Genome-Wide Identification and Expression Analysis of the CmHAK Gene Family in Melon (Cucumis melo L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genome Wide Identification of CmHAK in Melon
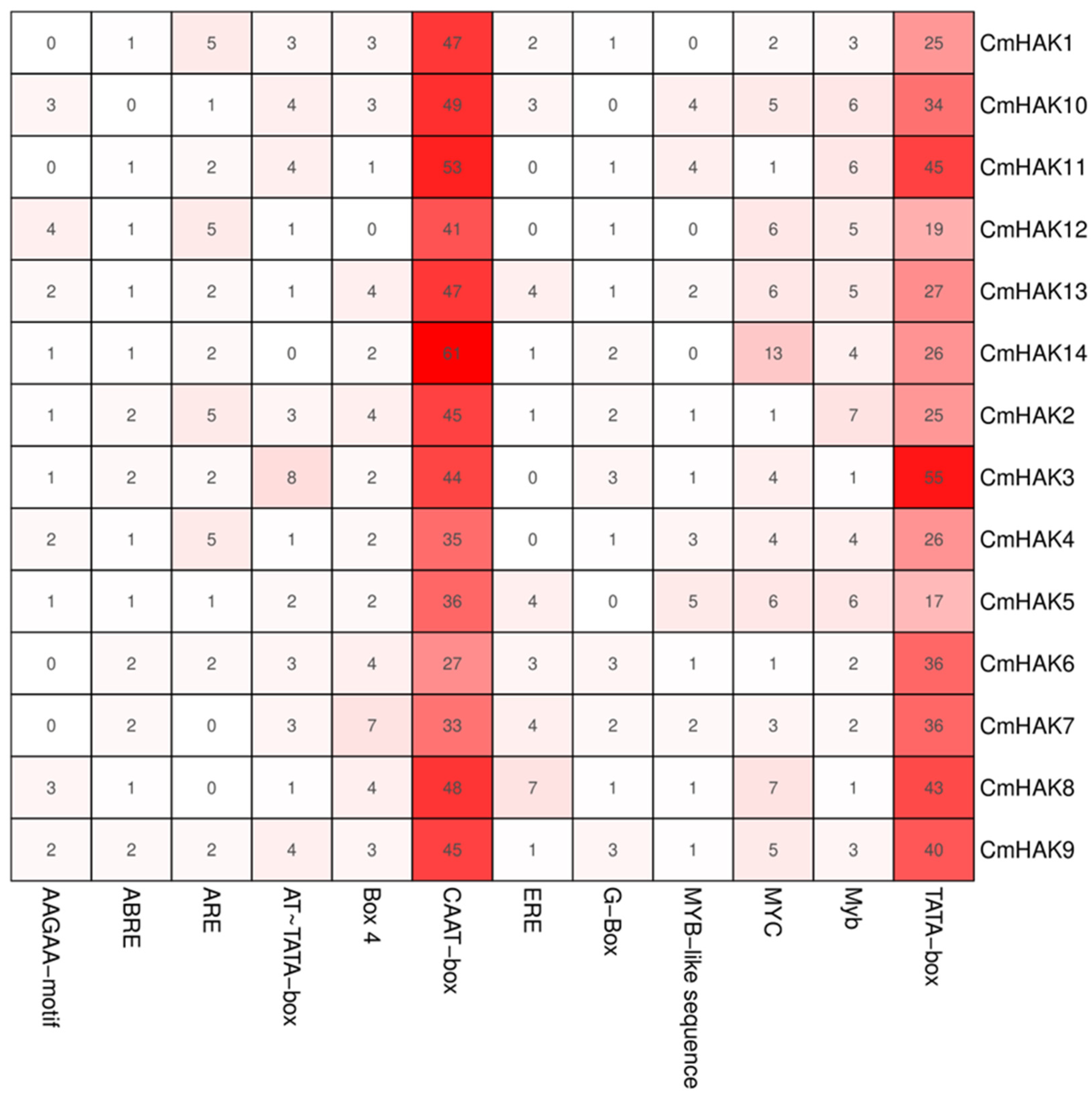
2.2. Classification, Structural, and Cis-Element of the Promoter Region Analysis of CmHAK Genes
2.3. Phylogenetic Tree of CmHAK Genes
2.4. Collinearity, Ka/Ks, and Expression Pattern Analysis
3. Results
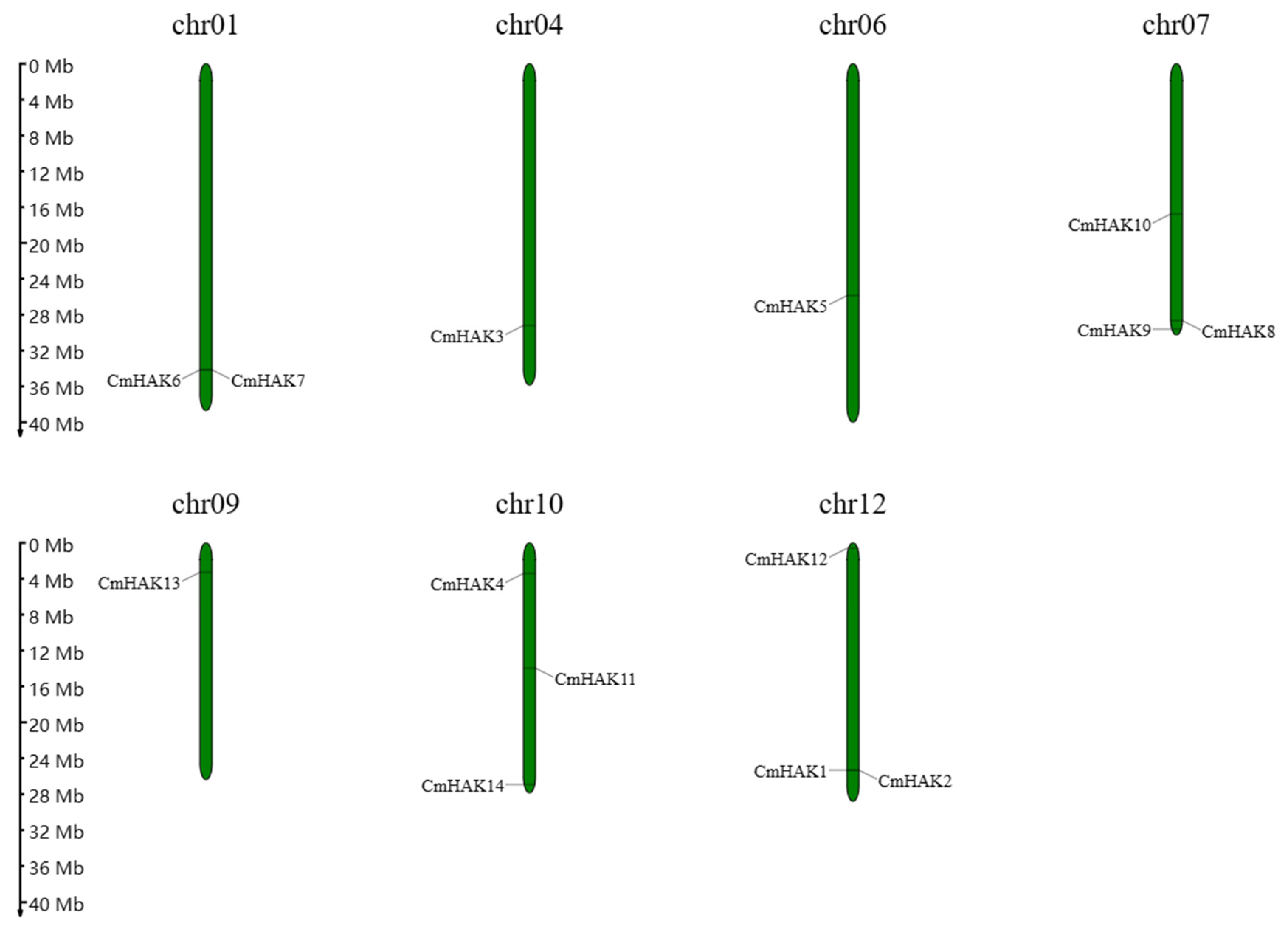
3.1. Identification of CmHAK Genes
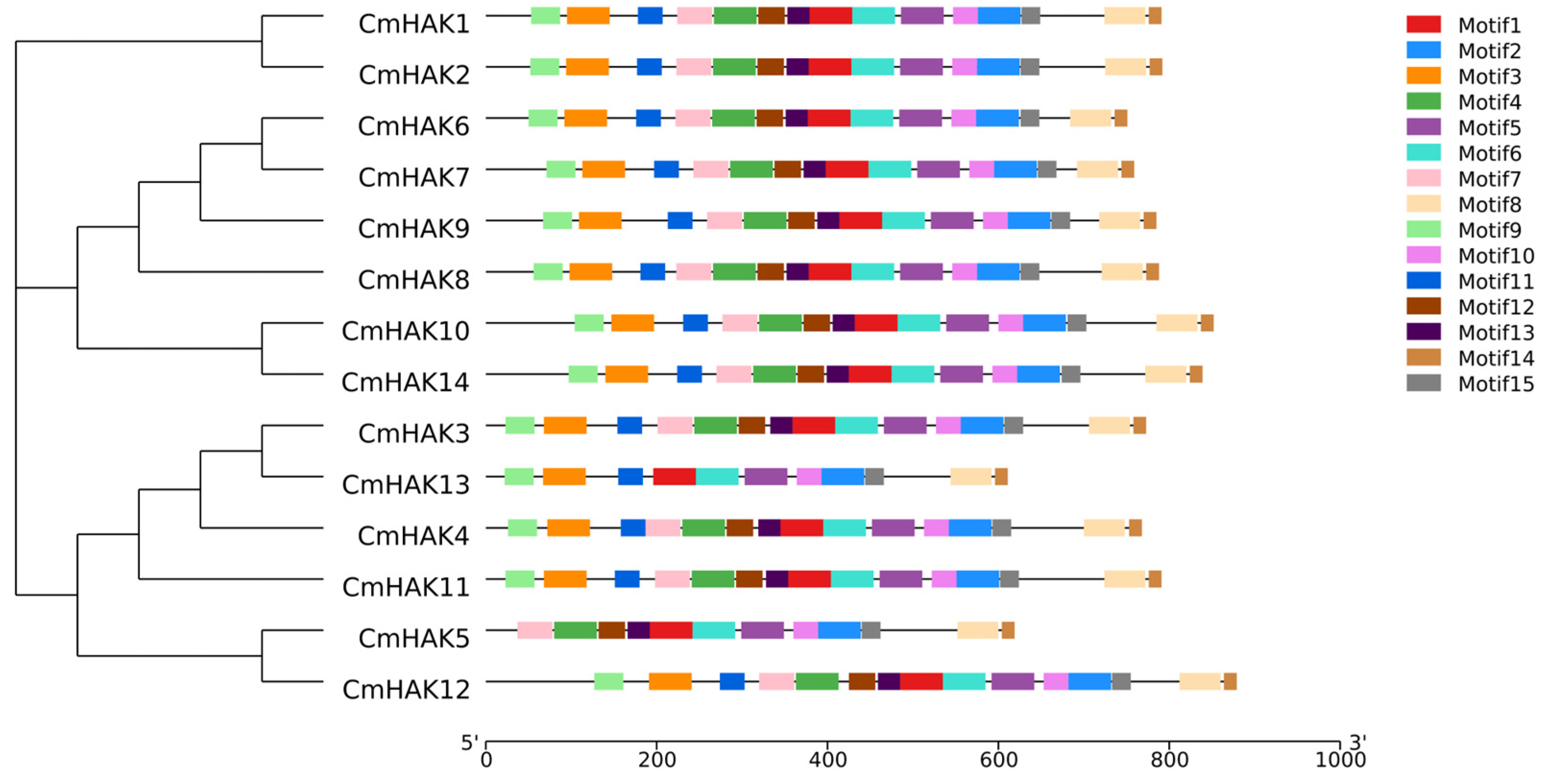
3.2. Classification and Structural Analysis of CmHAK Genes
3.3. Phylogenetic Analysis of CmHAK Genes
3.4. Collinearity and Ka/Ks Analysis of CmHAK
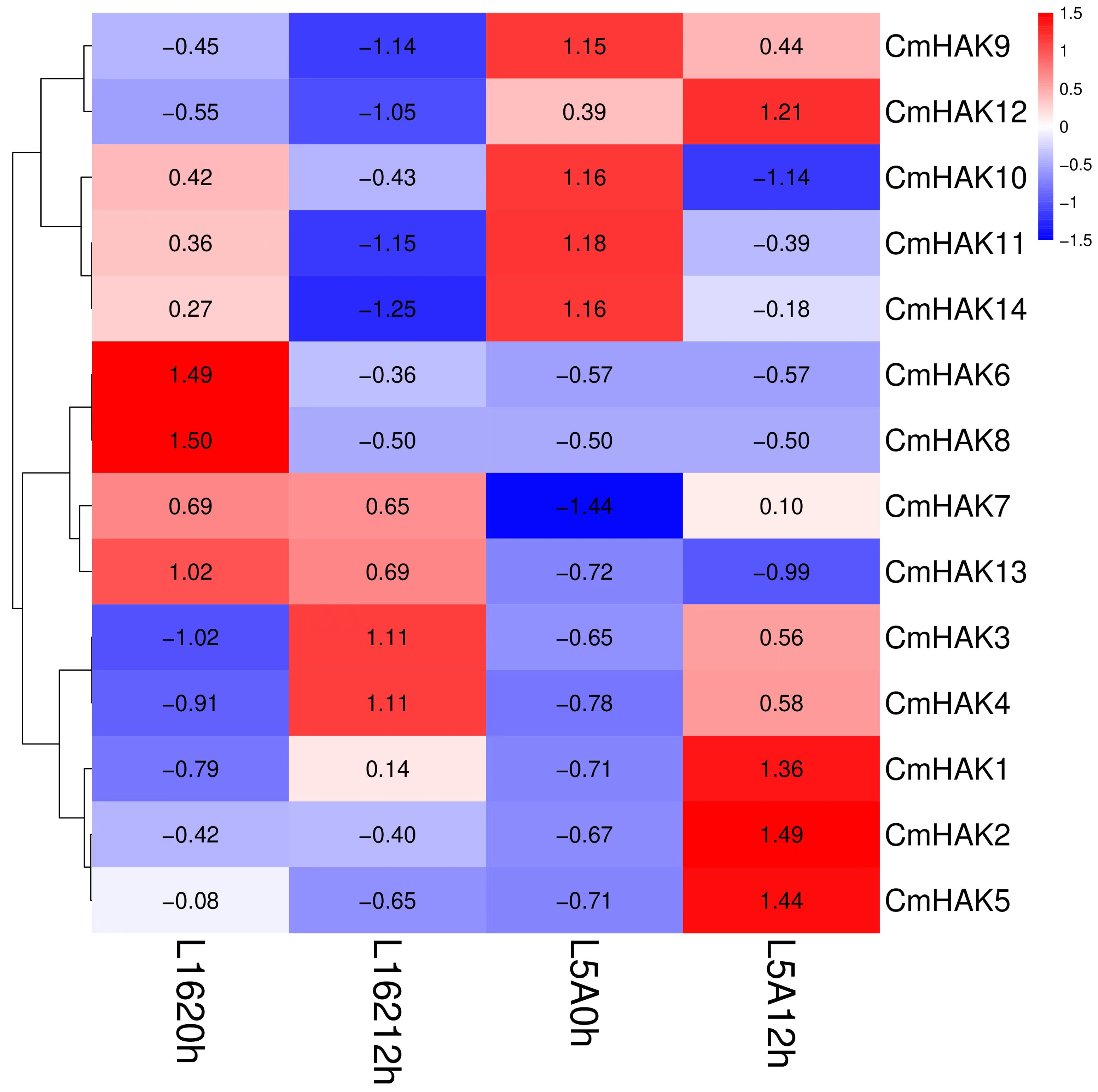
3.5. Gene Expression Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fatima, P.; Nadeem, M.; Hussain, A.; Kausar, T.; Rehman, A.; Siddique, T.; Kabir, K.; Noreen, S.; Nisar, R.; Fatima, H.; et al. Synergistic effect of microwave heating and thermosonication on the physicochemical and nutritional quality of muskmelon and sugarcane juice blend. Food Chem. 2023, 425, 136489. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Bi, Y.; Ackah, S.; Li, Z.; Li, B.; Wang, B.; Wang, Y.; Li, Y.; Prusky, D. Sodium silicate treatment accelerates biosynthesis and polymerization of suberin polyaliphatics monomers at wounds of muskmelon. Food Chem. 2023, 417, 135847. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, X.; Liu, S.; Zhu, X.; Xia, J. First report of muskmelon fruit rot caused by Fusarium sulawesiense in China. Plant Dis. 2023. [Google Scholar] [CrossRef] [PubMed]
- Vella, F.M.; Calandrelli, R.; Cautela, D.; Laratta, B. Natural antioxidant potential of melon peels for fortified foods. Foods 2023, 12, 2523. [Google Scholar] [CrossRef] [PubMed]
- Ankit, A.; Kamali, S.; Singh, A. Genomic & structural diversity and functional role of potassium (K) transport proteins in plants. Int. J. Biol. Macromol. 2022, 208, 844–857. [Google Scholar] [PubMed]
- Liang, M.; Gao, Y.; Mao, T.; Zhang, X.; Zhang, S.; Zhang, H.; Song, Z. Characterization and expression of KT/HAK/KUP transporter family genes in willow under potassium deficiency, drought, and salt stresses. BioMed Res. Int. 2020, 2020, 2690760. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, L.; Zhao, P.; Liu, Z.; Guo, S.; Li, Y.; Liu, H. Genome-wide identification, characterization and expression analysis of HAK genes and decoding their role in responding to potassium deficiency and abiotic stress in Medicago truncatula. PeerJ 2022, 10, e14034. [Google Scholar] [CrossRef]
- Yang, T.; Lu, X.; Wang, Y.; Xie, Y.; Ma, J.; Cheng, X.; Xia, E.; Wan, X.; Zhang, Z. HAK/KUP/KT family potassium transporter genes are involved in potassium deficiency and stress responses in tea plants (Camellia sinensis L.): Expression and functional analysis. BMC Genom. 2020, 21, 556. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Wei, Y.; Meng, J.; Zhong, C.; Fan, C. Characterization of HAK protein family in Casuarina equisetifolia and the positive regulatory role of CeqHAK6 and CeqHAK11 genes in response to salt tolerance. Front. Plant Sci. 2023, 13, 1084337. [Google Scholar] [CrossRef]
- Shen, L.; Fan, W.; Li, N.; Wu, Q.; Chen, D.; Luan, J.; Zhang, G.; Tian, Q.; Jing, W.; Zhang, Q.; et al. Rice potassium transporter OsHAK18 mediates phloem K loading and redistribution. Plant J. 2023, 116, 201–216. [Google Scholar] [CrossRef]
- Templalexis, D.; Tsitsekian, D.; Liu, C.; Daras, G.; Šimura, J.; Moschou, P.; Ljung, K.; Hatzopoulos, P.; Rigas, S. Potassium transporter TRH1/KUP4 contributes to distinct auxin-mediated root system architecture responses. Plant Physiol. 2022, 188, 1043–1060. [Google Scholar] [CrossRef] [PubMed]
- Amo, J.; Lara, A.; Martínez-Martínez, A.; Martínez, V.; Rubio, F.; Nieves-Cordones, M. The protein kinase SlCIPK23 boosts K+ and Na+ uptake in tomato plants. Plant Cell Environ. 2021, 44, 3589–3605. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Xiao, H.; Li, R.; Zeng, Y.; Gu, M.; Moran, N.; Yu, L.; Xu, G. Potassium transporter OsHAK18 mediates potassium and sodium circulation and sugar translocation in rice. Plant Physiol. 2023, kiad435. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Guo, J.; Ma, T.; Liu, S.; Zhou, Y.; Yang, X.; Li, Q.; Yu, K.; Wang, T.; He, S.; et al. The Sweet Potato K+ Transporter IbHAK11 Regulates K+ Deficiency and High Salinity Stress Tolerance by Maintaining Positive Ion Homeostasis. Plants 2023, 12, 2422. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Tang, Q.; Cai, J.; Xu, B.; Xu, G.; Yu, L. Rice OsHAK16 functions in potassium uptake and translocation in shoot, maintaining potassium homeostasis and salt tolerance. Planta 2019, 250, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Jiang, W.; Yan, M.; Zhang, A.; Liu, M.; Zhao, P.; Chen, X.; Tang, Z. Genome-wide characterization and expression analysis of HAK K+ transport family in Ipomoea. 3 Biotech 2021, 11, 3. [Google Scholar] [CrossRef]
- Cheng, X.; Liu, X.; Mao, W.; Zhang, X.; Chen, S.; Zhan, K.; Bi, H.; Xu, H. Genome-Wide Identification and Analysis of HAK/KUP/KT Potassium Transporters Gene Family in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2018, 19, 3969. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Du, W.; Tian, X.; Jiang, W.; Zhang, B.; Wang, Y.; Pang, Y. Genome-wide characterization and expression analysis of the HAK gene family in response to abiotic stresses in Medicago. BMC Genom. 2022, 23, 791. [Google Scholar] [CrossRef] [PubMed]
- Greiner, T.; Ramos, J.; Alvarez, M.C.; Gurnon, J.R.; Kang, M.; Van Etten, J.L.; Moroni, A.; Thiel, G. Functional HAK/KUP/KT-like potassium transporter encoded by chlorella viruses. Plant J. 2011, 68, 977–986. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, Y.; Wang, X.; Hong, Z.; Yang, A.; Liu, Y.; Yan, L.; He, Y.; Zhu, Z.; Wang, H. PACLOBUTRAZOL-RESISTANCE4 positively regulates cell expansion to promote tendril elongation in cucumber. Plant Physiol. 2023, 192, 2756–2767. [Google Scholar] [CrossRef]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The arabidopsis information resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, 1202–1210. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Mao, Q.; Ma, R.; Xu, J.; Yu, M. Genome-Wide Identification and Expression Analysis of the PpYUCCA Gene Family in Weeping Peach Trees (Prunus persica ‘Pendula’). Horticulturae 2022, 8, 878. [Google Scholar] [CrossRef]
- Wu, R.; Ran, K.; Zhao, S.; Cheng, F. Genome-Wide Identification of the Light-Harvesting Chlorophyll a/b Binding Protein Gene Family in Pyrus bretschneideri and Their Transcriptomic Features under Drought Stress. Horticulturae 2023, 9, 522. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Yang, D.C.; Meng, Y.Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020, 48, 1104–1113. [Google Scholar] [CrossRef]
- Hung, J.H.; Weng, Z. Sequence Alignment and Homology Search with BLAST and ClustalW. Cold Spring Harb. Protoc. 2016, 11. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.K.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef]
- Zhi, Q.; Hampton, C.R.; Shin, R.; Barkla, B.J.; White, P.J.; Schachtman, D.P. The high affinity K+ transporter AtHAK5 plays a physiological role in planta at very low K+ concentrations and provides a caesium uptake pathway in Arabidopsis. J. Exp. Bot. 2008, 59, 595–607. [Google Scholar]
- Desbrosses, G.; Josefsson, C.; Rigas, S.; Hatzopoulos, P.; Dolan, L. AKT1 and TRH1 are required during root hair elongation in Arabidopsis. J. Exp. Bot. 2003, 54, 781–788. [Google Scholar] [CrossRef] [PubMed]
- Vicente-Agullo, F.; Rigas, S.; Desbrosses, G.; Dolan, L.; Hatzopoulos, P.; Grabov, A. Potassium carrier TRH1 is required for auxin transport in Arabidopsis roots. Plant J. 2004, 40, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Gierth, M.; Mäser, P.; Schroeder, J.I. The potassium transporter AtHAK5 functions in K(+) deprivation-induced high-affinity K(+) uptake and AKT1 K(+) channel contribution to K(+) uptake kinetics in Arabidopsis roots. Plant Physiol. 2005, 137, 1105–1114. [Google Scholar] [CrossRef]
- Wang, Y.; Lü, J.; Chen, D.; Zhang, J.; Qi, K.; Cheng, R.; Zhang, H.; Zhang, S. Genome-wide identification, evolution, and expression analysis of the KT/HAK/KUP family in pear. Genome 2018, 61, 755–765. [Google Scholar] [CrossRef]
- Gupta, M.; Qiu, X.; Wang, L.; Xie, W.; Zhang, C.; Xiong, L.; Lian, X.; Zhang, Q. KT/HAK/KUP potassium transporters gene family and their whole-life cycle expression profile in rice (Oryza sativa). Mol. Genet. Genom. 2008, 280, 437–452. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, J.; Chen, Y.; Li, R.; Wang, H.; Wei, J. Genome-wide analysis and identification of HAK potassium transporter gene family in maize (Zea mays L.). Mol. Biol. Rep. 2012, 39, 8465–8473. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Elumalai, R.P.; Nagpal, P.; Reed, J.W. A mutation in the Arabidopsis KT2/KUP2 potassium transporter gene affects shoot cell expansion. Plant Cell 2002, 14, 119–131. [Google Scholar] [CrossRef]
- Rigas, S.; Debrosses, G.; Haralampidis, K.; Vicente-Agullo, F.; Feldmann, K.A.; Grabov, A.; Dolan, L.; Hatzopoulos, P. TRH1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. Plant Cell 2001, 13, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Arinaga, N.; Umezawa, T.; Katsura, S.; Nagamachi, K.; Tanaka, H.; Ohiraki, H.; Yamada, K.; Seo, S.-U.; Abo, M.; et al. Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 2013, 25, 609–624. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Kwak, J.M.; Uozumi, N.; Schroeder, J.I. AtKUP1: An Arabidopsis gene encoding high-affinity potassium transport activity. Plant Cell 1998, 10, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Zhang, S.; Hu, Y.; Wu, F.; Hu, Q.; Chen, G.; Cai, J.; Wu, T.; Moran, N.; Yu, L.; et al. The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 2014, 166, 945–959. [Google Scholar] [CrossRef]
- Takahashi, R.; Nishio, T.; Ichizen, N.; Takano, T. Cloning and functional analysis of the K+ transporter, PhaHAK2, from salt-sensitive and salt-tolerant reed plants. Biotechnol. Lett. 2007, 29, 501–506. [Google Scholar] [CrossRef]
- Maathuis, F.J.M. The role of monovalent cation transporters in plant responses to salinity. J. Exp. Bot. 2006, 57, 1137–1147. [Google Scholar] [CrossRef]
- Yang, T.; Feng, H.; Zhang, S.; Xiao, H.; Hu, Q.; Chen, G.; Xuan, W.; Moran, N.; Murphy, A.; Yu, L.; et al. The Potassium Transporter OsHAK5 Alters Rice Architecture via ATP-Dependent Transmembrane Auxin Fluxes. Plant Commun. 2020, 1, 1000–1005. [Google Scholar] [CrossRef]




| Name | Gene ID | Protein Number | Number of Amino Acid | Molecular Weight (Da) | PI | Instability Index | Aliphatic Index | Gravy | Number of Predicted TMRs |
|---|---|---|---|---|---|---|---|---|---|
| CmHAK1 | MELO3C002408.2 | MELO3C002408.2.1 | 790 | 88,244.04 | 6.93 | 38.39 | 109.38 | 0.40 | 12 |
| CmHAK2 | MELO3C002409.2 | MELO3C002409.2.1 | 791 | 88,627.52 | 8.65 | 36.49 | 109.20 | 0.35 | 12 |
| CmHAK3 | MELO3C009941.2 | MELO3C009941.2.1 | 772 | 86,450.17 | 8.69 | 37.60 | 105.76 | 0.285 | 12 |
| CmHAK4 | MELO3C012009.2 | MELO3C012009.2.1 | 767 | 86,166.87 | 8.06 | 39.72 | 108.23 | 0.308 | 12 |
| CmHAK5 | MELO3C014763.2 | MELO3C014763.2.1 | 618 | 68,789.20 | 9.15 | 45.90 | 115.74 | 0.532 | 10 |
| CmHAK6 | MELO3C016040.2 | MELO3C016040.2.1 | 750 | 84,726.73 | 6.39 | 36.08 | 113.67 | 0.286 | 12 |
| CmHAK7 | MELO3C016041.2 | MELO3C016041.2.1 | 758 | 85,178.40 | 8.94 | 41.16 | 107.10 | 0.250 | 12 |
| CmHAK8 | MELO3C017828.2 | MELO3C017828.2.1 | 787 | 86,897.46 | 8.92 | 27.54 | 109.43 | 0.393 | 12 |
| CmHAK9 | MELO3C017974.2 | MELO3C017974.2.1 | 784 | 88,060.25 | 6.96 | 33.00 | 103.38 | 0.197 | 12 |
| CmHAK10 | MELO3C019015.2 | MELO3C019015.2.1 | 851 | 94,840.64 | 5.48 | 35.32 | 108.57 | 0.341 | 12 |
| CmHAK11 | MELO3C020132.2 | MELO3C020132.2.1 | 790 | 88,054.72 | 6.97 | 38.73 | 110.53 | 0.381 | 12 |
| CmHAK12 | MELO3C020532.2 | MELO3C020532.2.1 | 878 | 99,060.22 | 7.94 | 37.14 | 108.42 | 0.393 | 12 |
| CmHAK13 | MELO3C021533.2 | MELO3C021533.2.1 | 610 | 68,685.45 | 8.68 | 34.01 | 106.89 | 0.259 | 8 |
| CmHAK14 | MELO3C026470.2 | MELO3C026470.2.1 | 838 | 93,447.33 | 6.24 | 39.95 | 106.78 | 0.36 | 12 |
| Sequence | Ka | Ks | Ka/Ks | p-Value (Fisher) | Length |
|---|---|---|---|---|---|
| CmHAK13-CmHAK3 | 0.142624 | 1.83892 | 0.077559 | 5.69E-225 | 1821 |
| CmHAK13-CmHAK4 | 0.201892 | 3.51955 | 0.0573628 | 0 | 1824 |
| CmHAK3-CmHAK4 | 0.171156 | 2.35486 | 0.0726821 | 0 | 2262 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, L.; Wang, H.; Leng, X.; Zhang, X.; Xiao, B.; Liu, H.; Xue, D.; Wang, Y.; Wu, C.; Wang, W. Genome-Wide Identification and Expression Analysis of the CmHAK Gene Family in Melon (Cucumis melo L.). Horticulturae 2023, 9, 1138. https://doi.org/10.3390/horticulturae9101138
Fu L, Wang H, Leng X, Zhang X, Xiao B, Liu H, Xue D, Wang Y, Wu C, Wang W. Genome-Wide Identification and Expression Analysis of the CmHAK Gene Family in Melon (Cucumis melo L.). Horticulturae. 2023; 9(10):1138. https://doi.org/10.3390/horticulturae9101138
Chicago/Turabian StyleFu, Lina, Huizhi Wang, Xifang Leng, Xinsheng Zhang, Baoying Xiao, Hui Liu, Dongxu Xue, Yangyang Wang, Chunyan Wu, and Wei Wang. 2023. "Genome-Wide Identification and Expression Analysis of the CmHAK Gene Family in Melon (Cucumis melo L.)" Horticulturae 9, no. 10: 1138. https://doi.org/10.3390/horticulturae9101138
APA StyleFu, L., Wang, H., Leng, X., Zhang, X., Xiao, B., Liu, H., Xue, D., Wang, Y., Wu, C., & Wang, W. (2023). Genome-Wide Identification and Expression Analysis of the CmHAK Gene Family in Melon (Cucumis melo L.). Horticulturae, 9(10), 1138. https://doi.org/10.3390/horticulturae9101138





