXP1 | AT4G33380 | CTGAGTTTGGACGACGTGAAG | 173 | 105.26 | 0.9979 |
| CCAGCTTTCTTCTTCCTCATCG | ||||||
| Expressed protein 2 (S-adenosyl-L-methionine-dependent methyltransferases) | EXP2 | AT2G32170 | CCGAGTATGGATGCTATTCTCCAG | 203 | 105.54 | 0.9939 |
| GATTGACTGCTCATTGTGAAGTCC | ||||||
| Serine/threonine-protein phosphatase PP2AA3 | PP2AA3 | AT1G13320 | GCCTGAAGATTGTGTTGCTCAC | 197 | 97.97 | 0.9959 |
| CTGCTATTCGTACTTCAGCCTCA | ||||||
| Polypyrimidine tract-binding protein 1 | PTB1 | AT3G01150 | TGCTGTCACTGTGGATGTGC | 142 | 106.44 | 0.9974 |
| TGCGTCTCTAGCAACTGCTG | ||||||
| Type 2a phosphatase activator tip41 | TIP41 | AT4G34270 | CGCAAACGCTCCATTCTCACTTC | 241 | 98.15 | 0.9944 |
| CCTGCTGAGAAGGTTTGCATCTG | ||||||
| Ubiquitin-conjugating enzyme 9 | UBC9 | AT4G27960 | TTGCTCTCAATCTGCTCGCTG | 175 | 119.79 | 0.9978 |
| TCCTATCGCAACCATCATAGCG | ||||||
| Mitosisprotein YLS8 | YLS8 | AT5G08290 | TGATTGGGATGAAACATGCATGC | 146 | 110.54 | 0.9908 |
| TTGTGGATGGATCGTACAGCTC | ||||||
| 3-ketoacyl-CoA synthase 11 | KCS11 | AT2G26640 | AGGAAGACTCGGACGGGAAGAT | 115 | 100.36 | 0.9982 |
| GACATGGGTAGAACAAGTGGTCCG |
| Ranking Order | Organs | Seeds in Different Developmental Stages | All Samples | |||
|---|---|---|---|---|---|---|
| Gene Name | Stability Value | Gene Name | Stability Value | Gene Name | Stability Value | |
| 1 | EXP1 | 0.175 | EXP1 | 0.155 | EXP1 | 0.070 |
| 2 | EXP2 | 0.268 | ROC3 | 0.158 | ACT2 | 0.262 |
| 3 | TUB6 | 0.300 | ACT2 | 0.257 | PP2AA3 | 0.375 |
| 4 | ACT2 | 0.346 | CYP2 | 0.399 | EXP2 | 0.376 |
| 5 | UBC9 | 0.422 | EF1-α | 0.416 | CYP2 | 0.443 |
| 6 | PP2AA3 | 0.504 | PP2AA3 | 0.424 | ROC3 | 0.499 |
| 7 | CYP2 | 0.505 | EXP2 | 0.434 | GAPC2 | 0.553 |
| 8 | GAPC2 | 0.535 | GAPC2 | 0.463 | EF1-α | 0.554 |
| 9 | TIP41 | 0.576 | YLS8 | 0.643 | UBC9 | 0.658 |
| 10 | ROC3 | 0.642 | UBC9 | 0.714 | YLS8 | 0.722 |
| 11 | YLS8 | 0.687 | PTB1 | 0.941 | TUB6 | 1.053 |
| 12 | EF1-α | 0.699 | TUB6 | 0.987 | PTB1 | 1.091 |
| 13 | TUA4 | 0.859 | TIP41 | 1.179 | TIP41 | 1.099 |
| 14 | PTB1 | 0.921 | TUA4 | 1.481 | TUA4 | 1.686 |
| Ranking Order | Organs | Seeds in Different Developmental Stages | All Samples | |||
|---|---|---|---|---|---|---|
| Gene Name | Stability Value | Gene Name | Stability Value | Gene Name | Stability Value | |
| 1 | TUB6 | 2.39 ± 0.44 | UBC9 | 2.23 ± 0.42 | EXP2 | 2.94 ± 0.64 |
| 2 | ROC3 | 2.91 ± 0.52 | CYP2 | 2.26 ± 0.51 | CYP2 | 3.06 ± 0.68 |
| 3 | EXP1 | 3.20 ± 0.73 | EXP2 | 2.37 ± 0.52 | EXP1 | 3.11 ± 0.72 |
| 4 | EXP2 | 3.23 ± 0.69 | GAPC2 | 2.40 ± 0.45 | GAPC2 | 3.28 ± 0.61 |
| 5 | GAPC2 | 3.65 ± 0.68 | YLS8 | 2.50 ± 0.51 | ROC3 | 3.47 ± 0.63 |
| 6 | EF1-α | 3.85 ± 0.65 | ACT2 | 2.62 ± 0.51 | UBC9 | 4.08 ± 0.77 |
| 7 | CYP2 | 4.00 ± 0.88 | EXP1 | 2.89 ± 0.67 | ACT2 | 4.26 ± 0.81 |
| 8 | UBC9 | 4.54 ± 0.88 | PTB1 | 3.57 ± 0.79 | YLS8 | 4.55 ± 0.93 |
| 9 | ACT2 | 4.84 ± 0.90 | ROC3 | 3.68 ± 0.66 | PP2AA3 | 5.15 ± 1.17 |
| 10 | TIP41 | 4.88 ± 1.13 | PP2AA3 | 4.66 ± 1.06 | PTB1 | 5.95 ± 1.36 |
| 11 | PP2AA3 | 5.57 ± 1.23 | EF1-α | 5.71 ± 1.03 | EF1-α | 6.05 ± 1.05 |
| 12 | YLS8 | 5.96 ± 1.24 | TUB6 | 6.05 ± 1.29 | TIP41 | 6.80 ± 1.56 |
| 13 | PTB1 | 6.11 ± 1.44 | TUA4 | 7.53 ± 1.67 | TUB6 | 8.39 ± 1.69 |
| 14 | TUA4 | 7.97 ± 1.43 | TIP41 | 9.04 ± 2.03 | TUA4 | 11.72 ± 2.35 |
| Ranking Order | Organs | Seeds in Different Developmental Stages | All Samples | |||
|---|---|---|---|---|---|---|
| Gene Name | Stability Value | Gene Name | Stability Value | Gene Name | Stability Value | |
| 1 | EXP1 | 1.57 | EXP1 | 2.21 | EXP1 | 1.97 |
| 2 | EXP2 | 2.11 | CYP2 | 2.83 | EXP2 | 2.45 |
| 3 | TUB6 | 2.71 | ROC3 | 3.44 | CYP2 | 2.99 |
| 4 | ACT2 | 6.00 | ACT2 | 3.66 | ACT2 | 3.60 |
| 5 | PP2AA3 | 6.31 | EXP2 | 4.12 | GAPC2 | 3.96 |
| 6 | ROC3 | 6.32 | UBC9 | 5.01 | ROC3 | 4.12 |
| 7 | UBC9 | 6.47 | GAPC2 | 5.24 | PP2AA3 | 5.69 |
| 8 | GAPC2 | 6.51 | YLS8 | 5.38 | UBC9 | 7.67 |
| 9 | CYP2 | 6.65 | PP2AA3 | 7.93 | EF1-α | 8.74 |
| 10 | TIP41 | 7.02 | EF1-α | 8.80 | YLS8 | 9.46 |
| 11 | EF1-α | 8.30 | PTB1 | 10.46 | PTB1 | 11.24 |
| 12 | YLS8 | 11.49 | TUB6 | 12.00 | TUB6 | 12.22 |
| 13 | TUA4 | 13.00 | TUA4 | 13.00 | TIP41 | 12.49 |
| 14 | PTB1 | 14.00 | TIP41 | 14.00 | TUA4 | 14.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, Q.; Zhou, G.-C.; Liu, S.-J.; Li, W.; Wang, Y.-L.; Xu, G.-Y.; Li, T.-F.; Meng, G.-Q.; Xue, J.-Y. Selection and Validation of Reference Genes for qRT-PCR Analysis of Gene Expression in Tropaeolum majus (Nasturtium). Horticulturae 2023, 9, 1176. https://doi.org/10.3390/horticulturae9111176
Tang Q, Zhou G-C, Liu S-J, Li W, Wang Y-L, Xu G-Y, Li T-F, Meng G-Q, Xue J-Y. Selection and Validation of Reference Genes for qRT-PCR Analysis of Gene Expression in Tropaeolum majus (Nasturtium). Horticulturae. 2023; 9(11):1176. https://doi.org/10.3390/horticulturae9111176
Chicago/Turabian StyleTang, Qing, Guang-Can Zhou, Si-Jie Liu, Wen Li, Yi-Lei Wang, Gao-Ying Xu, Teng-Fei Li, Guo-Qing Meng, and Jia-Yu Xue. 2023. "Selection and Validation of Reference Genes for qRT-PCR Analysis of Gene Expression in Tropaeolum majus (Nasturtium)" Horticulturae 9, no. 11: 1176. https://doi.org/10.3390/horticulturae9111176
APA StyleTang, Q., Zhou, G.-C., Liu, S.-J., Li, W., Wang, Y.-L., Xu, G.-Y., Li, T.-F., Meng, G.-Q., & Xue, J.-Y. (2023). Selection and Validation of Reference Genes for qRT-PCR Analysis of Gene Expression in Tropaeolum majus (Nasturtium). Horticulturae, 9(11), 1176. https://doi.org/10.3390/horticulturae9111176







