Abstract
Increased consumption of vegetables has been recommended worldwide as a part of a healthy diet; therefore, determining gene function among breeding materials is crucial for vegetable improvement to meet the sustainable development of new vegetable varieties. However, genetic transformation is time-consuming and laborious, which limits the exploration of gene function for various vegetable crops. Virus-Induced Gene Silencing (VIGS) can perform large-scale and rapid gene silencing in plants due to a reduction in the experimental period and its independence from the stable genetic transformation, providing an excellent opportunity for functional research. VIGS can accelerate model plant research and make it easier to analyze gene function and validation in vegetable crops. Moreover, with the advent of technologies such as virus-mediated heterologous protein expression and the development of CRISPR/Cas9 technology, virus-mediated genetic tools have ushered in a new era in genetics and crop improvement. This study summarizes recent achievements in VIGS and Virus-Induced Gene Editing (VIGE) in vegetables. We also identify several challenges in the current state of VIGS technology in vegetables, serving as a guide for future research.
1. Virus-Induced Gene Silencing (VIGS) System
Vegetables are grown worldwide and play an important role in the nutrition demands of humans’ daily diets, especially for providing vitamins, minerals, and dietary fiber, which have been strongly associated with human health. They can also be a major source of protein in poor regions. The continuous increase in human living standards, along with increasing demand for vegetable production and quality, make the improvement of molecular breeding technologies applied to vegetable breeding a need to achieve more efficient and sustainable crop production. This market demand for high-quality and more uniform products, together with global warming, oblige scientists to explore gene functions which are important for agronomic traits (e.g., disease, pest, or abiotic stress resistance) in vegetables. Traditional methods to study plant gene function include transgenic technology, gene knockout, gene-induced overexpression, and RNAi technology. These research methods have certain limitations, such as long research cycles, the need for genetic transformation, and low conversion efficiency, limiting rapid and efficient study of plant gene functions [1,2]. However, Virus-Induced Gene Silencing (VIGS) provides an alternative tool to investigate gene functional validation in vegetables.
VIGS is an effective method for switching off the expression of a gene. It was developed based on the mechanism of plants’ defenses against viruses, using RNA-mediated post-transcriptional gene silencing (PTGS) [3,4,5]. It has emerged rapidly as a key regulator of gene expression applicable to reverse genetics for plant gene functional studies. Plant scientists discovered gene silencing-related mechanisms while performing plant transformation experiments in which the introduction of a transgene into a desired genome resulted in the silencing of both the transgene and its homologous endogenes [6,7]. As a result of these first observations, plant geneticists and biologists developed this molecular biology approach to address not only gene silencing method questions, but also to explore the complexity of the biological pathways involved, as well as to demonstrate their multilayer relationships with one another. For instance, it is well documented that, after the virus infests a plant, viral transcription and replication in the plant cell cytoplasm lead to a double-stranded RNA (dsRNA), which is key in the VIGS process. dsRNA was cleaved into small interfering RNA (siRNA) by the Dicer or Dicer-like (DCL) nuclease, ranging from 21 to 24 integrated nucleotides, and siRNA binds to RNase in plants as a single strand to form an RNA-induced silencing complex (RISC). Further, the RNA-induced silencing complexes (RISCs) cleave to viral RNA in the cytoplasm in a nucleotide-specific manner, ultimately triggering the degradation of the targeted mRNA [8,9,10,11,12] (Figure 1). RNA silencing is an evolutionarily conserved RNA-mediated process [13], where sequence-specific eukaryotic gene silencing mechanisms are involved in numerous biological processes in plants and animals [14]. Therefore, virus infection has been proven as an efficient trigger of RNA silencing, turning VIGS into a powerful tool for gene function studies and vegetable improvement.
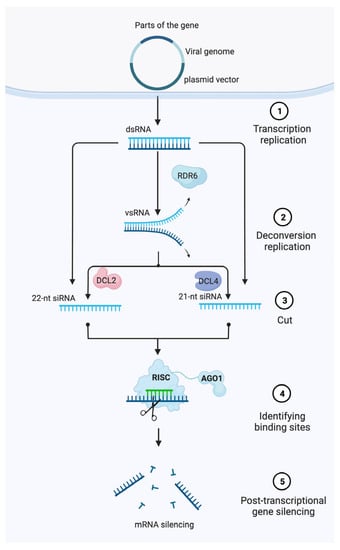
Figure 1.
Model of post-transcriptional gene silencing (PTGS)-mediated gene silencing in plants. First, a partial segment (~200–500 bp) homologous to the target gene of interest is cloned into a modified single/multipartite viral genome harbored within a plasmid vector. Then, Agrobacteria are used to transfect plant cells and transfer DNA from the binary vector into the nucleus where it is transiently expressed. Finally, dsRNA formed during virus replication are cleaved by DICER proteins to produce siRNAs that guide RISC and the local amplification of siRNAs that systemically spread to induce post-transcriptional gene silencing of a target mRNA. Abbreviations: RISC, RNA-induced silencing complex; siRNA, small-interfering RNA; dsRNA, double-stranded RNA; DCL, DICER-Like.
In this study, we (1) described the advantages of current VIGS applications in plants; (2) review the VIGS vectors successfully used to study gene function in vegetables; and (3) discuss the improvements in VIGS technology and its potential application in the future crop enhancement.
2. Advantages of VIGS
2.1. Transient Silence of VIGS
Transient silencing of VIGS to analyze plant endogenous gene function is a fast and effective reverse genetic tool in plant functional genomics. This is possible since phenotypic changes induced by the down-regulation of endogenous plant genes can be detected in a short period of time [1,15,16]. The effective silencing time and the effectiveness of viral vectors differ depending on the viral vector and the target plant infested [2,17,18]. For instance, photobleaching in leaves, stems, axillary buds, and sepals of Tobacco Rattle Virus (TRV)-based VIGS system, using phytoene desaturase (PDS) as a reporter gene, infiltrated tobacco (Nicotiana benthamiana) plants appeared 10 days after TRV infestation [19]. Another observation was that the percentage of white tissue in leaves decreased 28 days after infestation [5]. A similar experiment in tomato reported that the transient silencing response of TRV–PDS sprayed on 4-week-old tomato seedlings showed symptoms of photobleaching caused by Silene latifolia (Sl) PDS gene silencing after ~8 weeks of leaf inoculation. The systemic photobleaching persisted throughout the experiment for 4 months after the inoculation [19].
In another experiment to assess silencing by Agrobacterium-mediated barley stripe mosaic virus (BSMV) VIGS, a 370 bp PDS (NbPDS) fragment from N.benthamiana was cloned into pCa-cbLIC to generate pCa-cb:NbPDS370. Then, the four-leaf stage of N. benthamiana was infiltrated with Agrobacterium mixtures containing pCaBS-a, pCaBS-b, and pCa-cb: NbPDS370. Leaves infiltrated with virus to elicit PDS silencing developed a mottled photobleaching phenotype on the fifth or sixth leaves at 9 to 10 dpi, and, about 5 days later (15 dpi), larger (and more uniform) white PDS silencing areas were observed at the 6- to 8-leaf-stage. Furthermore, PDS silencing was most pronounced at 30 to 45 dpi, with larger and more apparent areas of photobleaching on many stems and petioles [20].
2.2. VIGS Overcomes Functional Redundancy
Determination of gene function is particularly problematic when studying large gene families because two or more genes could perform the same function, either by gene copy duplication or a higher ploidy level. The inactivation of one of these genes has little or no effect on the phenotypic appearance; thus, gene redundancy limits the ability to experimentally assess the contributions of individual genes. However, VIGS can overcome this gene function redundancy by constructing the viral vector carrying highly conserved regions of the target gene family and potentially knocking off all the family members [1,21]. One example is the heat shock protein 90 (HSP90) that belongs to a large gene family of transcription factors that control fundamental processes of plant development. An insertion of the highly conserved coding sequence of the HSP90 gene family into the Potato Virus X (PVX) viral vector silenced all HSP90 mRNAs and was confirmed by protein blotting in tomato. Lack of HSP90 protein led to stunted development and leaf deformation plant phenotypes. Therefore, the use of VIGS technology allowed us to demonstrate how HSP90 protein likely had a key role in tomato growth and development [2].
VIGS can also overcome the redundancy issue in polyploid species. Cabbage (Brassica rapa L.) is a globally significant vegetable crop (71 million tonnes per year) [22], where its ploidy level, high gene duplication rate, and long growth cycle have posed challenges for stable genetic transformation, greatly limiting study at the gene functional level. To overcome this challenge, a VIGS system of cabbage was constructed using the Tomato Yellow Leaf Curl Virus (TYLCV) viral vector. This tailored molecular biology approach allowed us to demonstrate, for example, how the gene Basic Helix–Loop–Helix transcription factor, BcbHLHpol, regulates pollen development and the fact that it is likely activated at low temperatures as an essential step in meiosis [23].
VIGS was also used to investigate the role of transcription factors (TFs) synchronized with the expression of genes related to programmed cell death (PCD) during PCD and salt stress. Knockdown mutants of these TFs were generated in tobacco by modifying the TRV and utilizing VIGS to produce knockout mutants of these TFs in tobacco. Results of knockdown mutant tobacco cells confirmed the influence of two TFs during PCD. In addition, the knockout insertion mutants and overexpression lines indicated the role of ERF109 in conferring salt tolerance in Arabidopsis [24].
2.3. VIGS Overcomes Conditional Constraints
CRISPR/Cas9 technology is widely used for gene validation by performing gene knockouts at the DNA level. Although it is a powerful technology, it may not be suitable when investigating essential genes that have been shown to be plantlet lethal (in the knockout stage) during the regeneration of plant transformation [21]. The main advantage of VIGS is that it can effectively down-regulate the expression of those same essential genes and can provide a better understanding of gene effects’ influence on the phenotype, primarily by taking advantage of post-translation regulation impacts of reducing protein level expression [1,2,25]. Another benefit is that, because the knockdown regulation is temporary, it can return to normal growth and seed production and does not retain the virus or vector components [26].
One example that highlights the power of VIGS as a tool to study temporarily inhibited gene expression is the Proliferating Cell Nuclear Antigen (PCNA), which is essential for host cell growth and development. For PCNA, most mutations are lethal and difficult to retrieve. Therefore, gene functional verification cannot be performed by transgenic silencing. PCNA is an important component in the replication and repair machinery involved in nucleic acid metabolism [27]. PCNA contributes to the persistent DNA polymerase δ and DNA polymerase ε synthesis factor that attaches the polymerase catalytic unit to the DNA template for rapid and sustained DNA synthesis. Knockout of PCNA in plants by CRISPR/Cas9 methods leads to death during regeneration, providing only partial information on the gene function due to the scarcity of phenotypes [28]. In contrast, using VIGS technology to silence the PCNA gene in tomato permitted the screening of the whole set of individuals tested. This essay resulted in severely stunted growth of infested tomato plants with the VIGS–PCNA viral vector, in contrast to no morphological effects observed in an empty vector plant test with VIGS–GFP as the reported gene. This proved the importance of the virus-induced gene silencing technology in demonstrating the causality of the PCNA gene in tomatoes [29].
2.4. Disadvantages VIGS
When performing a comparison of VIGS technologies, their main disadvantages are that most viruses used for VIGS have a limited number of hosts, and the virus–host combination seems to be a crucial factor in determining the efficacy of silencing. Some of the viruses used in VIGS can cause symptoms that might mask the phenotype caused by the silencing of the target gene. Moreover, many viruses do not infect the growing points or floral parts of plants, especially the seed, precluding gene silencing in these tissues [30].
3. VIGS Applications in Vegetable Plants
To date, many plant viruses have been successfully modified as VIGS vectors to induce targeted gene silencing in host vegetable plants (Table 1), such as tobacco mosaic virus (TMV), PVX, and TRV. Among them, TRV is especially widely used in Solanaceae vegetables, and gene silencing can be effectively induced by constructing recombinant TRV virus vectors [10,31,32].
TRV vector has been successfully applied in several plant organs (leaf, root, and flower), affecting key aspects of plant nutritional growth and reproductive stages [5]. Recently, studies have shown that this same technology can be applied to fruits, for example, tomato or pepper [33]. In tomato, the characteristic bleaching phenotype after TRV–PDS injection was obtained and those symptoms expanded, infesting peduncles at the tomato fruit developmental stage. Gene silencing was confirmed at the molecular level by qPCR. In pepper, an optimized TRV vector was developed using a Viral Silencing suppressor of RNA silencing (VSR). pTRV2-C2b-CaCCS vector was constructed, targeting a key gene in capsanthin/capsorubin biosynthesis that achieved high efficiency of calcium-activated chloride channels’ (CaCCS) protein silencing [33]. Another example of studying fruit organs was the silencing of the tomato ethylene (EIN3)-binding F-box genes. SlEBF1 and SlEBF2 have been reported to negatively regulate ethylene signaling, causing constitutive ethylene-related symptoms, fertility defects, growth decline, plant senescence acceleration, and fruit ripening [34]. Altogether, these examples show the impact this molecular gene silencing advancement can have, to better explain gene function validation throughout the whole vegetable life cycle.

Table 1.
Overview of the characteristics of VIGS applied in vegetable crops.
Table 1.
Overview of the characteristics of VIGS applied in vegetable crops.
| Viral Vectors | Host Range | Virus Symptoms | Features | Reference |
|---|---|---|---|---|
| TRV | Solanaceae, Asteraceae, Leguminosae, etc. More than 12 families and 60 species | Minor | The VIGS expression system has been successfully established in a wide range of hosts, while the effectiveness in cucurbits needs further validation. | [33,35,36,37] |
| ALSV | Solanaceae, Leguminosae, Cucurbitaceae, Brassicaceae, etc. | No symptoms | Long-term effective induction of stable virus-induced gene silencing, but the expression of the viral genome needs to be processed by a dedicated protease, limiting its application. | [38,39,40] |
| TRSV | Leguminosae, Cucurbitaceae, etc. | Minor | Silencing efficiency was high in both model plants and crops, but the infestation feasibility of TRSV’s infestation clones in watermelon was not confirmed. | [41,42,43] |
| CGMMV | Cucurbitaceae | Minor | CGMMV is a single RNA virus, and, although it is easy to manipulate, the silencing effect is limited to the vicinity of leaf veins. | [12,44] |
| ToLCV | Solanaceae | Variable | The vector is able to replicate, in different plant species, and efficiently silences PCNA isogenes in the host plant. | [29] |
| PVX | Solanaceae | Moderate | The vector is more stable than TMV-based vectors, but the virus is excluded from the host’s growth sites or hyphal tissues. | [45,46,47] |
TRV: Tobacco brittle virus; ALSV: Apple latent spherical virus; TRSV: Tobacco ringspot virus; CGMMV: Cucumber green mottled mosaic virus; ToLCV: Tomato curly leaf virus; PVX: Potato X virus.
VIGS applications in vegetables have been challenged by the host-range reduced diversity. As a matter of fact, TRV’s host-range reduced diversity has restricted the ability to test gene silencing effects in the Cucurbitaceae family. The discovery and modification ability of plant viruses has allowed using a broader host range of target vegetables, e.g., apple latent spherical virus (ALSV), tobacco ringspot virus (TRSV), cucumber green mottle mosaic virus (CGMMV), and tomato leaf curl virus (ToLCV). ALSV has a wide range of vegetable hosts, including the Solanaceae, the Cucurbitaceae, and the Fabaceae families, most of which have shown no viral symptoms. At the same time, this viral vector was shown to effectively induce stable virus-induced gene silencing in a wide range of vegetable plants and it has been shown to possess long-lasting effects. For example, in pea (Pisum sativum L.), a 300 bp fragment of a PDS gene from soybean plants was inserted into ALSV-RNA2 vectors, and the resulting viruses (soyPDS-ALSV) were inoculated into primary leaves of pea plants. Inoculated pea plants initiated the development of white spots on the third trifoliate true leaf at 10 to 14 dpi and then showed highly uniform white photobleached phenotype in the fourth or fifth true leaves, indicating the PDS gene was silenced. The PDS silencing on these plants persisted for a month [48]. This caused pea death after one month due to the lack of photosynthesis ability. Similar results were obtained when ALSV-CuPDS and ALSV-CuSU vectors were used to infect Cucurbitaceae plants, including pumpkin (Cucurbita maxima L.) having mRNA 76% lower expression levels in the leaf tissues compared to controls after infection [49]. However, one of the disadvantages of the ALSV vector comes from its gene expression strategy of the virus genome. As the proteins encoded by the ALSV genome are expressed by polyprotein synthesis followed by proteolytic processing, it is necessary to ligate target sequences in the frame to the cloning sites of the ALSV vector. This necessity makes it difficult to apply an ALSV vector for high throughput functional genomics, as reported by other vectors [1,2,50].
Another viral vector with a wide host in vegetables is the tobacco ringspot virus (TRSV), a single-stranded positive-sense polyadenylated RNA molecules. This viral vector was first applied to cucurbits and legumes, having silenced all plants with new white leaves, petioles, and even tendrils being almost completely white [51]. Furthermore, the silencing phenotype of the PDS gene was stable and persisted for approximately 1 month [49]. Recent studies have shown the role of the soybean mid–late flow protein gene (GmLATE) in soybean by infesting plants with the VIGS system of TRSV. Researchers concluded that the silencing of GmLATE reduced the expression of flowering-related genes and the arrest of flower development in soybean [52]. They also demonstrated that the silencing effect of this virus vector can remain effective until the reproductive growth stages.
Unlike double-stranded RNA viruses, CGMMV is a positive-sense single-stranded RNA virus with a limited host range that turned out to be able to infect cucurbits [12]. In recent reports, the photobleaching caused by the infection of CGMMV-PDS vector was observed on the third leaf of melon and gourd, and the fifth true leaf of cucumbers. The stability of the photobleaching was variable in watermelon, melon, and cucumber plants at 32, 20, and 39 days, respectively. However, the remaining challenge is that the silencing effect is not as evident in the whole tissue as shown in Liu et al. [12], where the photobleaching phenotype was constrained at the vicinity of leaf veins. The positive side of this technique is the relative easiness of genetic manipulation of the virus vector, making this technique widely utilized in functional genomics on the Cucurbitaceae crop family.
ToLCV and PVX are two additional virus vectors with a wide host range that can be used to verify the gene function in tomatoes. The ToLCV vector belongs to the genus begomoviridae of the family Geminiviridae and was used to silence the PCNA endogenous gene in tomato, resulting in substantial stunting of the plant growth. Interestingly, the vector’s silencing effectiveness was enhanced with the inclusion of a mutation in the silencing suppressor Open Reading Frame (ORF) AC2 [29]. As for PVX, it has been used as a VIGS virus in the Solanaceous genus for a long time [53]. For instance, it was used to study the role SlymiR157 has during the ripening process in tomato. Pre-SlymiR157 was cloned into a PVX-based VIGS vector to produce a PVX/pre-SlymiR157, obtaining a PVX able to efficiently deliver pre-SlymiR157 into fruits [54]. The results of this study corroborated the correlation between pre-SlymiR157 presence and the delay in the ripening (DR) phenotype, thus concluding that SlymiR157 was the main contributor to the tomato fruit ripening [54].
4. The Function Expansion and Application of Viral Vector
4.1. Virus-Induced Transcriptional Gene Silencing System (VITGS)
RNA silencing is a conserved defense mechanism in plants against external invading entities, such as viruses, that regulate the expression of various genes [55]. It can occur via two distinct pathways: post-transcriptional gene silencing (PTGS), that represses the translation of RNA targets; and transcriptional gene silencing (TGS), which involves DNA methylation at cytosine sites.
Transcriptional gene silencing (TGS) has an important epigenetic marker in the form of DNA cytosine methylation which controls gene expression and plays a key role in genome defense mechanism [56,57,58]. An epigenetic mechanism drives small RNAs at the transcriptional level, leading to DNA methylation and resulting in endogenous gene silencing [59]. The siRNA-guided epigenetic modification in the host genome is termed RNA-directed DNA methylation (RdDM). Moreover, DNA methylation can be maintained for many generations [60], but its maintenance is directly dependent on the cytosine sequences in the target region and is associated with the different type of DNA methyltransferases [61].
VIGS has been shown to be a successful technique for RNA silencing-mediated knockdown of target genes in plants based on PTGS by siRNAs. Additionally, siRNAs may also trigger TGS by directing the RdDM machinery to induce methylation of the corresponding DNA sequence in the nucleus. The target gene could be transcriptionally silenced as a result of cytosine residues in the promoter of the gene being methylated (Figure 2) [56]. Several viral vectors have already been implicated in TGS, including PVX, TRV, Cucumber mosaic virus (CMV), and ALSV, as already mentioned. Recent results indicate that virus-induced TGS (VITGS) is equally effective for both exogenous and endogenous genes for gene silencing, showing its potential [61]. Another important aspect is the heritability stability of VITGS, as it can be inherited for several generations, although little is known about its pervasiveness and efficiency. As a matter of fact, to our knowledge, the VITGS application in vegetables has not yet been reported. This technique has drawn a lot of attention to the scientific community and may represent a valuable advancement in this field in the near future, even if it produces genetically modified vegetable products without altering the genome.
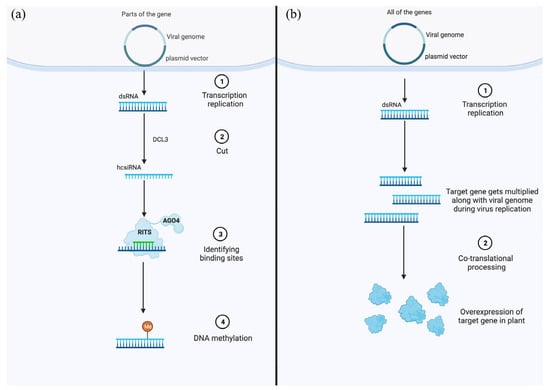
Figure 2.
Models of virus-induced transcriptional gene silencing system and virus-induced gene overexpression. (a) For VITGS, the RNA-mediated transcriptional gene silencing (TGS) can be utilized to target the gene promoter in DNA through small RNAs, which resulted in DNA methylation of specific targeted promoter sites. (b) For VIGO, the gene of interest must contain a full-length mRNA sequence (with start and stop codons) and be placed downstream of a promoter and upstream from a terminator sequence. Alternatively, protease cleavage sites can be incorporated on either side of the coding sequence to ensure excision of a functional protein from the viral genome during replication.
4.2. Virus-Induced Gene Overexpression (VIGO)
Virus Induced Gene Overexpression (VIGO) is used to transiently overexpress genes by carrying part of the sequence of VIGS- or TGS-target endogenous genes, thus triggering RNA-mediated silencing of target gene expression. VIGO vectors contain a full-length coding sequence (CDS) of the targeted gene of interest, inserted in-frame within the viral coding region [62]. The target gene is translated, along with the viral genome, during viral replication inside infected cells, resulting in a high level of synthesized proteins. However, due to the limited carrying capacity, large fragments of the target gene may not be expressed by the viral vector [63].
Researchers have applied this technology to the field of vegetables and developed several VIGO vectors. VIGO was improved by inserting a subgenomic RNA promoter from a related tobacco virus, obtaining a more stable TMV vector and enabling the expression of foreign genes in the plant. Green fluorescent proteins (GFP) are often used as reporter genes, inserted downstream of the promoter sequence, and their expression is translocated throughout the infected plant [64]. Similarly, GFP may be used as a traceable marker for functional genomics analysis on vegetable fruits from early developmental stages to the full ripening process, provided that it has the capacity to efficiently translocate and replicate in fruit as a signaling marker [36,65]. Similarly, a TRV- or TRSV-based expression vector could simultaneously express GFP in the infected plants to be used for virus component tracking. However, previous reports have shown that GFP expression is temporally correlated with VIGS effects and may reduce the infection efficiency [41,66]. Alternative uses (other than overexpression of foreign genes) utilized VIGO vectors to validate the function of endogenous genes in a variety of plants [67]. For example, a bean pod mottle virus (BPMV)-based vector was developed to investigate the sensitivity of the function of the gene GmCaM4 to salt stress effects or to study several soybean disease infection. One observation was that overexpression of GmCaM4 in soybean provided greater resistance to three different diseases and increased tolerance to salinity conditions [67]. Recently, the expression of GFP and iLOV, and their co-expression with the target gene, has been studied. In addition, there have been studies on the expression effects of GFP with different fragment sizes in hosts carrying the VIGS vector [11,68]. Cheuk and Houde (2017) changed the components of BSMV to confirm that different amounts of components had different cargo capacities [69]. This research can allow changes in virus vectors, so that they can carry at least two gene fragments, which would permit more gene functions to be determined. When we build vectors, we can put a target gene and a marker gene in the vector, or two genes that produce different phenotypes, allowing their simultaneously silencing. In this regard, we can use VIGS technology to study more gene functions [21]. Currently, in vegetables, there are only a few scientific reports of endogenous genes tested using VIGO in vegetables. This aspect may be due to the limited carrying capacity of the virus vector.
4.3. Virus-Induced Genome Editing (VIGE)
CRISPR/Cas is the overall simplest and most well-studied system. It requires a single protein, Cas9, which is guided by paired trans-activating crRNA (tracrRNA) and crRNA molecules to introduce site-specific double-stranded breaks (DSBs) into a target DNA sequence during the interference stage. An sgRNA engineered from a dual tracrRNA means that a crRNA molecule directs Cas9 to the target site. Then, Cas9 utilizes two distinct nuclease domains, HNH and RuvC-like, to cleave both strands of the target DNA, generating sequence-specific DSBs. This triggers two DNA repair systems, nonhomologous end-joining (NHEJ) and homology-directed repair (HDR) [70]. By using CRISPR/Cas technology, specific sequences at the specific target locations in the genome can be deleted, replaced, or inserted to accurately design target genes and generate novel traits [71,72]. This technique enables editing of crops at high speeds and, thus, it possesses great potential in shaping novel genetic makeup of vegetable crops. Current CRISPR/Cas approaches in vegetables have mainly focused on the delivery of the editing machinery by transformation technologies. However, nearly all methods rely on the tissue culture, requiring a lot of time and being genotype-dependent. Recent studies highlight the potential use of viral vectors to deliver components of CRISPR/Cas reactions into plant cells for genome editing, a strategy known as Virus-Induced Genome Editing (VIGE) [73]. VIGE aids in avoiding using tissue culture for genome editing by delivering transgenes directly to the meristem or the egg cell. In the last decade, VIGE systems have been developed and used for a range of host plants, with excellent outcomes in genome editing (Table 2).

Table 2.
VIGE overview of subsequent carrier characteristics available in vegetables.
VIGE vectors can be classified into two categories according to their cargo capacity and the reagents that may be delivered (Figure 3). The first category is VIGE vectors that express an sgRNA (a single RNA molecule that contains both the custom-designed short crRNA sequence fused to the tracrRNA sequence), infecting plants to stably express the Cas9 to enable the editing of target genes. Nevertheless, this approach typically results in low frequencies of gene editing in somatic cells of the infected plants. The recovery of mutant progeny is rare, therefore limiting its utility. Recent studies demonstrated that the mobile RNA element fusing to the sgRNA facilitates the guide RNA to enter the meristem, producing heritable changes, thus overcoming the deficiency of the stable transformation pathway and acquiring gene-edited offspring [73]. The second category includes VIGE vectors that deliver both Cas9 and sgRNA, which are spread systemically into the plant. One example used Sonchus yellow net rhabdovirus (SYNV) that stably carried ~5 kb of exogenous sequences in its genome, obtaining the expression of Cas9 and sgRNA simultaneously [45,76,77]. However, this category has seen its application reduced due to a smaller host range of this virus. Remarkably, Li and his colleagues developed a new virus vector using the tomato spotted wilt virus (TSWV), that stably carried Cas9-, Cas12-, or Cas-derived base editors together with multiple guide RNAs in various host plants, including tomato, different peppers, and peanut cultivars [75]. Although this strategy did not provide stable gene-edited offspring, their work is a notable improvement towards the use of VIGE and TSWV-based CRISPR–Cas as delivery systems for vegetable breeding. The main limiting aspects to consider for its application are as follows: How can we achieve stable heritable offspring? When expressing the viral vector, could it be possible for the Cas nuclease mRNAs and its derivatives to reach the germline cells in the meristems, perhaps with the help of other mobile elements? If these answers were attained, transgene-free and tissue culture-free genome-engineered plants would be possible (Figure 3).
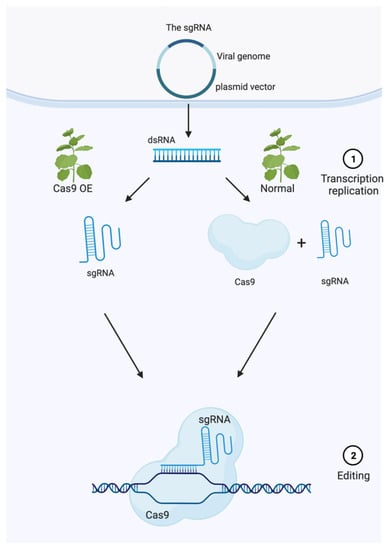
Figure 3.
Model of Virus-Induced Genome Editing. VIGE vectors can be classified into two categories according to their cargo capacity and the reagents that may be delivered. The first category includes VIGE vectors that express a sgRNA and infected plants that stably express Cas9. Viral vectors could efficiently deliver sgRNAs into plant cells to enable the editing of target genes. The second category includes VIGE vectors that deliver both Cas9 and sgRNA, which spread systemically in plants.
5. Future Directions
5.1. Viral Silencing Inhibitors That Increase VIGS Efficiency
In plants, traditional gene functional verification relies on genetic transformation technology, and most of the genetic transformations of various species are unstable. Therefore, virus-induced gene silencing technology is suitable for plant functional genomics research [1,78]. VIGS offers a fast substitute to knock down genes of interest by sequence-specific RNA degradation processes [79]. After infecting plants, viruses produce double-stranded RNA (dsRNA) with a length of 21 to 30 nucleotides during virus replication in the cytoplasm of plant cells, which is processed into siRNA by DCL (DCL2/3/4). The loading siRNA is incorporated into different RISC complexes described in the model, including RNA-induced transcriptional gene silencing complex (RITS). It directs chromatin methylation and siRNA/miRNA-dependent RNA-induced silencing complexes, leading to the transcription of target mRNAs, along with cleavage and translation arrest. For virus-encoded RNA-dependent RNA polymerase (RdRP), secondary siRNAs are produced in the amplification loop by RDR and its cofactors (FX, SGS3, etc.) [80].
RNA silencing is a major antiviral defense in plants [80]. To counteract this antiviral defense, most plant viruses have evolved silencing suppressor proteins, targeting different steps of the antiviral silencing pathway. Several viral suppressors of RNA silencing (VSRs) have been identified from almost all plant virus genera. VSRs efficiently inhibit host antiviral responses by interacting with the key components of cellular silencing machinery. In general, VSR can be classified into three categories: (1) binding long dsRNA inhibits Dicer processing; (2) binding and sequestration of siRNA duplexes prevents RISC assembly; and (3) direct targeting of effectors blocks amplification of antiviral silencing [81].
VSRs have been found to boost the efficiency of VIGS by temporarily inhibiting the RNA silencing machine of host plants, facilitating the transmission of RNA viruses in plants (Figure 4) [81]. Furthermore, VSR genes may develop independently in each virus family as viruses continue to adapt to host RNA silencing immunity [33]. They are surprisingly diverse both within and between populations, with no apparent sequence homology. The VSR protein encoded by many viruses interacts with effectors that block RNA silencing pathways [82,83], such as DICER, dsRNA, siRNA, RNA-induced silencing complex (RISC), or systemic signals [33,84,85].
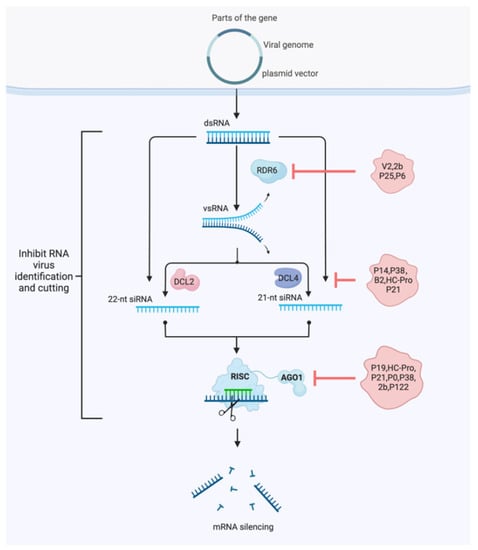
Figure 4.
Model of the relationship between antiviral RNA silencing and silencing suppressors in plants. Viral-silencing suppressors can disrupt these pathways at multiple points, thereby preventing the assembly of different effectors or inhibiting their actions. The points at which certain VSRs (i.e., P14, P38, B2, V2, 2b, P19, HC-Pro, P21, and P0) interact with the silencing pathways are depicted.
Among the VSRs, the P19 protein encoded by tomato bushy stunt virus (TBSV) is a powerful VSR that inhibits RNA interference (RNAi) by forming homodimers that bind to siRNA produced by Dicer endonuclease. Sequestration of siRNAs by P19 prevents RISC from being programmed by these molecules, inhibiting the endonuclease activity of RISC and interfering with the degradation of the RNA corresponding to the siRNA [85]. Meanwhile, researchers demonstrated that P19 has been used to enhance the expression of recombinant proteins in plants. Exogenous expression of p19, for instance, increased the infectivity of TRV viral vectors carrying the gene of green fluorescent protein (GFP) and led to a GFP significant increase expression. In a similar experimental design, a P19-deleted TBSV vector was used to infect tobacco. Deletion of P19 in the viral vector resulted in low-level expression of GFP, which was significantly restored when a separate P19 construct was infiltrated into the same leaf [85].
5.2. The Derivation and Future Development Direction of VIGS
Over the past two decades, VIGS technology has advanced significantly, largely due to the creation of new VIGS vectors that could infect wider hosts, facilitate multiple cloning sites for homologous recombination, and improve Agrobacterium/viral vector immunization methods. The primary VIGS techniques for examining the gene function of vegetable crops at the present moment are gene silencing and the co-expression of heterologous viral RNA silencing inhibitors (VSRs), that extends the duration of the silencing effect.
The use of VIGO to transiently overexpress target genes regulating biological processes is rather sparse compared to the application of VIGS. To better utilize VIGO, exploring more target genes is a possible future research direction [63]. Similarly, regarding VITGS, low levels of RdDM is one of the factors that affects silencing efficiency. Increasing RdDM levels through the use of mutant plants that increase 24 nt siRNA production may be a future research direction [85].
On the other hand, VIGE compared to traditional VIGS have two main advantages. Firstly, traditional VIGS uses target gene fragments to generate siRNA, allowing the silencing of the corresponding gene. However, it can also lead to non-specific silencing, particularly for highly homologous genes. In contrast, CRISPR-/Cas9-based VIGE allows the targeting of specific genes that result in gene knockout after NHEJ repair of DSBs. Thus, CRISPR-/Cas9-based VIGE can be used to study the functional validation of individual genes. Secondly, VIGS requires cloning of fragments of target genes by PCR, while VIGE requires only a 20 bp sgRNA tailor-designed for the target sequence, providing an effective high-throughput platform for genome-wide gene function analysis. In addition, viruses are excluded during plant regeneration, and progeny plants do not carry any virus fragments. Therefore, mutant plants can be regenerated from systemic tissues without antibiotic selection and further genetic transformation. Moreover, regenerated mutant plants have the benefit of not possessing additional T-DNA insertions other than Cas9. Finally, VIGE progeny plants are not generally required to be genotyped, nor self- or back-crossed.
The biggest obstacle to the development of VIGE systems is the limitation of the size of the inserts that can be delivered and retained by viral vectors. However, studies have shown that the co-expression of VSRs can increase the expression level of foreign genes and increase the load of foreign genes inserted into viral vectors. This feature would allow the expression of larger proteins or a larger number of proteins in plants. By using viral vectors to deliver CRISPR/Cas9 constructs, the time and resource allocation needed to regenerate plants can be saved. On the other hand, VIGE is still at an early developmental phase, and most research objects are limited to tobacco or Arabidopsis. There is still work to be carried out on how to optimize this system for vegetable crops; in fact, one aspect to consider would be that there are model plants and non-model crops (e.g., melon (Cucumis melo L.) and cucumber (Cucumis sativus L.) which are difficult to transform, which hampers the possibility of developing reverse genetic studies for crop improvement.
6. Conclusions
In conclusion, VIGS and the derived VIGE are promising technical means at the service of molecular breeding. Solving meaningful challenges, such as overcoming heritable non-transgenic mutations and inducing gene overexpression, will provide unprecedented opportunities for future functional genomics research and plant breeding efforts in vegetable crops.
Author Contributions
Conceptualization, X.X. and Y.H.; methodology, W.S.; software, S.C.; validation, X.X. and Y.H.; formal analysis, W.S.; investigation, Z.W.; resources, J.S.; data curation, W.S.; writing—original draft preparation, Z.W.; writing—review and editing, Z.W. and E.D.M.; visualization, C.Y.; supervision, C.Y.; project administration, J.S.; funding acquisition, J.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Department of Science and Technology of Zhejiang Province (2022C02032), the Key Research and Development Project of Zhejiang Province (2021C02042), and the Major Science and Technology Project of Plant Breeding in Zhejiang Province (2021C02065-3).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Burch Smith, T.M.; Anderson, J.C.; Martin, G.B.; Dinesh Kumar, S.P. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 2004, 39, 734–746. [Google Scholar] [CrossRef]
- Lu, R.; Martin Hernandez, A.M.; Peart, J.R.; Malcuit, I.; Baulcombe, D.C. Virus-induced gene silencing in plants. Methods 2003, 30, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Li, Y.; An, W.; Jiao, E.; Zhao, J.; Liu, L.; Qin, K.; Cao, Y. Applications of virus-induced gene silencing for analysis of gene function in Solanaceae species. J. Henan Agric. Sci. 2018, 47, 8–19. [Google Scholar]
- Sijen, T.; Kooter, J.M. Post-transcriptional gene-silencing: RNAs on the attack or on the defense? Bioessays 2000, 22, 520–531. [Google Scholar] [CrossRef]
- Ratcliff, F.; Martin Hernandez, A.M.; Baulcombe, D.C. Technical advance: Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 2001, 25, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.C.; Mendell, J.T. microRNAs in vertebrate physiology and human disease. Annu. Rev. Genom. Hum. Genet. 2007, 8, 215–239. [Google Scholar] [CrossRef]
- Scholthof, H.B.; Scholthof, K.G. Plant virology: An RNA treasure trove. Trends Plant Sci. 2023, 223, 1360–1385. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, N.; Dasaradhi, P.; Mohmmed, A.; Malhotra, P.; Bhatnagar, R.K.; Mukherjee, S.K. RNA interference: Biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 2003, 67, 657–685. [Google Scholar] [CrossRef]
- Ramanna, H.; Ding, X.S.; Nelson, R.S. Rationale for developing new virus vectors to analyze gene function in grasses through virus-induced gene silencing. Virus-Induc. Gene Silenc. Methods Protoc. 2013, 975, 15–32. [Google Scholar]
- Robertson, D. VIGS vectors for gene silencing: Many targets, many tools. Annu. Rev. Plant Biol. 2004, 55, 495–519. [Google Scholar] [CrossRef]
- Scofield, S.R.; Nelson, R.S. Resources for virus-induced gene silencing in the grasses. Plant Physiol. 2009, 149, 152–157. [Google Scholar] [CrossRef]
- Senthil-Kumar, M.; Mysore, K.S. Virus-induced gene silencing can persist for more than 2 years and also be transmitted to progeny seedlings in Nicotiana benthamiana and tomato. Plant Biotechnol. J. 2011, 9, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Cerutti, H.; Ma, X.; Msanne, J.; Repas, T. RNA-mediated silencing in algae: Biological roles and tools for analysis of gene function. Eukaryot. Cell 2011, 10, 1164–1172. [Google Scholar] [CrossRef] [PubMed]
- Kalantidis, K.; Schumacher, H.T.; Alexiadis, T.; Helm, J.M. RNA silencing movement in plants. Biol. Cell 2008, 100, 13–26. [Google Scholar] [CrossRef]
- Ramegowda, V.; Mysore, K.S.; Senthil-Kumar, M. Virus-induced gene silencing is a versatile tool for unraveling the functional relevance of multiple abiotic-stress-responsive genes in crop plants. Front. Plant Sci. 2014, 5, 323. [Google Scholar] [CrossRef] [PubMed]
- Senthil Kumar, M.; Anand, A.; Uppalapati, S.R.; Mysore, K.S. Virus-induced gene silencing and its applications. CABI Rev. 2008, 3, 11. [Google Scholar] [CrossRef]
- Baulcombe, D.C. Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol. 1999, 2, 109–113. [Google Scholar] [CrossRef]
- Sasaki, S.; Yamagishi, N.; Yoshikawa, N. Efficient virus-induced gene silencing in apple, pear and Japanese pear using Apple latent spherical virus vectors. Plant Methods 2011, 7, 15. [Google Scholar] [CrossRef]
- Fu, D.; Zhu, B.; Zhu, H.; Zhang, H.; Xie, Y.; Jiang, W.; Zhao, X.; Luo, Y. Enhancement of virus-induced gene silencing in tomato by low temperature and low humidity. Mol. Cells 2006, 21, 153–160. [Google Scholar]
- Yuan, C.; Li, C.; Yan, L.; Jackson, A.O.; Liu, Z.; Han, C.; Yu, J.; Li, D. A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS ONE 2011, 6, e26468. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, H.; Han, R. Advances in the application of virus-induced gene silencing in plants. Am. J. Plant Sci. 2019, 10, 1649–1661. [Google Scholar] [CrossRef]
- Anusha, M.; Roy, K.; Maiti, A.K. Feeding efficiency of predatory spiders on Myzus persicae (Sulzer). Indian J. Entomol. 2022, 84, 129–131. [Google Scholar]
- Liu, T.; Li, Y.; Zhang, C.; Duan, W.; Huang, F.; Hou, X. Basic helix-loop-helix transcription factor BcbHLHpol functions as a positive regulator of pollen development in non-heading Chinese cabbage. Funct. Integr. Genom. 2014, 14, 731–739. [Google Scholar] [CrossRef]
- Bahieldin, A.; Atef, A.; Edris, S.; Gadalla, N.O.; Ali, H.M.; Hassan, S.M.; Al Kordy, M.A.; Ramadan, A.M.; Makki, R.M.; Al Hajar, A.S. Ethylene responsive transcription factor ERF109 retards PCD and improves salt tolerance in plant. BMC Plant Biol. 2016, 16, 216. [Google Scholar] [CrossRef] [PubMed]
- Lange, M.; Yellina, A.L.; Orashakova, S.; Gadalla, N.O.; Ali, H.M.; Hassan, S.M.; Al Kordy, M.A.; Ramadan, A.M.; Makki, R.M.; Al Hajar, A.S. Virus-induced gene silencing (VIGS) in plants: An overview of target species and the virus-derived vector systems. Virus-Induc. Gene Silenc. Methods Protoc. 2013, 80, 975. [Google Scholar]
- Kurreck, J. RNA interference: From basic research to therapeutic applications. Angew. Chem. Int. Ed. 2009, 48, 1378–1398. [Google Scholar] [CrossRef]
- Lee, C.C.; Wang, J.W.; Leu, W.M.; Huang, Y.T.; Huang, Y.W.; Hsu, Y.H.; Meng, M. Proliferating cell nuclear antigen suppresses RNA replication of Bamboo mosaic virus through an interaction with the viral genome. J. Virol. 2019, 93, e00961-19. [Google Scholar] [CrossRef]
- Kelman, Z. PCNA: Structure, functions and interactions. Oncogene 1997, 14, 629–640. [Google Scholar] [CrossRef]
- Pandey, P.; Choudhury, N.R.; Mukherjee, S.K. A geminiviral amplicon (VA) derived from Tomato leaf curl virus (ToLCV) can replicate in a wide variety of plant species and also acts as a VIGS vector. Virol. J. 2009, 6, 152. [Google Scholar] [CrossRef]
- Waterhouse, P.M.; Helliwell, C.A. Exploring plant genomes by RNA-induced gene silencing. Nat. Rev. Genet. 2003, 4, 29–38. [Google Scholar] [CrossRef]
- Zhou, P.; Peng, J.; Zeng, M.; Wu, L.; Fan, Y.; Zeng, L. Virus-induced gene silencing (VIGS) in Chinese narcissus and its use in functional analysis of NtMYB3. Hortic. Plant J. 2021, 7, 565–572. [Google Scholar] [CrossRef]
- Zhang, J.; Tian, J.; Tai, D.; Li, K.; Zhu, Y.; Yao, Y. An optimized TRV-based virus-induced gene silencing protocol for Malus crabapple. Plant Cell Tissue Organ Cult. 2016, 126, 499–509. [Google Scholar] [CrossRef]
- Zhou, Y.; Deng, Y.; Liu, D.; Wang, H.; Zhang, X.; Liu, T.; Wang, J.; Li, Y.; Ou, L.; Liu, F.; et al. Promoting virus-induced gene silencing of pepper genes by a heterologous viral silencing suppressor. Plant Biotechnol. J. 2021, 19, 2398–2400. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, Y.; Pirrello, J.; Regad, F.; Bouzayen, M.; Deng, W.; Li, Z. Silencing Sl-EBF1 and Sl-EBF2 expression causes constitutive ethylene response phenotype, accelerated plant senescence, and fruit ripening in tomato. J. Exp. Bot. 2010, 61, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Ding, B.; Fei, Z.; Wang, Y. Comprehensive transcriptome analyses reveal tomato plant responses to tobacco rattle virus-based gene silencing vectors. Sci. Rep. 2017, 7, 9771. [Google Scholar] [CrossRef]
- Quadrana, L.; Rodriguez, M.C.; Lopez, M.; Bermudez, L.; Nunes Nesi, A.; Fernie, A.R.; Descalzo, A.; Asis, R.; Rossi, M.; Asurmendi, S.; et al. Coupling virus-induced gene silencing to exogenous Green Fluorescence Protein expression provides a highly efficient system for functional genomics in Arabidopsis and across all stages of tomato fruit development. Plant Physiol. 2011, 156, 1278–1291. [Google Scholar] [CrossRef]
- Liu, E.; Page, J.E. Optimized cDNA libraries for virus-induced gene silencing (VIGS) using tobacco rattle virus. Plant Methods 2008, 4, 5. [Google Scholar] [CrossRef]
- Yamagishi, N.; Yoshikawa, N. Efficient virus-induced gene silencing system in pumpkin (Cucurbita maxima) using apple latent spherical virus vector. J. Virol. Methods 2022, 301, 114456. [Google Scholar] [CrossRef]
- Yaegashi, H.; Yamatsuta, T.; Takahashi, T.; Li, C.; Isogai, M.; Kobori, T.; Ohki, S.; Yoshikawa, N. Characterization of virus-induced gene silencing in tobacco plants infected with apple latent spherical virus. Arch. Virol. 2007, 152, 1839–1849. [Google Scholar] [CrossRef] [PubMed]
- Devani, R.S.; Kute, A.; John, S.; Adhikari, S.; Sinha, S.; Banerjee, A.K. Development of a Virus-Induced Gene Silencing System for Dioecious Coccinia grandis. Mol. Biotechnol. 2020, 62, 412–422. [Google Scholar] [CrossRef]
- Zhao, F.; Lim, S.; Igori, D.; Yoo, R.H.; Kwon, S.Y.; Moon, J.S. Development of tobacco ringspot virus-based vectors for foreign gene expression and virus-induced gene silencing in a variety of plants. Virology 2016, 492, 166–178. [Google Scholar] [CrossRef]
- Fang, L.; Wei, X.; Liu, L.; Zhou, L.; Tian, Y.; Geng, C.; Li, X. A tobacco ringspot virus-based vector system for gene and microRNA function studies in cucurbits. Plant Physiol. 2021, 186, 853–864. [Google Scholar] [CrossRef]
- Siddiqui, S.A.; Sarmiento, C.; Kiisma, M.; Koivumaki, S.; Lemmetty, A.; Truve, E.; Lehto, K. Effects of viral silencing suppressors on tobacco ringspot virus infection in two Nicotiana species. J. Gen. Virol. 2008, 89, 1502–1508. [Google Scholar] [CrossRef]
- Bi, X.; Guo, H.; Li, X.; Zheng, L.; An, M.; Xia, Z.; Wu, Y. A novel strategy for improving watermelon resistance to cucumber green mottle mosaic virus by exogenous boron application. Mol. Plant Pathol. 2022, 23, 1361–1380. [Google Scholar] [CrossRef] [PubMed]
- Roshan, P.; Kulshreshtha, A.; Kumar, S.; Purohit, R.; Hallan, V. AV2 protein of tomato leaf curl Palampur virus promotes systemic necrosis in Nicotiana benthamiana and interacts with host Catalase2. Sci. Rep. 2018, 8, 1273. [Google Scholar] [CrossRef]
- Uranga, M.; Aragones, V.; Selma, S.; Vazquez Vilar, M.; Orzaez, D.; Daros, J.A. Efficient Cas9 multiplex editing using unspaced sgRNA arrays engineering in a Potato virus X vector. Plant J. 2021, 106, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Ariga, H.; Toki, S.; Ishibashi, K. Potato Virus X Vector-Mediated DNA-Free Genome Editing in Plants. Plant Cell Physiol. 2020, 61, 1946–1953. [Google Scholar] [CrossRef]
- Igarashi, A.; Yamagata, K.; Sugai, T.; Takahashi, Y.; Sugawara, E.; Tamura, A.; Yaegashi, H.; Yamagishi, N.; Takahashi, T.; Isogai, M.; et al. Apple latent spherical virus vectors for reliable and effective virus-induced gene silencing among a broad range of plants including tobacco, tomato, Arabidopsis thaliana, cucurbits, and legumes. Virology 2009, 386, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Peele, C.; Jordan, C.V.; Muangsan, N.; Turnage, M.; Egelkrout, E.; Eagle, P.; Hanley Bowdoin, L.; Robertson, D. Silencing of a meristematic gene using geminivirus-derived vectors. Plant J. 2001, 27, 357–366. [Google Scholar] [CrossRef]
- Lu, R.; Malcuit, I.; Moffett, P.; Ruiz, M.T.; Peart, J.; Wu, A.J.; Rathjen, J.P.; Bendahmane, A.; Day, L.; Baulcombe, D.C. High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 2003, 22, 5690–5699. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, Y.; Wang, X.; Hong, Z.; Yang, A.; Liu, Y.; Yan, L.; He, Y.; Zhu, Z.; Wang, H. PACLOBUTRAZOL-RESISTANCE4 positively regulates cell expansion to promote tendril elongation in cucumber. Plant Physiol. 2023, 192, 2756–2767. [Google Scholar] [CrossRef]
- Shin, S.Y.; Park, M.R.; Kim, H.S.; Moon, J.S.; Lee, H.J. Virus-induced gene silencing shows that LATE FLOWERING plays a role in promoting flower development in soybean. Plant Growth Regul. 2023, 99, 229–239. [Google Scholar] [CrossRef]
- Faivre Rampant, O.; Gilroy, E.M.; Hrubikova, K.; Hein, I.; Millam, S.; Loake, G.J.; Birch, P.; Taylor, M.; Lacomme, C. Potato virus X-induced gene silencing in leaves and tubers of potato. Plant Physiol. 2004, 134, 1308–1316. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Kong, J.; Lai, T.; Manning, K.; Wu, C.; Wang, Y.; Qin, C.; Li, B.; Yu, Z.; Zhang, X. Tuning LeSPL-CNR expression by SlymiR157 affects tomato fruit ripening. Sci. Rep. 2015, 5, 7852. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D.C.; Chapman, S.; Santa Cruz, S. Jellyfish green fluorescent protein as a reporter for virus infections. Plant J. 1995, 7, 1045–1053. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.; Ratcliff, F.; Baulcombe, D.C. RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr. Biol. 2001, 11, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, A.; Inaba, J.I.; Kasai, M.; Shimura, H.; Masuta, C. RNA-mediated epigenetic modifications of an endogenous gene targeted by a viral vector: A potent gene silencing system to produce a plant that does not carry a transgene but has altered traits. Plant Signal. Behav. 2011, 6, 1090–1093. [Google Scholar] [CrossRef][Green Version]
- Kon, T.; Yoshikawa, N. Induction and maintenance of DNA methylation in plant promoter sequences by apple latent spherical virus-induced transcriptional gene silencing. Front. Microbiol. 2014, 5, 595. [Google Scholar] [CrossRef]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef]
- Liu, J.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.J.; Hammond, S.M.; Joshua Tor, L.; Hannon, G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004, 305, 1437–1441. [Google Scholar] [CrossRef]
- Khan, A.H.; Akram, A.; Saeed, M.; Ur Rahman, M.; Ur Rehman, A.; Mansoor, S.; Amin, I. Establishment of transcriptional gene silencing targeting the promoter regions of GFP, PDS, and PSY genes in cotton using Virus-Induced Gene Silencing. Mol. Biotechnol. 2023, 65, 1052–1061. [Google Scholar] [CrossRef]
- Li, J.; Yu, X.; Zhang, C.; Li, N.; Zhao, J. The application of CRISPR/Cas technologies to Brassica crops: Current progress and future perspectives. Abiotech 2022, 3, 146–161. [Google Scholar] [CrossRef]
- Paudel, L.; Kerr, S.; Prentis, P.; Tanurdžić, M.; Papanicolaou, A.; Plett, J.M.; Cazzonelli, C.I. Horticultural innovation by viral-induced gene regulation of carotenogenesis. Hortic. Res. 2022, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Nian, H.; Yang, S.; Zhang, X. Development of foreign gene expression strategy in plant virus vector. Northwest J. Bot. 2002, 22, 1268–1274. [Google Scholar]
- Ling, Q.; Sadali, N.M.; Soufi, Z.; Zhou, Y.; Huang, B.; Zeng, Y.; Rodriguez Concepcion, M.; Jarvis, R.P. The chloroplast-associated protein degradation pathway controls chromoplast development and fruit ripening in tomato. Nat. Plants 2021, 7, 655–666. [Google Scholar] [CrossRef] [PubMed]
- Klimczak, R.R.; Koerber, J.T.; Dalkara, D.; Flannery, J.G.; Schaffer, D.V. A novel adeno-associated viral variant for efficient and selective intravitreal transduction of rat Müller cells. PLoS ONE 2009, 4, 7467. [Google Scholar] [CrossRef]
- Rao, S.S.; El-Habbak, M.H.; Havens, W.M.; Singh, A.; Zheng, D.; Vaughn, L.; Haudenshield, J.S.; Hartman, G.L.; Korban, S.S.; Ghabrial, S.A. Overexpression of GmCaM4 in soybean enhances resistance to pathogens and tolerance to salt stress. Mol. Plant Pathol. 2014, 15, 145–160. [Google Scholar] [CrossRef]
- Holzberg, S.; Brosio, P.; Gross, C.; Pogue, G.P. Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 2002, 30, 315–327. [Google Scholar] [CrossRef]
- Cheuk, A.; Houde, M. A new barley stripe mosaic virus allows large protein overexpression for rapid function analysis. Plant Physiol. 2018, 176, 1919–1931. [Google Scholar] [CrossRef]
- Li, X.; Yang, L.; Chen, L.L. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, F.; Gao, G. CRISPR-based therapeutic genome editing: Strategies and in vivo delivery by AAV vectors. Cell 2020, 181, 136–150. [Google Scholar] [CrossRef]
- Zou, R.; Marin Gonzalez, A.; Liu, Y.; Liu, H.; Shen, L.; Dveirin, R.K.; Luo, J.; Kalhor, R.; Ha, T. Massively parallel genomic perturbations with multi-target CRISPR interrogates Cas9 activity and DNA repair at endogenous sites. Nat. Cell Biol. 2022, 24, 1433–1444. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, S.; Li, X.; Zhang, R.; Li, J. Virus-induced gene editing and its applications in plants. Int. J. Mol. Sci. 2022, 23, 10202. [Google Scholar] [CrossRef] [PubMed]
- Dahan Meir, T.; Filler Hayut, S.; Melamed Bessudo, C.; Bocobza, S.; Czosnek, H.; Aharoni, A.; Levy, A.A. Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 2018, 95, 5–16. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, X.; Liu, H.; Li, Z. Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nat. Plants 2020, 6, 773–779. [Google Scholar] [CrossRef]
- Peng, X.; Ma, X.; Lu, S.; Li, Z. A versatile plant rhabdovirus-based vector for gene silencing, miRNA expression and depletion, and antibody production. Front. Plant Sci. 2021, 11, 627880. [Google Scholar] [CrossRef]
- Wang, Q.; Ma, X.; Qian, S.; Zhou, X.; Sun, K.; Chen, X.; Zhou, X.; Jackson, A.O.; Li, Z. Rescue of a plant negative-strand RNA virus from cloned cDNA: Insights into enveloped plant virus movement and morphogenesis. PLoS Pathog. 2015, 11, 1005223. [Google Scholar] [CrossRef]
- Cantó Pastor, A.; Mollá-Morales, A.; Ernst, E.; Dahl, W.; Zhai, J.; Yan, Y.; Meyers, B.; Shanklin, J.; Martienssen, R. Efficient transformation and artificial mi RNA gene silencing in Lemna minor. Plant Biol. 2015, 17, 59–65. [Google Scholar] [CrossRef]
- Jauvion, V.; Elmayan, T.; Vaucheret, H. The conserved RNA trafficking proteins HPR1 and TEX1 are involved in the production of endogenous and exogenous small interfering RNA in Arabidopsis. Plant Cell 2010, 22, 2697–2709. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ding, S.W. Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 2006, 60, 503–531. [Google Scholar] [CrossRef]
- Brigneti, G.; Voinnet, O.; Li, W.; Ji, L.; Ding, S.; Baulcombe, D.C. Retracted: Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 1998, 17, 6739–6746. [Google Scholar] [CrossRef] [PubMed]
- Kasschau, K.D.; Xie, Z.; Allen, E.; Llave, C.; Chapman, E.J.; Krizan, K.A.; Carrington, J.C. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 2003, 4, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Moissiard, G.; Voinnet, O. Viral suppression of RNA silencing in plants. Mol. Plant Pathol. 2004, 5, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Pendon, J.A.; Ding, S.W. Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Microbiol. 2008, 46, 303–326. [Google Scholar] [CrossRef]
- Chiong, K.T.; Cody, W.B.; Scholthof, H.B. RNA silencing suppressor-influenced performance of a virus vector delivering both guide RNA and Cas9 for CRISPR gene editing. Sci. Rep. 2021, 11, 6769. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).