Physiological, Transcriptomic and Metabolomic Response of Basil (O. basilicum Linn. var. pilosum (Willd.) Benth.) to Red and Blue Light
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Light Treatment
2.2. Analysis of Photosynthetic Parameters and Chlorophyll Fluorescence
2.3. Determination of Photosynthetic Pigment and Antioxidant Activity of Leaf Extracts
2.4. RNA Extraction and Illumina Sequencing
2.5. Transcriptomic Analysis
2.6. Extraction and Detection of Metabolites
2.7. Metabolomic Analysis
2.8. Integrative Analysis of Metabolomic and Transcriptomic Datasets
3. Results
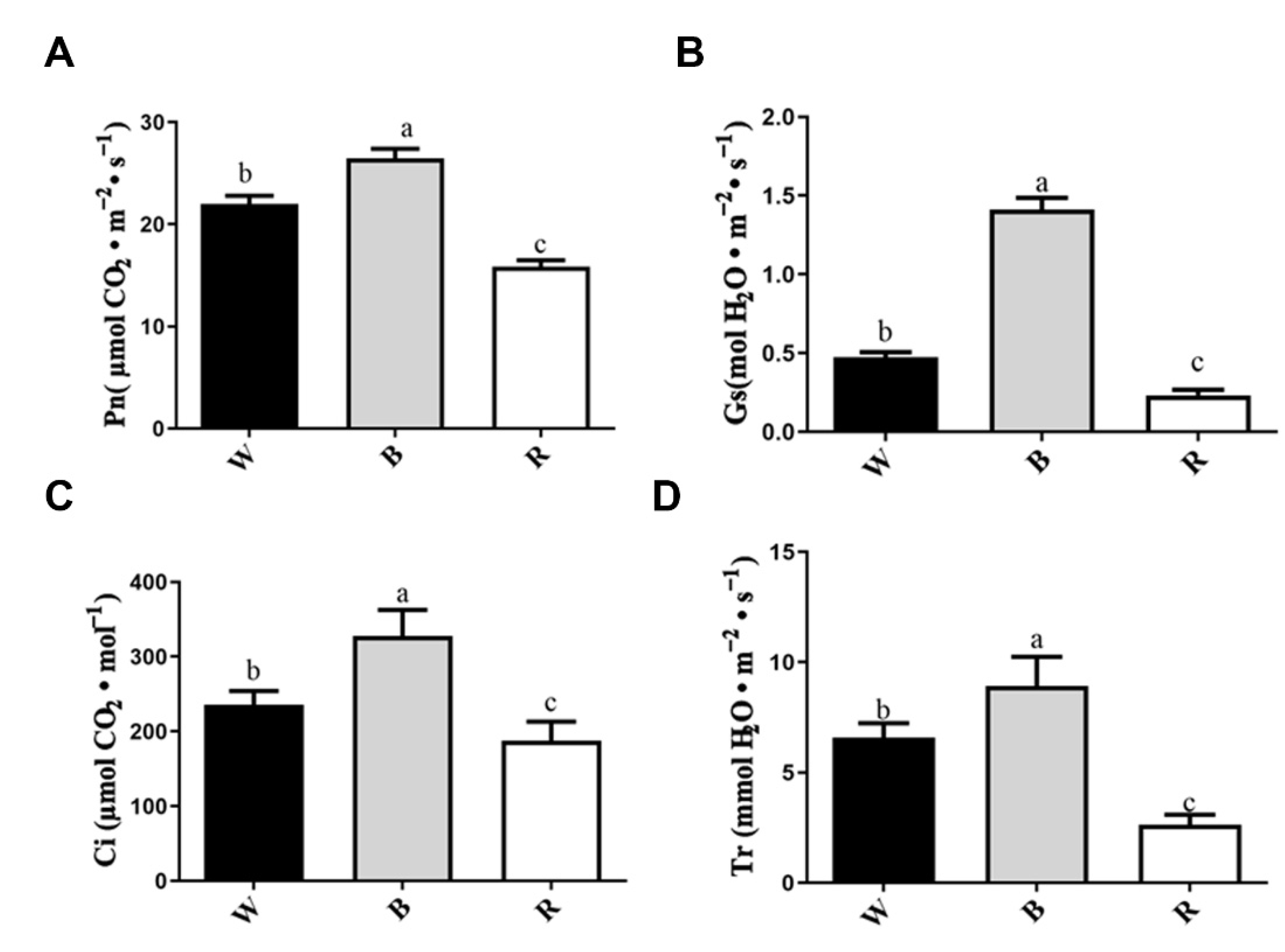
3.1. Plant Morphology and Growth Characteristics
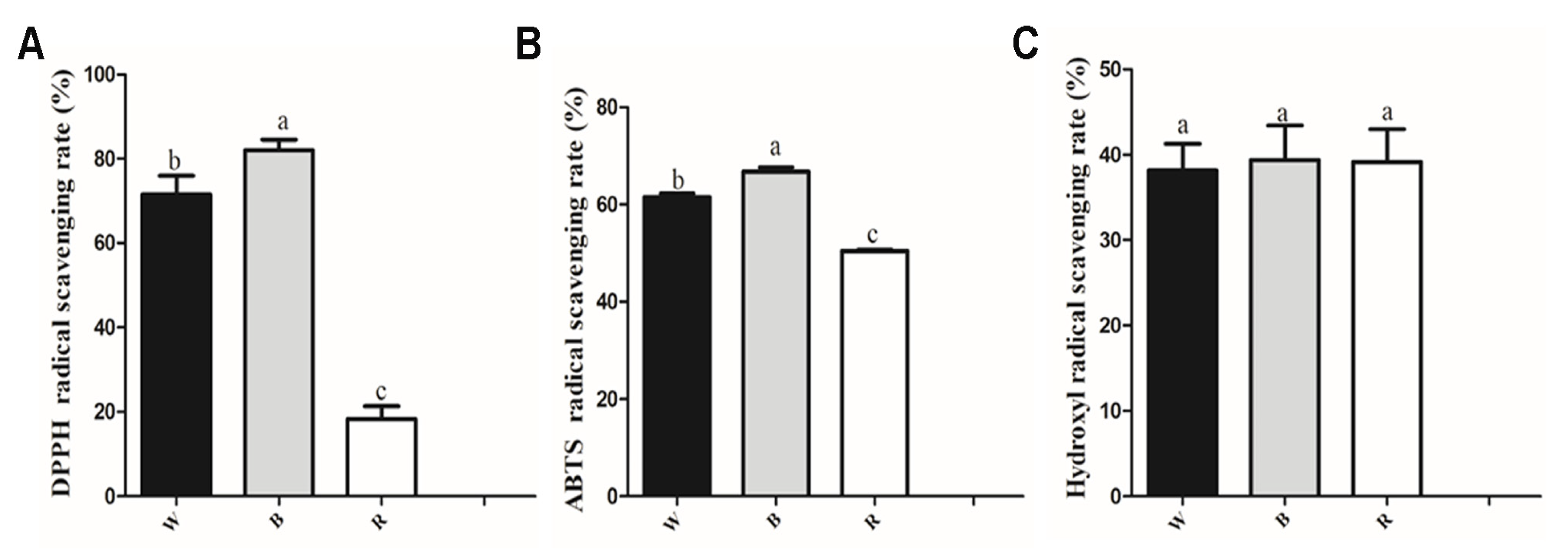
3.2. Antioxidant Activity Analysis of Leaf Extracts
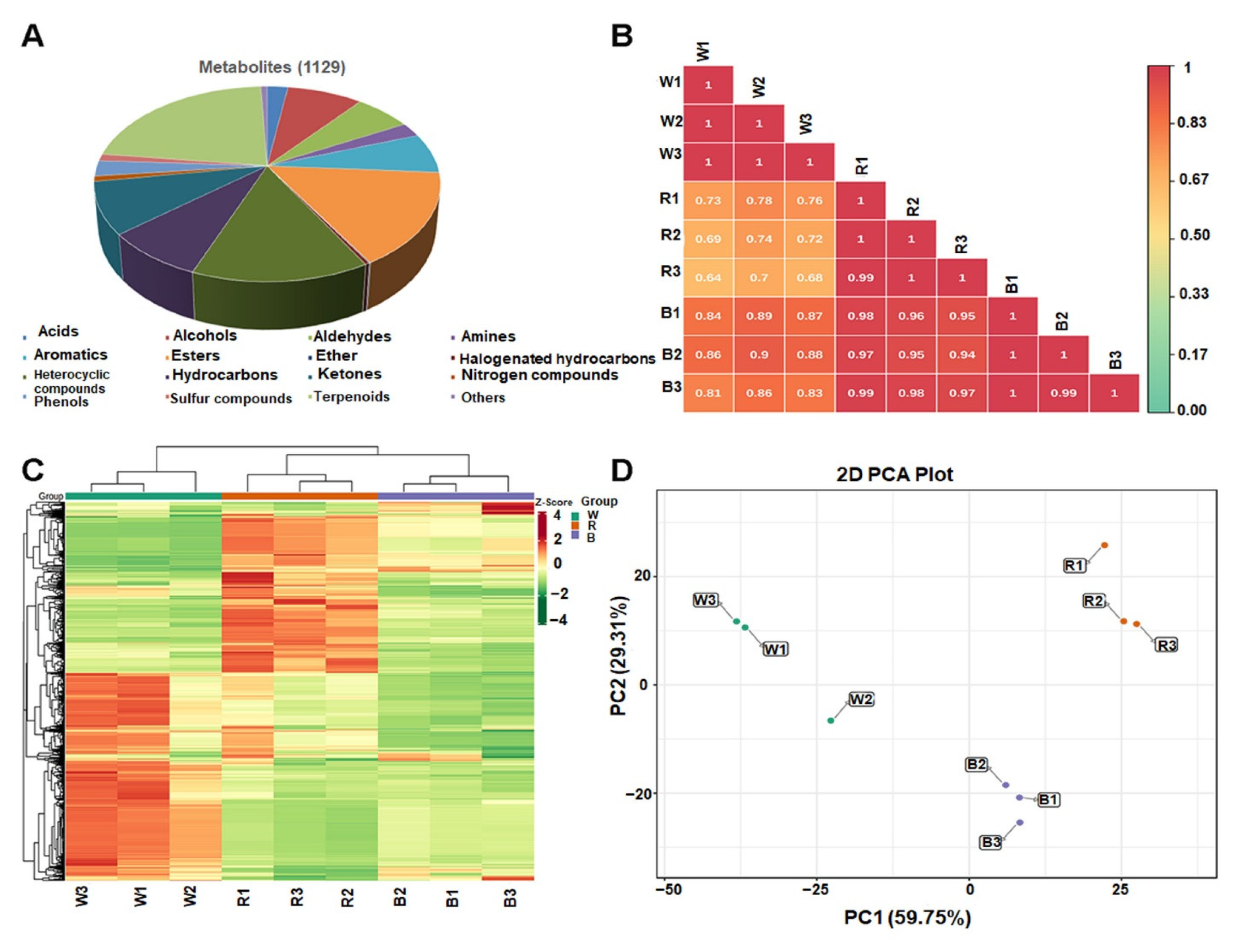
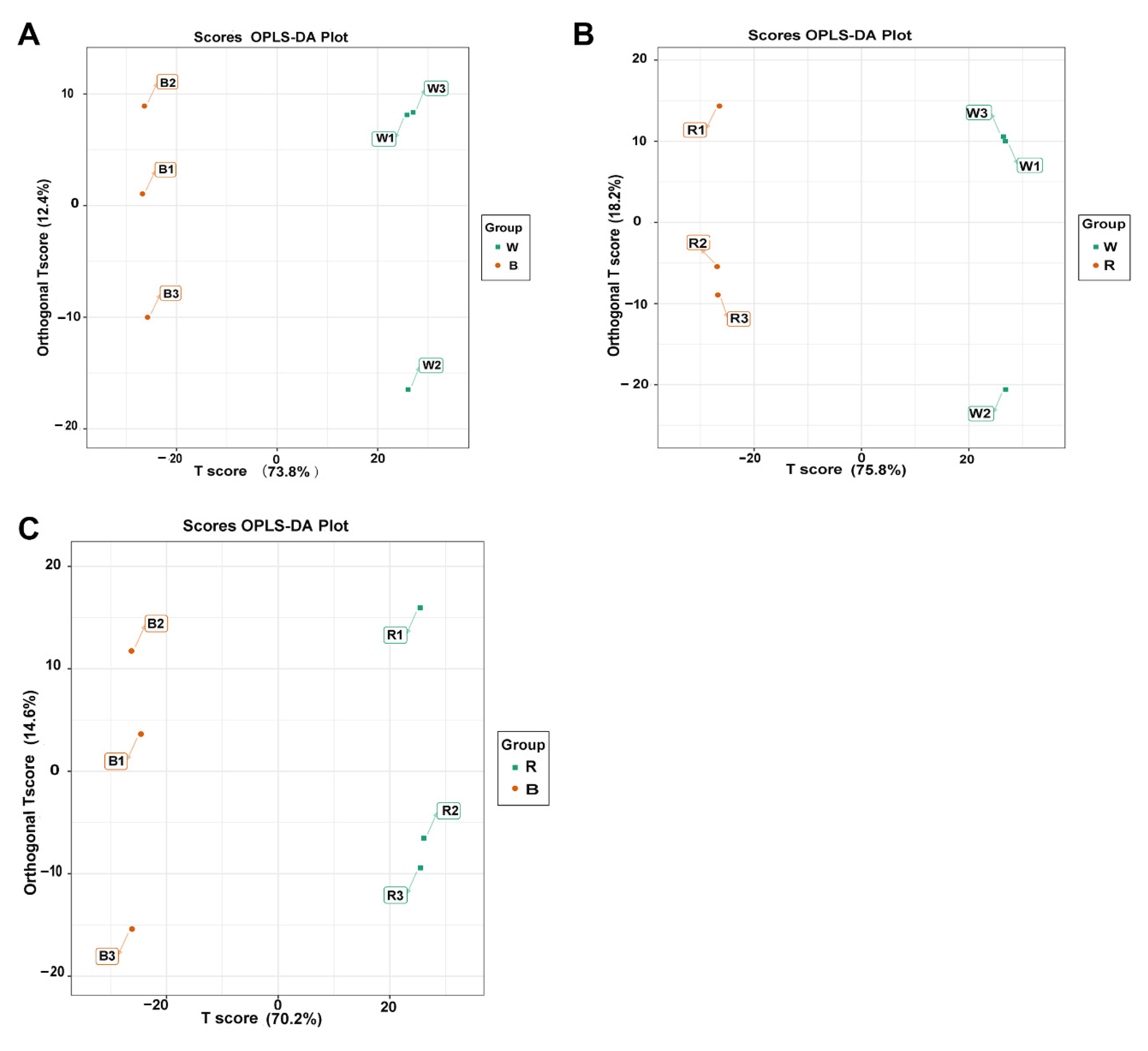
3.3. Metabolite Profiling of the Leaves via GC-MS
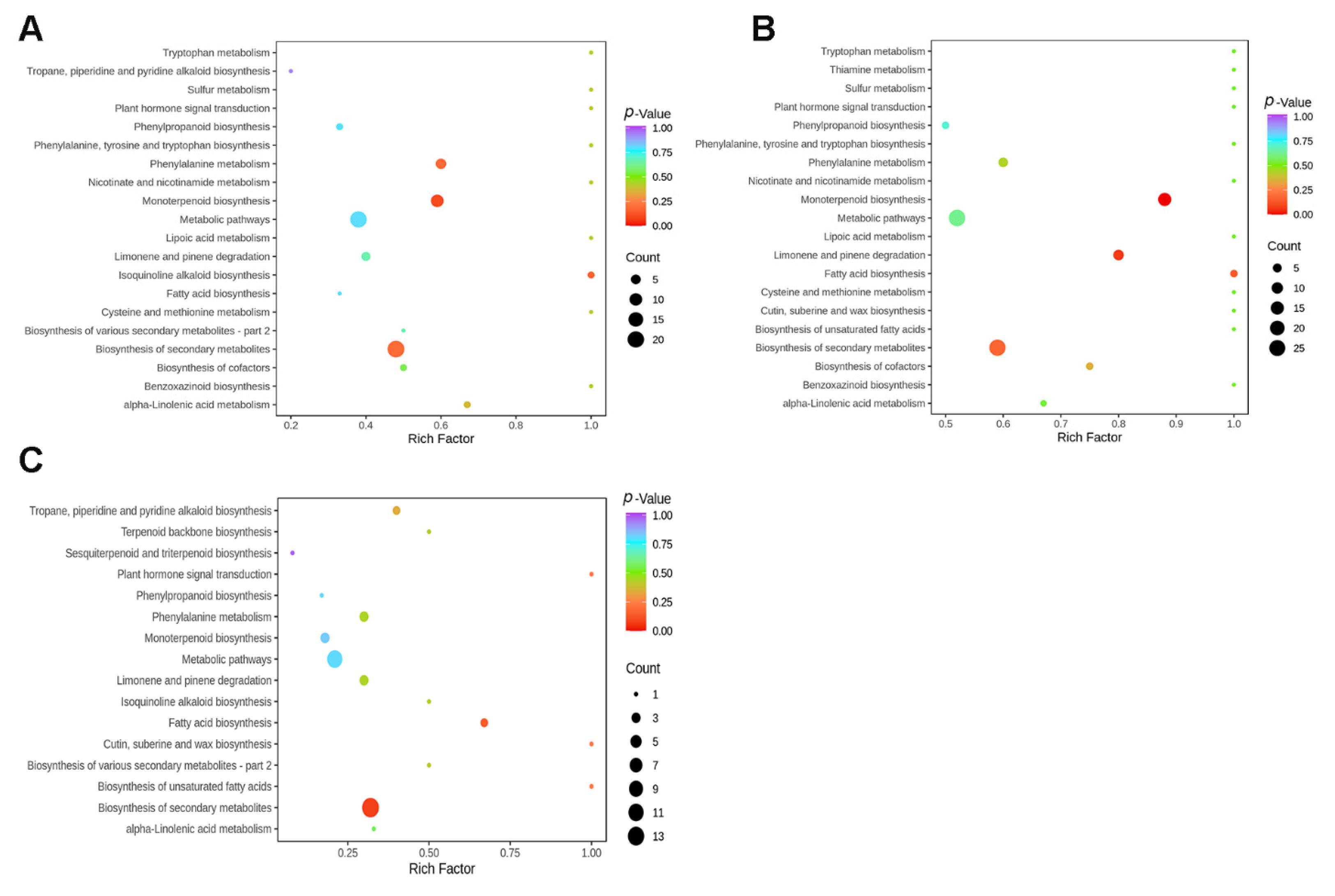
3.4. Pathway Enrichment Analysis of Differentially Accumulated Metabolites (DAMs)
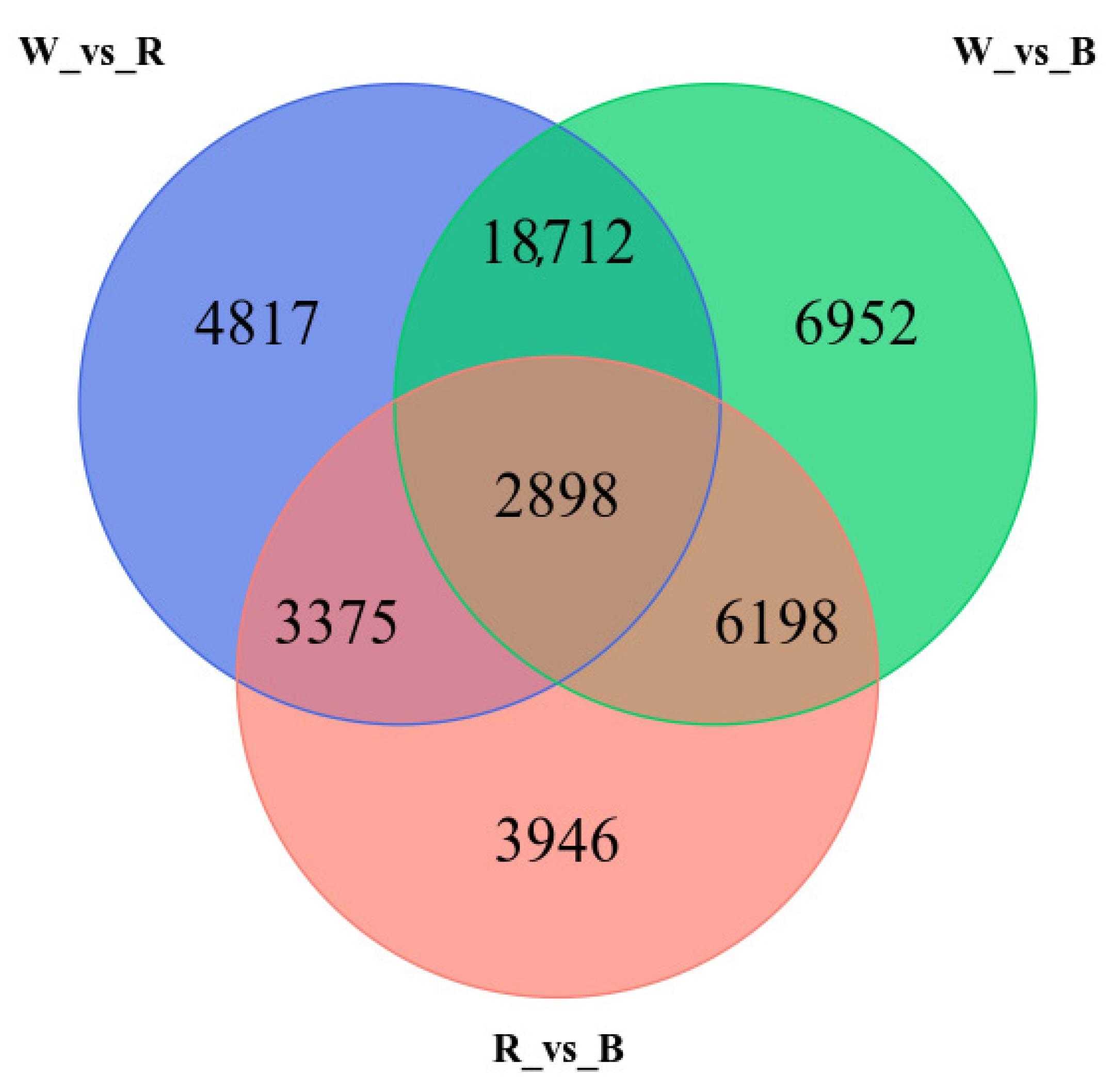
3.5. Annotation of Transcripts and Differentially Expressed Genes (DEGs)
3.6. Dynamic Transcriptome Analysis in Response to Light Spectra
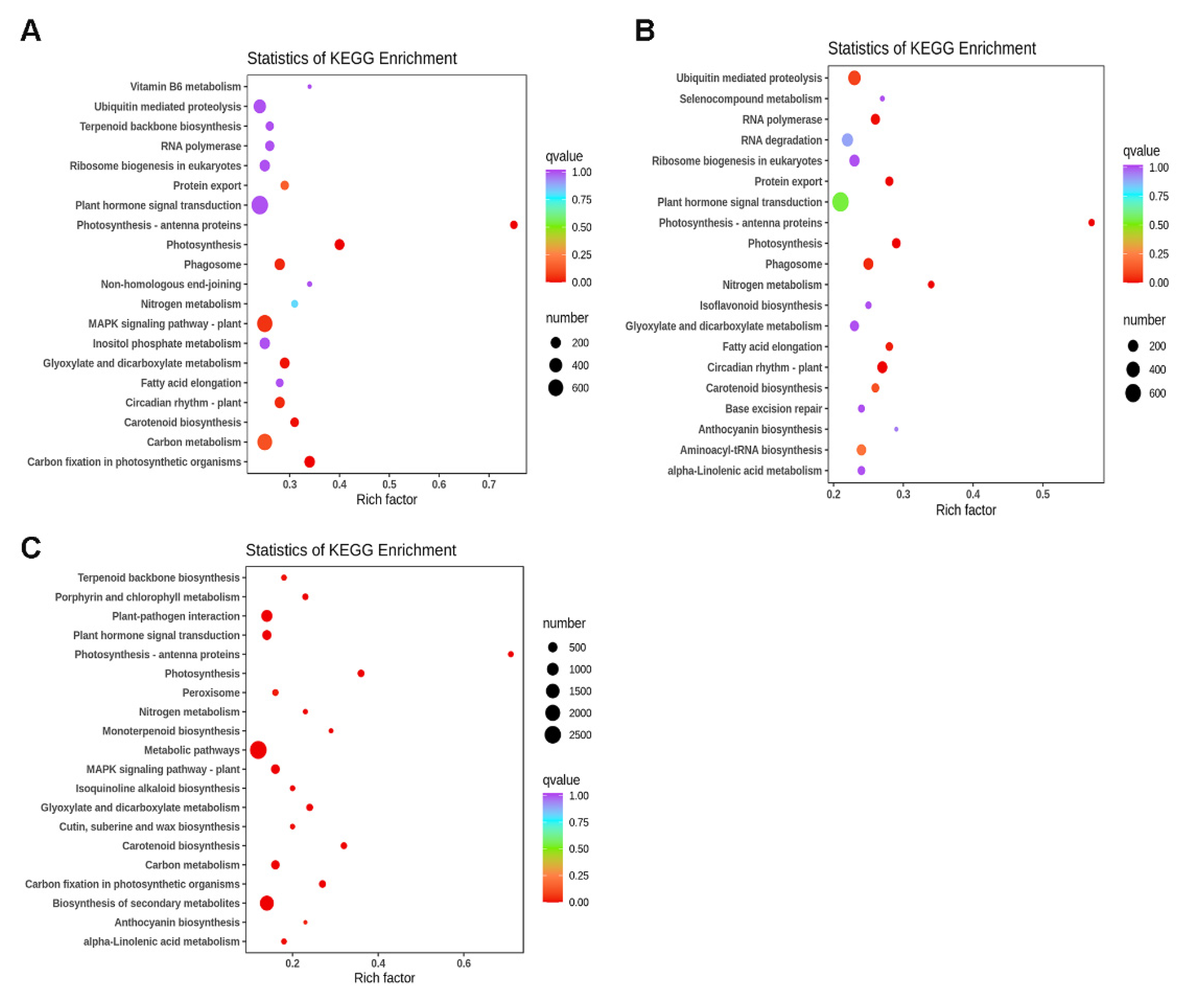
3.7. GO and KEGG Pathway Enrichment Analysis of DEGs
3.8. Association Analysis of DAMs and DEGs
4. Discussion
4.1. Changes in the Physiology and Antioxidant Capacity of Leaf Extracts
4.2. Involvement of Key Metabolites in Response to Light Spectra
4.3. Key Genes Associated with the Light Response in Basil
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deng, X.W.; Quail, P.H. Signalling in light-controlled development. Semin. Cell Dev. Biol. 1999, 10, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.; Singh, D.; Lingwan, M.; Yadukrishnan, P.; Masakapalli, S.K.; Datta, S. Light signaling and UV-B-mediated plant growth regulation. J. Integr. Plant Biol. 2020, 62, 1270–1292. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, W.; Tong, Y.; Yang, Q. Leaf morphology, photosynthetic performance, chlorophyll fluorescence, stomatal development of lettuce (Lactuca Sativa L.) exposed to different ratios of red light to blue light. Front. Plant Sci. 2016, 7, 250. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; You, W.; Gong, Y.; El Hajjami, M.; Liang, W.; Xu, J.; Poetsch, A. Transcriptomic and proteomic choreography in response to light quality variation reveals key adaption mechanisms in marine Nannochloropsis oceanica. Sci. Total Environ. 2020, 720, 137667. [Google Scholar] [CrossRef]
- Suetsugu, N.; Wada, M. Evolution of three LOV blue light receptor families in green plants and photosynthetic stramenopiles: Phototropin, ZTL/FKF1/LKP2 and aureochrome. Plant Cell Physiol. 2013, 54, 8–23. [Google Scholar] [CrossRef]
- Jenkins, G.I. The UV-B photoreceptor UVR8: From structure to physiology. Plant Cell 2014, 26, 21–37. [Google Scholar] [CrossRef]
- Paik, I.; Huq, E. Plant photoreceptors: Multi-functional sensory proteins and their signaling networks. Semin. Cell Dev. Biol. 2019, 92, 114–121. [Google Scholar] [CrossRef]
- Chaves, I.; Pokorny, R.; Byrdin, M.; Hoang, N.; Ritz, T.; Brettel, K.; Essen, L.-O.; van der Horst, G.T.J.; Batschauer, A.; Ahmad, M. The cryptochromes: Blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 2011, 62, 335–364. [Google Scholar] [CrossRef]
- Chen, M.; Chory, J. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 2011, 21, 664–671. [Google Scholar] [CrossRef]
- Liu, J.; van Iersel, M.W. Photosynthetic physiology of blue, green, and red light: Light intensity effects and underlying mechanisms. Front. Plant Sci. 2021, 12, 619987. [Google Scholar] [CrossRef]
- Muneer, S.; Kim, E.J.; Park, J.S.; Lee, J.H. Influence of green, red and blue light emitting diodes on multiprotein complex proteins and photosynthetic activity under different light intensities in lettuce leaves (Lactuca Sativa L.). Int. J. Mol. Sci. 2014, 15, 4657–4670. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Xia, K.; Wu, Q.; Su, N.; Cui, J. Effects of light quality on the chloroplastic ultrastructure and photosynthetic characteristics of cucumber seedlings. Plant Growth Regul. 2014, 73, 227–235. [Google Scholar]
- Hoenecke, M.E.; Bula, R.J.; Tibbitts, T.W. Importance of “blue” photon levels for Lettuce seedlings grown under red-light-emitting diodes. HortScience 1992, 27, 427–430. [Google Scholar] [CrossRef]
- Goins, G.D.; Yorio, N.C.; Sanwo, M.M.; Brown, C.S. Photomorphogenesis, photosynthesis, and seed yield of wheat plants grown under red light-emitting diodes (LEDs) with and without supplemental blue lighting. J. Exp. Bot. 1997, 48, 1407–1413. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Qin, L.; Chong, E.L.C.; Choong, T.-W.; Lee, S.K. Plant growth and photosynthetic characteristics of Mesembryanthemum crystallinum grown aeroponically under different blue- and red-LEDs. Front. Plant Sci. 2017, 8, 361. [Google Scholar] [CrossRef]
- Sergejeva, D.; Alsina, I.; Duma, M.; Dubova, L.; Berzina, K. Evaluation of different lighting sources on the growth and chemical composition of lettuce. Agron. Res. 2018, 16. [Google Scholar] [CrossRef]
- Shimizu, H.; Saito, Y.; Nakashima, H.; Miyasaka, J.; Ohdoi, K. Light environment optimization for lettuce growth in plant factory. IFAC Proc. Vol. 2011, 44, 605–609. [Google Scholar] [CrossRef]
- Kobayashi, K.; Amore, T.; Lazaro, M. Light-emitting diodes (LEDs) for miniature hydroponic lettuce. Opt. Photon. J. 2013, 3, 74–77. [Google Scholar] [CrossRef]
- Sæbø, A.; Krekling, T.; Appelgren, M. Appelgren light quality affects photosynthesis and leaf anatomy of birch plantlets in vitro. Plant Cell Tissue Organ Cult. 1995, 41, 177–185. [Google Scholar] [CrossRef]
- Miao, L.; Zhang, Y.; Yang, X.; Xiao, J.; Zhang, H.; Zhang, Z.; Wang, Y.; Jiang, G. Colored light-quality selective plastic films affect anthocyanin content, enzyme activities, and the expression of flavonoid genes in strawberry (Fragaria × Ananassa) Fruit. Food Chem. 2016, 207, 93–100. [Google Scholar] [CrossRef]
- Wang, C.; Qiu, H.; Chen, Y.; Xu, Y.; Shan, F.; Li, H.; Yan, C.; Ma, C. Red light regulates metabolic pathways of soybean hypocotyl elongation and thickening. Environ. Exp. Bot. 2022, 199, 104890. [Google Scholar] [CrossRef]
- Mao, P.; Li, Q.; Li, Y.; Xu, Y.; Yang, Q.; Bian, Z.; Wang, S.; He, L.; Xu, Z.; Zheng, Y.; et al. The beneficial functions of blue light supplementary on the biosynthesis of glucosinolates in pakchoi (Brassica Rapa L. ssp. Chinensis) under greenhouse conditions. Environ. Exp. Bot. 2022, 197, 104834. [Google Scholar] [CrossRef]
- Chen, X.; Cai, W.; Xia, J.; Yu, H.; Wang, Q.; Pang, F.; Zhao, M. Metabolomic and transcriptomic analyses reveal that blue light promotes chlorogenic scid synthesis in strawberry. J. Agric. Food Chem. 2020, 68, 12485–12492. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.M.; Noga, G.; Hunsche, M. High blue light improves acclimation and photosynthetic recovery of pepper plants exposed to UV stress. Environ. Exp. Bot. 2015, 109, 254–263. [Google Scholar] [CrossRef]
- Calderón Bravo, H.; Vera Céspedes, N.; Zura-Bravo, L.; Muñoz, L.A. Basil seeds as a novel food, source of nutrients and functional ingredients with beneficial properties: A review. Foods 2021, 10, 1467. [Google Scholar] [CrossRef]
- Srivastava, S.; Lal, R.K.; Maurya, R.; Mishra, A.; Yadav, A.K.; Pandey, G.; Rout, P.K.; Chanotiya, C.S. Chemical diversity of essential oil among basil genotypes (Ocimum Viride Willd.) across the years. Ind. Crops Prod. 2021, 173, 114153. [Google Scholar] [CrossRef]
- Prakash, P.; Gupta, N. Therapeutic uses of Ocimum sanctum Linn (Tulsi) with a note on eugenol and its pharmacological actions: A short review. Indian J. Physiol. Pharmacol. 2005, 49, 125–131. [Google Scholar]
- Ghasemzadeh, A.; Ashkani, S.; Baghdadi, A.; Pazoki, A.; Jaafar, H.Z.E.; Rahmat, A. Improvement in flavonoids and phenolic acids production and pharmaceutical quality of sweet basil (Ocimum Basilicum L.) by ultraviolet-B irradiation. Molecules 2016, 21, 1203. [Google Scholar] [CrossRef]
- Nazir, M.; Asad Ullah, M.; Mumtaz, S.; Siddiquah, A.; Shah, M.; Drouet, S.; Hano, C.; Abbasi, B.H. Interactive effect of melatonin and UV-C on phenylpropanoid metabolite production and antioxidant potential in callus cultures of purple basil (Ocimum Basilicum L. var.s Purpurascens). Molecules 2020, 25, 1072. [Google Scholar] [CrossRef]
- Nadeem, M.; Abbasi, B.H.; Younas, M.; Ahmad, W.; Zahir, A.; Hano, C. LED-enhanced biosynthesis of biologically active ingredients in callus cultures of Ocimum basilicum. J. Photochem. Photobiol. B. 2019, 190, 172–178. [Google Scholar] [CrossRef]
- Pennisi, G.; Blasioli, S.; Cellini, A.; Maia, L.; Crepaldi, A.; Braschi, I.; Spinelli, F.; Nicola, S.; Fernandez, J.A.; Stanghellini, C.; et al. Unraveling the role of red:blue LED lights on resource use efficiency and nutritional properties of indoor grown sweet basil. Front. Plant Sci. 2019, 10, 305. [Google Scholar] [CrossRef]
- Larsen, D.H.; Li, H.; Shrestha, S.; Verdonk, J.C.; Nicole, C.C.S.; Marcelis, L.F.M.; Woltering, E.J. Lack of blue light regulation of antioxidants and chilling tolerance in basil. Front. Plant Sci. 2022, 13, 852654. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-W.; Li, S.-K.; Wu, W.-J. The main chemical composition and in vitro antifungal activity of the essential oils of Ocimum basilicum Linn. var. pilosum (Willd.) Benth. Molecules 2009, 14, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Han, T.; Yang, L.; Wang, Q.; Zhao, Y.; Jiang, D.; Ruan, X. The essential roles of OsFtsH2 in developing the chloroplast of rice. BMC Plant Biol. 2021, 21, 445. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zheng, P.-C.; Liu, P.-P.; Song, X.-W.; Guo, F.; Li, Y.-Y.; Ni, D.-J.; Jiang, C.-J. Novel insight into the role of withering process in characteristic flavor formation of teas using transcriptome analysis and metabolite profiling. Food Chem. 2019, 272, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hua, J.; Yu, Q.; Li, J.; Wang, J.; Deng, Y.; Yuan, H.; Jiang, Y. Widely targeted metabolomic analysis reveals dynamic changes in non-volatile and volatile metabolites during green tea processing. Food Chem. 2021, 363, 130131. [Google Scholar] [CrossRef]
- Li, L.; Kong, Z.; Huan, X.; Liu, Y.; Liu, Y.; Wang, Q.; Liu, J.; Zhang, P.; Guo, Y.; Qin, P. Transcriptomics integrated with widely targeted metabolomics reveals the mechanism underlying grain color formation in wheat at the grain-filling stage. Front. Plant Sci. 2021, 12, 757750. [Google Scholar] [CrossRef] [PubMed]
- Eduardo, Z.; Jianxin, Z. Role of zeaxanthin in blue light photoreception and the modulation of light-CO2 interactions in guard cells. J. Exp. Bot. 1998, 49, 433–442. [Google Scholar]
- Sharkey, T.D.; Raschke, K. Effect of light quality on stomatal opening in leaves of Xanthium strumarium L. Plant Physiol. 1981, 68, 1170–1174. [Google Scholar] [CrossRef]
- Kinoshita, T.; Doi, M.; Suetsugu, N.; Kagawa, T.; Wada, M.; Shimazaki, K. Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 2001, 414, 656–660. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, T.; Fang, S.; Zhou, M.; Qin, J. Responses of morphology, gas exchange, photochemical activity of photosystem II, and antioxidant balance in Cyclocarya paliurus to light spectra. Front. Plant Sci. 2018, 9, 1704. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Shao, Q.; Xu, M.; Li, S.; Wu, M.; Tan, X.; Su, L. Effects of light quality on morphology, enzyme activities, and bioactive compound contents in Anoectochilus roxburghii. Front. Plant Sci. 2017, 8, 857. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, Q.; Xin, Y.; Mei, Z.; Gao, A.; Liu, W.; Yu, L.; Chen, X.; Chen, Z.; Wang, N. Analyses of the photosynthetic characteristics, chloroplast ultrastructure, and transcriptome of apple (Malus Domestica) grown under red and blue lights. BMC Plant Biol. 2021, 21, 483. [Google Scholar] [CrossRef]
- Izzo, L.G.; Mele, B.H.; Vitale, L.; Vitale, E.; Arena, C. The role of monochromatic red and blue light in tomato early photomorphogenesis and photosynthetic traits. Environ. Exp. Bot. 2020, 179, 104195. [Google Scholar] [CrossRef]
- Miao, Y.; Chen, Q.; Qu, M.; Gao, L.; Hou, L. Blue light alleviates “red Light syndrome” by regulating chloroplast ultrastructure, photosynthetic traits and nutrient accumulation in cucumber plants. Sci. Hortic. 2019, 257, 108680. [Google Scholar] [CrossRef]
- Zheng, L.; Van Labeke, M.-C. Chrysanthemum morphology, photosynthetic efficiency and antioxidant capacity are differentially modified by light quality. J. Plant Physiol. 2017, 213, 66–74. [Google Scholar] [CrossRef]
- Fukuda, N.; Ajima, C.; Yukawa, T.; Olsen, J.E. Antagonistic action of blue and red light on shoot elongation in petunia depends on gibberellin, but the effects on flowering are not generally linked to gibberellin. Environ. Exp. Bot. 2016, 121, 102–111. [Google Scholar] [CrossRef]
- Lau, O.S.; Deng, X.W. Plant hormone signaling lightens up: Integrators of light and hormones. Curr. Opin. Plant Biol. 2010, 13, 571–577. [Google Scholar] [CrossRef]
- Kurepin, L.V.; Emery, R.J.N.; Pharis, R.P.; Reid, D.M. Uncoupling light quality from light irradiance effects in helianthus annuus shoots: Putative roles for plant hormones in leaf and internode growth. J. Exp. Bot. 2007, 58, 2145–2157. [Google Scholar] [CrossRef]
- Kurepin, L.V.; Pharis, R.P. Light signaling and the phytohormonal regulation of shoot growth. Plant Sci. 2014, 229, 280–289. [Google Scholar] [CrossRef]
- Samuolienė, G.; Sirtautas, R.; Brazaitytė, A.; Duchovskis, P. LED lighting and seasonality effects antioxidant properties of baby leaf lettuce. Food Chem. 2012, 134, 1494–1499. [Google Scholar] [CrossRef]
- Kapoor, S.; Raghuvanshi, R.; Bhardwaj, P.; Sood, H.; Saxena, S.; Chaurasia, O.P. Influence of light quality on growth, secondary metabolites production and antioxidant activity in callus culture of Rhodiola imbricata. Edgew. J. Photochem. Photobiol. B 2018, 183, 258–265. [Google Scholar] [CrossRef]
- Lanfer-Marquez, U.M.; Barros, R.M.C.; Sinnecker, P. Antioxidant activity of chlorophylls and their derivatives. Food Res. Int. 2005, 38, 885–891. [Google Scholar] [CrossRef]
- Eggersdorfer, M.; Wyss, A. Carotenoids in human nutrition and health. Arch. Biochem. Biophys. 2018, 652, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Sgherri, C.; Pinzino, C.; Navari-Izzo, F.; Izzo, R. Contribution of major lipophilic antioxidants to the antioxidant activity of basil extracts: An EPR study. J. Sci. Food Agric. 2011, 91, 1128–1134. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Li, C.; Li, D.; Wang, J.; Yang, L.; Qu, A.; Wu, Q. Physiological, metabolic and transcriptional responses of basil (Ocimum basilicum Linn. var. pilosum (Willd.) Benth.) to heat stress. Agronomy 2022, 12, 1434. [Google Scholar]
- Xiao, Y.; Chu, L.; Zhang, Y.; Bian, Y.; Xiao, J.; Xu, D. HY5: A pivotal regulator of light-dependent development in higher plants. Front. Plant Sci. 2021, 12, 800989. [Google Scholar] [CrossRef]
- Fiorucci, A.-S.; Michaud, O.; Schmid-Siegert, E.; Trevisan, M.; Allenbach Petrolati, L.; Çaka Ince, Y.; Fankhauser, C. Shade suppresses wound-induced leaf repositioning through a mechanism involving PHYTOCHROME KINASE SUBSTRATE (PKS) genes. PLoS Genet. 2022, 18, e1010213. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, B.; Su, J.; Liao, J.; Lin, C.; Oka, Y. Cryptochromes orchestrate transcription regulation of diverse blue light responses in plants. Photochem. Photobiol. 2017, 93, 112–127. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Q.; Ye, R.; Duan, J.; Lin, D.; Jia, Y.; Dang, F.; Han, T. Physiological, Transcriptomic and Metabolomic Response of Basil (O. basilicum Linn. var. pilosum (Willd.) Benth.) to Red and Blue Light. Horticulturae 2023, 9, 1172. https://doi.org/10.3390/horticulturae9111172
Wu Q, Ye R, Duan J, Lin D, Jia Y, Dang F, Han T. Physiological, Transcriptomic and Metabolomic Response of Basil (O. basilicum Linn. var. pilosum (Willd.) Benth.) to Red and Blue Light. Horticulturae. 2023; 9(11):1172. https://doi.org/10.3390/horticulturae9111172
Chicago/Turabian StyleWu, Qingfei, Rigui Ye, Jingmian Duan, Duo Lin, Yuru Jia, Fengfeng Dang, and Tiantian Han. 2023. "Physiological, Transcriptomic and Metabolomic Response of Basil (O. basilicum Linn. var. pilosum (Willd.) Benth.) to Red and Blue Light" Horticulturae 9, no. 11: 1172. https://doi.org/10.3390/horticulturae9111172
APA StyleWu, Q., Ye, R., Duan, J., Lin, D., Jia, Y., Dang, F., & Han, T. (2023). Physiological, Transcriptomic and Metabolomic Response of Basil (O. basilicum Linn. var. pilosum (Willd.) Benth.) to Red and Blue Light. Horticulturae, 9(11), 1172. https://doi.org/10.3390/horticulturae9111172




