CRISPR-Cas Genome Editing for Horticultural Crops Improvement: Advantages and Prospects
Abstract
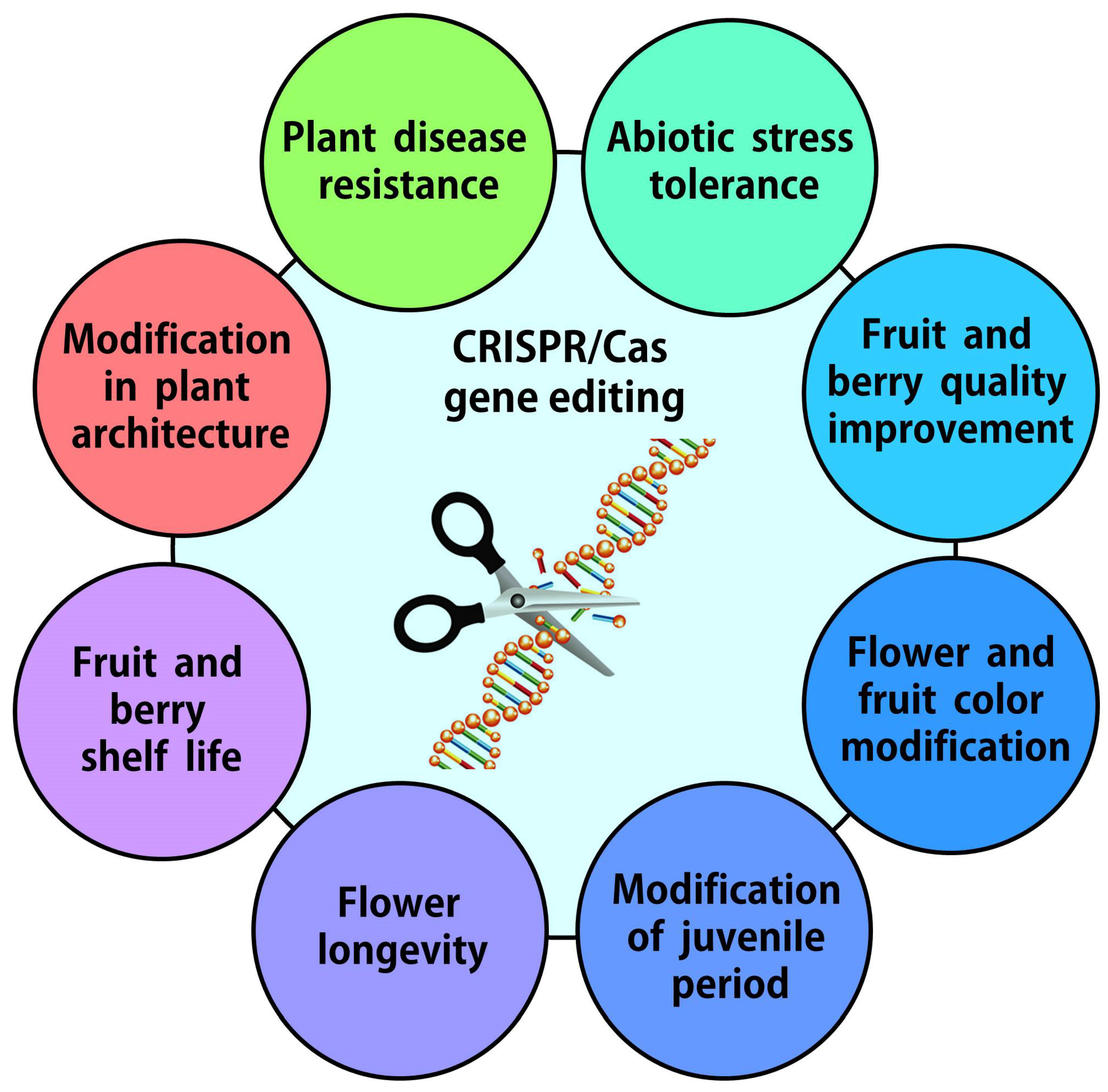
1. Introduction
2. Methods of Delivery of CRISPR/Cas Components to Plant Cells and Optimization of Genome Editing Conditions
3. Increasing the Resistance of Horticultural Plants to Biotic and Abiotic Stresses
4. Changing the Agronomic Traits of Fruit and Berry Plants Using Genome Editing
5. Changing Flower Color and Shape, Flowering Time, and Flower Longevity
6. Limitations in the Use of CRISPR/Cas9 in Genome Editing of Horticultural Plants and Further Prospects
7. Legal Regulation of Growing Plants Produced by Genome Editing Technology
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Smith, J.; Grizot, S.; Arnould, S.; Duclert, A.; Epinat, J.C.; Chames, P.; Prieto, J.; Redondo, P.; Blanco, F.J.; Bravo, J.; et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006, 34, e149. [Google Scholar] [CrossRef]
- Petolino, J.F. Genome editing in plants via designed zinc finger nucleases. In Vitro Cell. Dev. Biol. 2015, 51, 1–8. [Google Scholar] [CrossRef]
- Malzahn, A.; Lowder, L.; Qi, Y. Plant genome editing with TALEN and CRISPR. Cell Biosci. 2017, 7, 21. [Google Scholar] [CrossRef]
- Jansen, R.; Embden, J.D.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. [Google Scholar] [CrossRef]
- Mojica, F.J.; Díez-Villaseñor, C.; García-Martínez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Mojica, F.J.; Díez-Villaseñor, C.; García-Martínez, J.; Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 2009, 155, 733–740. [Google Scholar] [CrossRef]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Nonhomologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Scully, R.; Panday, A.; Elango, R.; Willis, N.A. DNA double-strandbreak repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019, 20, 698–714. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Koonin, E.V. Annotation and classification of CRISPR-Cas systems. Methods Mol. Biol. 2015, 1311, 47–75. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef]
- Barrangou, R.; Horvath, P. A decade of discovery: CRISPR functions and applications. Nat. Microbiol. 2017, 2, 17092. [Google Scholar] [CrossRef]
- Chylinski, K.; Makarova, K.S.; Charpentier, E.; Koonin, E.V. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014, 42, 6091–6105. [Google Scholar] [CrossRef] [PubMed]
- Qin, R.; Li, J.; Li, H.; Zhang, Y.; Liu, X.; Miao, Y.; Zhang, X.; Wei, P. Developing a highly efficient and wildly adaptive CRISPR-SaCas9 toolset for plant genome editing. Plant Biotechnol. J. 2019, 17, 706–708. [Google Scholar] [CrossRef]
- Zeng, D.; Li, X.; Huang, J.; Li, Y.; Cai, S.; Yu, W.; Li, Y.; Huang, Y.; Xie, X.; Gong, Q.; et al. Engineered Cas9 variant tools expand targeting scope of genome and base editing in rice. Plant Biotechnol. J. 2020, 18, 1348–1350. [Google Scholar] [CrossRef] [PubMed]
- Malzahn, A.A.; Tang, X.; Lee, K.; Ren, Q.; Sretenovic, S.; Zhang, Y.; Chen, H.; Kang, M.; Bao, Y.; Zheng, X.; et al. Application of CRISPR-Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis. BMC Biol. 2019, 17, 9. [Google Scholar] [CrossRef]
- Paul, B.; Montoya, G. CRISPR-Cas12a: Functional overview and applications. Biomed. J. 2020, 43, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cheng, Y.; Fang, H.; Roberts, N.; Zhang, L.; Vakulskas, C.A.; Niedz, R.P.; Culver, J.N.; Qi, Y. Highly efficient genome editing in plant protoplasts by ribonucleoprotein delivery of CRISPR Cas12a nucleases. Front. Genome Ed. 2022, 4, 780238. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Y.; Ye, J.; Cao, X.; Xu, S.; Chen, B.; An, H.; Jiao, Y.; Zhang, F.; Yang, X.; et al. Establishing CRISPR/Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant Biotechnol. J. 2019, 17, 1185–1187. [Google Scholar] [CrossRef]
- Malnoy, M.; Viola, R.; Jung, M.H.; Koo, O.J.; Kim, S.; Kim, J.S.; Velasco, R.; Kanchiswamy, C.N. DNA-Free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef]
- Pessina, S.; Lenzi, L.; Perazzolli, M.; Campa, M.; Dalla Costa, L.; Urso, S.; Valè, G.; Salamini, F.; Velasco, R.; Malnoy, M. Knockdown of MLO genes reduces susceptibility to powdery mildew in grapevine. Hortic. Res. 2016, 3, 16016. [Google Scholar] [CrossRef]
- Peng, A.; Chen, S.; Lei, T.; Xu, L.; He, Y.; Wu, L.; Yao, L.; Zou, X. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 2017, 15, 1509–1519. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, J.N.; Ntui, V.O.; Ron, M.; Muiruri, S.K.; Britt, A.; Tripathi, L. CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun. Biol. 2019, 2, 46. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Zheng, X.; Huang, Y.; Ye, J.; Chen, P.; Zhang, C.; Zhao, F.; Xie, Z.; Zhang, S.; Wang, N. Genome sequencing and CRISPR/Cas9 gene editing of an early flowering Mini-Citrus (Fortunella hindsii). Plant Biotechnol. J. 2019, 17, 2199–2210. [Google Scholar] [CrossRef]
- Charrier, A.; Vergne, E.; Dousset, N.; Richer, A.; Petiteau, A.; Chevreau, E. Efficient targeted mutagenesis in apple and first time edition of pear using the CRISPR-Cas9 system. Front. Plant Sci. 2019, 10, 40. [Google Scholar] [CrossRef]
- Shao, X.; Wu, S.; Dou, T.; Zhu, H.; Hu, C.; Huo, H.; He, W.; Deng, G.; Sheng, O.; Bi, F.; et al. Using CRISPR/Cas9 genome editing system to create MaGA20ox2 gene-modified semi-dwarf banana. Plant Biotechnol. J. 2020, 18, 17–19. [Google Scholar] [CrossRef]
- Hu, C.; Sheng, O.; Deng, G.; He, W.; Dong, T.; Yang, Q.; Dou, T.; Li, C.; Gao, H.; Liu, S. CRISPR/Cas9-mediated genome editing of MaACO1 (aminocyclopropane-1-carboxylate oxidase1) promotes the shelf life of banana fruit. Plant Biotechnol. J. 2021, 19, 654–656. [Google Scholar] [CrossRef] [PubMed]
- Varkonyi-Gasic, E.; Wang, T.; Cooney, J.; Jeon, S.; Voogd, C.; Douglas, M.J.; Pilkington, S.M.; Akagi, T.; Allan, A.C. Shy Girl, a kiwifruit suppressor of feminization, restricts gynoecium development via regulation of cytokinin metabolism and signalling. New Phytol. 2021, 230, 1461–1475. [Google Scholar] [CrossRef] [PubMed]
- Alquézar, B.; Bennici, S.; Carmona, L.; Gentile, A.; Peña, L. Generation of transfer-DNA-free base-edited citrus plants. Front. Plant Sci. 2022, 13, 835282. [Google Scholar] [CrossRef]
- Ren, C.; Lin, Y.; Liang, Z. CRISPR/Cas genome editing in grapevine: Recent advances, challenges and future prospects. Fruit Res. 2022, 2, 7. [Google Scholar] [CrossRef]
- Ren, F.; Ren, C.; Zhang, Z.; Duan, W.; Lecourieux, D.; Li, S.; Liang, Z. Efficiency optimization of CRISPR/Cas9-mediated targeted mutagenesis in grape. Front. Plant Sci. 2019, 10, 612. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Liu, Y.; Guo, Y.; Duan, W.; Fan, P.; Li, S.; Liang, Z. Optimizing the CRISPR/Cas9 system for genome editing in grape by using grape promoters. Hortic. Res. 2021, 8, 52. [Google Scholar] [CrossRef]
- Yin, K.; Han, T.; Liu, G.; Chen, T.; Wang, Y.; Yu, A.Y.; Liu, Y. A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci. Rep. 2015, 5, 14926. [Google Scholar] [CrossRef]
- Ali, Z.; Abul-faraj, A.; Li, L.; Ghosh, N.; Piatek, M.; Mahjoub, A.; Aouida, M.; Piatek, A.; Baltes, N.J.; Voytas, D.F.; et al. Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol. Plant. 2015, 8, 1288–1291. [Google Scholar] [CrossRef]
- Ali, Z.; Eid, A.; Ali, S.; Mahfouz, M.M. Pea early-browning virus-mediated genome editing via the CRISPR/Cas9 system in Nicotiana benthamiana and Arabidopsis. Virus Res. 2018, 244, 333–337. [Google Scholar] [CrossRef]
- Wu, S.; Zhu, H.; Liu, J.; Yang, Q.; Shao, X.; Bi, F.; Hu, S.; Huo, H.; Chen, K.; Yi, G. Establishment of a PEG-mediated protoplast transformation system based on DNA and CRISPR/Cas9 ribonucleoprotein complexes for banana. BMC Plant Biol. 2020, 20, 425. [Google Scholar] [CrossRef]
- Mercx, S.; Tollet, J.; Magy, B.; Navarre, C.; Boutry, M. Gene inactivation by CRISPR-Cas9 in Nicotiana tabacum BY-2 suspension cells. Front. Plant Sci. 2016, 7, 40. [Google Scholar] [CrossRef]
- Tsutsui, H.; Higashiyama, T. pKAMA-ITACHI vectors for highly efficient CRISPR/Cas9-mediated gene knockout in Arabidopsis thaliana. Plant Cell Physiol. 2017, 58, 46–56. [Google Scholar] [CrossRef]
- Dutt, M.; Mou, Z.; Zhang, X.; Tanwir, S.E.; Grosser, J.W. Efficient CRISPR/Cas9 genome editing with Citrus embryogenic cell cultures. BMC Biotechnol. 2020, 20, 58. [Google Scholar] [CrossRef]
- Kaur, N.; Alok, A.; Shivani; Kaur, N.; Pandey, P.; Awasthi, P.; Tiwari, S. CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Funct. Integr. Genom. 2018, 18, 89–99. [Google Scholar] [CrossRef]
- Wilson, F.M.; Harrison, K.; Armitage, A.D.; Simkin, A.J.; Harrison, R.J. CRISPR/Cas9-mediated mutagenesis of phytoene desaturase in diploid and octoploid strawberry. Plant Methods 2019, 15, 45. [Google Scholar] [CrossRef] [PubMed]
- Malabarba, J.; Chevreau, E.; Dousset, N.; Veillet, F.; Moizan, J.; Vergne, E. New strategies to overcome present CRISPR/Cas9 limitations in apple and pear: Efficient dechimerization and base editing. Int. J. Mol. Sci. 2020, 22, 319. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tu, M.; Wang, Y.; Yin, W.; Zhang, Y.; Wu, H.; Gu, Y.; Li, Z.; Xi, Z.; Wang, X. Whole genome sequencing reveals rare off-target mutations in CRISPR/Cas9-edited grapevine. Hortic. Res. 2021, 8, 114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wu, S.; Hu, C.; Yang, Q.; Dong, T.; Sheng, O.; Deng, G.; He, W.; Dou, T.; Li, C.; et al. Increased mutation efficiency of CRISPR/Cas9 genome editing in banana by optimized construct. Peer J. 2022, 10, e12664. [Google Scholar] [CrossRef]
- Pompili, V.; Dalla Costa, L.; Piazza, S.; Pindo, M.; Malnoy, M. Reduced fire blight susceptibility in apple cultivars using a high-efficiency CRISPR/Cas9-FLP/FRT-based gene editing system. Plant Biotechnol. J. 2020, 18, 845–858. [Google Scholar] [CrossRef]
- Nishitani, C.; Hirai, N.; Komori, S.; Wada, M.; Okada, K.; Osakabe, K.; Yamamoto, T.; Osakabe, Y. Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 2016, 6, 31481. [Google Scholar] [CrossRef]
- Zhou, H.; Bai, S.; Wang, N.; Sun, X.; Zhang, Y.; Zhu, J.; Dong, C. CRISPR/Cas9-Mediated Mutagenesis of MdCNGC2 in apple callus and VIGS-mediated silencing of MdCNGC2 in fruits improve resistance to Botryosphaeria dothidea. Front. Plant Sci. 2020, 11, 575477. [Google Scholar] [CrossRef]
- Naim, F.; Dugdale, B.; Kleidon, J.; Brinin, A.; Shand, K.; Waterhouse, P.; Dale, J. Gene editing the phytoene desaturase alleles of Cavendish banana using CRISPR/Cas9. Transgen. Res. 2018, 27, 451–460. [Google Scholar] [CrossRef]
- Ntui, V.O.; Tripathi, J.N.; Tripathi, L. Robust CRISPR/Cas9 mediated genome editing tool for banana and plantain (Musa spp.). Curr. Plant Biol. 2020, 21, 100128. [Google Scholar] [CrossRef]
- Kaur, N.; Alok, A.; Shivani, K.P.; Kaur, N.; Awasthi, P.; Chaturvedi, S.; Pandey, P.; Pandey, A.; Pandey, A.K.; Tiwari, S. CRISPR/Cas9 directed editing of lycopene epsilon-cyclase modulates metabolic flux for beta-carotene biosynthesis in banana fruit. Metab. Eng. 2020, 59, 76–86. [Google Scholar] [CrossRef]
- Tripathi, J.N.; Ntui, V.O.; Shah, T.; Tripathi, L. CRISPR/Cas9-mediated editing of DMR6 orthologue in banana (Musa spp.) confers enhanced resistance to bacterial disease. Plant Biotechnol. J. 2021, 19, 1291–1293. [Google Scholar] [CrossRef] [PubMed]
- Omori, M.; Yamane, H.; Osakabe, K.; Osakabe, Y.; Tao, R. Targeted mutagenesis of CENTRORADIALIS using CRISPR/Cas9 system through the improvement of genetic transformation efficiency of tetraploid highbush blueberry. J. Hortic. Sci. Biotechnol. 2021, 96, 153–156. [Google Scholar] [CrossRef]
- Fister, A.S.; Landherr, L.; Maximova, S.N.; Guiltinan, M.J. Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 enhances defense response in Theobroma cacao. Front. Plant Sci. 2018, 9, 268. [Google Scholar] [CrossRef] [PubMed]
- Breitler, J.C.; Dechamp, E.; Campa, C.; Rodrigues, L.A.Z.; Guyot, R.; Marraccini, P.; Etienne, H. CRISPR/Cas9-mediated efficient targeted mutagenesis has the potential to accelerate the domestication of Coffea canephora. PCTOC 2018, 134, 383–394. [Google Scholar] [CrossRef]
- Jia, H.; Orbovic, V.; Jones, J.B.; Wang, N. Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4:dCsLOB1.3 infection. Plant Biotechnol. J. 2016, 14, 1291–1301. [Google Scholar] [CrossRef]
- Jia, H.; Xu, J.; Orbovic, V.; Zhang, Y.; Wang, N. Editing citrus genome via SaCas9/sgRNA system. Front. Plant Sci. 2017, 8, 2135. [Google Scholar] [CrossRef]
- Jia, H.; Zhang, Y.; Orbovic, V.; Xu, J.; White, F.F.; Jones, J.B.; Wang, N. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 2017, 15, 817–823. [Google Scholar] [CrossRef]
- Jia, H.; Omar, A.; Orbović, V.; Wang, N. Biallelic editing of the LOB1 promoter via CRISPR/Cas9 creates canker-resistant ‘Duncan’ grapefruit. Phytopathology 2022, 112, 308–314. [Google Scholar] [CrossRef]
- Ren, C.; Liu, X.; Zhang, Z.; Wang, Y.; Duan, W.; Li, S.; Liang, Z. CRISPR/ Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitis vinifera L.). Sci. Rep. 2016, 6, 32289. [Google Scholar] [CrossRef]
- Osakabe, Y.; Liang, Z.; Ren, C.; Nishitani, C.; Osakabe, K.; Wada, M.; Komori, S.; Malnoy, M.; Velasco, R.; Poli, M.; et al. CRISPR-Cas9-mediated genome editing in apple and grapevine. Nat. Protoc. 2018, 13, 2844–2863. [Google Scholar] [CrossRef]
- Nakajima, I.; Ban, Y.; Azuma, A.; Onoue, N.; Moriguchi, T.; Yamamoto, T.; Toki, S.; Endo, M. CRISPR/Cas9-mediated targeted mutagenesis in grape. PLoS ONE 2017, 12, 0177966. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tu, M.; Wang, D.; Liu, J.; Li, Y.; Li, Z.; Wang, Y.; Wang, X. CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 2018, 16, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Wan, D.Y.; Guo, Y.; Cheng, Y.; Hu, Y.; Xiao, S.; Wang, Y.; Wen, Y. CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Hortic. Res. 2020, 7, 116. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y.; Jiao, Y.T.; Wang, Y.T.; Zhang, N.; Wang, B.B.; Liu, R.Q.; Yin, X.; Xu, Y.; Liu, G.T. CRISPR/Cas9-mediated VvPR4b editing decreases downy mildew resistance in grapevine (Vitis vinifera L.). Hortic. Res. 2020, 7, 149. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Guo, Y.; Kong, J.; Lecourieux, F.; Dai, Z.; Li, S.; Liang, Z. Knockout of VvCCD8 gene in grapevine affects shoot branching. BMC Plant Biol. 2020, 20, 47. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, S.; Li, D.; Zhang, Q.; Li, L.; Zhong, C.; Liu, Y.; Huang, H. Optimized paired-sgRNA/Cas9 cloning and expression cassette triggers high-efficiency multiplex genome editing in kiwifruit. Plant Biotechnol. J. 2018, 16, 1424–1433. [Google Scholar] [CrossRef]
- Varkonyi-Gasic, E.; Wang, T.; Voogd, C.; Jeon, S.; Drummond, R.S.M.; Gleave, A.P.; Allan, A.C. Mutagenesis of kiwifruit CENTRORADIALIS-like genes transforms a climbing woody perennial with long juvenility and axillary flowering into a compact plant with rapid terminal flowering. Plant Biotechnol. J. 2019, 17, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, L.; Lu, L.; Liu, J.; Yi, H.; Wu, J. An efficient CRISPR/Cas9 system for simultaneous editing two target sites in Fortunella hindsii. Hortic. Res. 2022, 9, uhac064. [Google Scholar] [CrossRef]
- Hooghvorst, I.; López-Cristoffanini, C.; Nogués, S. Efficient knockout of phytoene desaturase gene using CRISPR/Cas9 in melon. Sci. Rep. 2019, 9, 17077. [Google Scholar] [CrossRef]
- Liu, B.; Santo Domingo, M.; Mayobre, C.; Martín-Hernández, A.M.; Pujol, M.; Garcia-Mas, J. Knock-out of CmNAC-NOR affects melon climacteric fruit ripening. Front. Plant Sci. 2022, 13, 878037. [Google Scholar] [CrossRef]
- Giordano, A.; Santo Domingo, M.; Quadrana, L.; Pujol, M.; Martín-Hernández, A.M.; Garcia-Mas, J. CRISPR/Cas9 gene editing uncovers the roles of constitutive triple response 1 and repressor of silencing 1 in melon fruit ripening and epigenetic regulation. J. Exp. Bot. 2022, 73, 4022–4033. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, S.; Peng, A.; Xie, Z.; He, Y.; Zou, X. CRISPR/Cas9-mediated editing of CsWRKY22 reduces susceptibility to Xanthomonas citri subsp. citri in Wanjincheng orange (Citrus sinensis (L.) Osbeck). Plant Biotechnol. Rep. 2019, 13, 501–510. [Google Scholar] [CrossRef]
- Brewer, S.E.; Chambers, A.H. CRISPR/Cas9-mediated genome editing of Phytoene desaturase in Carica papaya L. J. Hortic. Sci. Biotechnol. 2022, 97, 580–592. [Google Scholar] [CrossRef]
- Pang, H.; Yan, Q.; Zhao, S.; He, F.; Xu, J.; Qi, B.; Zhang, Y. Knockout of the S-acyltransferase gene, PbPAT14, confers the dwarf yellowing phenotype in first generation pear by ABA accumulation. Int. J. Mol. Sci. 2019, 20, 6347. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Wu, S.; Tian, L. Effective genome editing and identification of a regiospecific gallic acid 4-O-glycosyltransferase in pomegranate (Punica granatum L.). Hortic. Res. 2019, 6, 123. [Google Scholar] [CrossRef] [PubMed]
- Miller, S. Gene Editing of Red Raspberry (Rubus ideaus L.) with CRISPR/Cas9 Knocking out F3’H. Master’s Thesis, Norwegian University of Life Sciences, Ås, Norway, 2019. Available online: http://hdl.handle.net/11250/2608590 (accessed on 13 May 2019).
- Zhou, J.; Wang, G.; Liu, Z. Efficient genome-editing of wild strawberry genes, vector development, and validation. Plant Biotechnol. J. 2018, 16, 1868–1877. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Jia, M.; Wei, L.; Mao, W.; Abbasi, U.A.; Zhao, Y.; Chen, Y.; Cao, M.; Zhang, K.; Dai, Z.; et al. CRISPR/Cas9-introduced single and multiple mutagenesis in strawberry. J. Genet. Genom. 2018, 45, 685–687. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Luo, H.; Li, Y.; Liu, Z.; Kang, C. Genetic modulation of RAP alters fruit coloration in both wild and cultivated strawberry. Plant Biotechnol. J. 2020, 18, 1550–1561. [Google Scholar] [CrossRef]
- Pi, M.; Hu, S.; Cheng, L.; Zhong, R.; Cai, Z.; Liu, Z.; Yao, J.-L.; Kang, C. The MADS-box gene FveSEP3 plays essential roles in flower organogenesis and fruit development in woodland strawberry. Hortic. Res. 2021, 8, 247. [Google Scholar] [CrossRef]
- Martin-Pizarro, C.; Trivino, J.C.; Pose, D. Functional analysis of TM6 MADS-box gene in the octoploid strawberry by CRISPR/Cas9 directed mutagenesis. J. Exp. Bot. 2019, 70, 885–895. [Google Scholar] [CrossRef]
- Zhang, F.; LeBlanc, C.; Irish, V.F.; Jacob, Y. Rapid and efficient CRISPR/Cas9 gene editing in Citrus using the YAO promoter. Plant Cell Rep. 2017, 36, 1883–1887. [Google Scholar] [CrossRef]
- Huang, X.; Wang, Y.; Xu, J.; Wang, N. Development of multiplex genome editing toolkits for citrus with high efficacy in biallelic and homozygous mutations. Plant Mol. Biol. 2020, 104, 297–307. [Google Scholar] [CrossRef]
- Tian, S.; Jiang, L.; Gao, Q. Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 2017, 36, 399–406. [Google Scholar] [CrossRef]
- Walawage, S.L.; Zaini, P.A.; Mubarik, M.S.; Martinelli, F.; Balan, B.; Caruso, T.; Leslie, C.A.; Dandekar, A.M. Deploying genome editing tools for dissecting the biology of nut trees. Front. Sustain. Food Syst. 2019, 3, 100. [Google Scholar] [CrossRef]
- Chang, Y.; Song, X.; Zhang, Q.; Zhang, P.; Lei, X.; Pei, D. Robust CRISPR/Cas9 mediated gene editing of JrWOX11 manipulated adventitious rooting and vegetative growth in a nut tree species of walnut. Sci. Hortic. 2022, 303, 111199. [Google Scholar] [CrossRef]
- Tian, S.; Jiang, L.; Cui, X.; Zhang, J.; Guo, S.; Li, M.; Zhang, H.; Ren, Y.; Gong, G.; Zong, M.; et al. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 2018, 37, 1353–1356. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Q.; Yang, X.; Xu, J.; Liu, G.; Yao, X.; Ren, R.; Xu, J.; Lou, L. CRISPR/Cas9-mediated mutagenesis of Clpsk1 in watermelon to confer resistance to Fusarium oxysporum f. sp. niveum. Plant Cell Rep. 2020, 39, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, J.; Guo, S.; Tian, S.; Zhang, J.; Ren, Y.; Li, M.; Gong, G.; Zhang, H.; Xu, Y. CRISPR/Cas9-mediated mutagenesis of ClBG1 decreased seed size and promoted seed germination in watermelon. Hortic. Res. 2021, 8, 70. [Google Scholar] [CrossRef]
- Chang, J.; Guo, Y.; Yan, J.; Zhang, Z.; Yuan, L.; Wei, C.; Zhang, Y.; Ma, J.; Yang, J.; Zhang, X.; et al. The role of watermelon caffeic acid O-methyltransferase (ClCOMT1) in melatonin biosynthesis and abiotic stress tolerance. Hortic. Res. 2021, 8, 210. [Google Scholar] [CrossRef]
- Kishi-Kaboshi, M.; Aida, R.; Sasaki, K. Generation of gene-edited Chrysanthemum morifolium using multicopy transgenes as targets and markers. Plant Cell Physiol. 2017, 58, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Kui, L.; Chen, H.; Zhang, W.; He, S.; Xiong, Z.; Zhang, Y.; Yan, L.; Zhong, C.; He, F.; Chen, J.; et al. Building a genetic manipulation tool box for orchid biology: Identification of constitutive promoters and application of CRISPR/Cas9 in the orchid, Dendrobium officinale. Front. Plant Sci. 2017, 7, 2036. [Google Scholar] [CrossRef] [PubMed]
- Tasaki, K.; Higuchi, A.; Watanabe, A.; Sasaki, N.; Nishihara, M. Effects of knocking out three anthocyanin modification genes on the blue pigmentation of gentian flowers. Sci. Rep. 2019, 9, 15831. [Google Scholar] [CrossRef] [PubMed]
- Tasaki, K.; Yoshida, M.; Nakajima, M.; Higuchi, A.; Watanabe, A.; Nishihara, M. Molecular characterization of an anthocyanin-related glutathione S-transferase gene in Japanese gentian with the CRISPR/Cas9 system. BMC Plant Biol. 2020, 20, 370. [Google Scholar] [CrossRef]
- Takahashi, S.; Yoshida, C.; Takahashi, H.; Nishihara, M. Isolation and functional analysis of EPHEMERAL1-LIKE (EPH1L) genes involved in flower senescence in cultivated Japanese gentians. Int. J. Mol. Sci. 2022, 23, 5608. [Google Scholar] [CrossRef]
- Watanabe, K.; Kobayashi, A.; Endo, M.; Sage-Ono, K.; Toki, S.; Ono, M. CRISPR/Cas9-mediated mutagenesis of the dihydroflavonol-4-reductase-B (DFR-B) locus in the Japanese morning glory Ipomoea (Pharbitis) nil. Sci. Rep. 2017, 7, 10028. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Oda-Yamamizo, C.; Sage-Ono, K.; Ohmiya, A.; Ono, M. Alteration of flower colour in Ipomoea nil through CRISPR/Cas9-mediated mutagenesis of carotenoid cleavage dioxygenase 4. Transgenic Res. 2018, 27, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, K.; Watanabe, K.; Ono, M. CRISPR/Cas9-mediated mutagenesis of the EPHEMERAL1 locus that regulates petal senescence in Japanese morning glory. Plant Physiol. Biochem. 2018, 131, 53–57. [Google Scholar] [CrossRef]
- Yan, R.; Wang, Z.; Ren, Y.; Li, H.; Liu, N.; Sun, H. Establishment of efficient genetic transformation systems and application of CRISPR/Cas9 genome editing technology in Lilium pumilum DC. Fisch. and Lilium longiflorum white heaven. Int. J. Mol. Sci. 2019, 20, 2920. [Google Scholar] [CrossRef]
- Zhang, B.; Yang, X.; Yang, C.; Li, M.; Guo, Y. Exploiting the CRISPR/Cas9 system for targeted genome mutagenesis in Petunia. Sci. Rep. 2016, 6, 20315. [Google Scholar] [CrossRef]
- Subburaj, S.; Chung, S.J.; Lee, C.; Ryu, S.-M.; Kim, D.H.; Kim, J.-S.; Bae, S.; Lee, G.-J. Site-directed mutagenesis in Petunia x hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 2016, 35, 1535–1544. [Google Scholar] [CrossRef]
- Xu, J.; Kang, B.C.; Naing, A.H.; Bae, S.J.; Kim, J.-S.; Kim, H.; Kim, C.K. CRISPR/Cas9-mediated editing of 1-aminocyclopropane-1-carboxylate oxidase1 enhances Petunia flower longevity. Plant Biotechnol. J. 2020, 18, 287–297. [Google Scholar] [CrossRef]
- Zhang, B.; Xu, X.; Huang, R.; Yang, S.; Li, M.; Guo, Y. CRISPR/Cas9-mediated targeted mutation reveals a role for AN4 rather than DPL in regulating venation formation in the corolla tube of Petunia hybrida. Hortic. Res. 2021, 8, 116. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Kao, T. CRISPR/Cas9-mediated knockout of PiSSK1 reveals essential role of S-locus F-box protein containing SCF complexes in recognition of non-self S-RNases during cross-compatible pollination in self-incompatible Petunia inflata. Plant Reprod. 2018, 31, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.-G.; Wu, F.-H.; Yuan, Y.-H.; Chen, Y.-R.; Lin, C.-S. High-efficiency CRISPR/Cas-based editing of Phalaenopsis orchid MADS genes. Plant Biotechnol. J. 2020, 18, 889–891. [Google Scholar] [CrossRef] [PubMed]
- Nitarska, D.; Boehm, R.; Debener, T.; Lucaciu, R.C.; Halbwirth, H. First genome edited poinsettias: Targeted mutagenesis of flavonoid 3’-hydroxylase using CRISPR/Cas9 results in a colour shift. PCTOC 2021, 147, 49–60. [Google Scholar] [CrossRef]
- Su, S.; Xiao, W.; Guo, W.; Yao, X.; Xiao, J.; Ye, Z.; Wang, N.; Jiao, K.; Lei, M.; Peng, Q.; et al. The cycloidea-radialis module regulates petal shape and pigmentation, leading to bilateral corolla symmetry in Torenia fournieri (Linderniaceae). New Phytol. 2017, 215, 1582–1593. [Google Scholar] [CrossRef] [PubMed]
- Nishihara, M.; Higuchi, A.; Watanabe, A.; Tasaki, K. Application of the CRISPR/Cas9 system for modification of flower color in Torenia fournieri. BMC Plant Biol. 2018, 18, 331. [Google Scholar] [CrossRef]
- Gottwald, T.R.; Graham, J.H.; Schubert, T.S. Citrus canker: The pathogen and its impact. Plant Health Prog. 2002, 3, 1–34. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, J.L.; Jia, H.G.; Sosso, D.; Li, T.; Frommer, W.B.; Yang, B.; White, F.F.; Wang, N.; Jones, J.B. Lateral organ boundaries1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA 2014, 111, 521–529. [Google Scholar] [CrossRef]
- Jia, H.; Orbović, V.; Wang, N. CRISPR-LbCas12a-mediated modification of citrus. Plant Biotechnol. J. 2019, 17, 1928–1937. [Google Scholar] [CrossRef]
- Leisen, T.; Bietz, F.; Werner, J.; Wegner, A.; Schaffrath, U.; Scheuring, D.; Willmund, F.; Mosbach, A.; Scalliet, G.; Hahn, M. CRISPR/Cas with ribonucleoprotein complexes and transiently selected telomere vectors allows highly efficient marker-free and multiple genome editing in Botrytis cinerea. PLoS Pathog. 2020, 16, e1008326. [Google Scholar] [CrossRef] [PubMed]
- Khadgi, A. Uncovering genetic and transcriptomic regulation of prickle development in red raspberry. A dissertation presented to the Faculty of the Graduate School of Cornell University in partial fulfillment of the requirements for the Degree of Doctor of Philosophy. Cornell Theses Diss. 2020, 12, 158. [Google Scholar] [CrossRef]
- Ge, Z.; Zheng, L.; Zhao, Y.; Jiang, J.; Zhang, E.J.; Liu, T.; Gu, H.; Qu, L.J. Engineered xCas9 and SpCas9-NG variants broaden PAM recognition sites to generate mutations in Arabidopsis plants. Plant Biotechnol. J. 2019, 17, 1865–1867. [Google Scholar] [CrossRef] [PubMed]
- Hua, K.; Tao, X.; Han, P.; Wang, R.; Zhu, J.K. Genome engineering in rice using Cas9 variants that recognize NG PAM sequences. Mol. Plant 2019, 12, 1003–1014. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Luo, J.; Xu, M.; Li, S.; Zhang, J.; Li, H.; Yan, L.; Zhao, Y.; Xia, L. Plant genome editing using xCas9 with expanded PAM compatibility. J. Genet. Genom. 2019, 46, 277–280. [Google Scholar] [CrossRef]
- Sretenovic, S.; Yin, D.; Levav, A.; Selengut, J.D.; Mount, S.M.; Qi, Y. Expanding plant genome-editing scope by an engineered iSpyMacCas9 system that targets A-rich PAM sequences. Plant Commun. 2021, 2, 100101. [Google Scholar] [CrossRef]
- Han, X.; Yang, Y.; Han, X.; Ryner, J.T.; Ahmed, ·E.A.H.; Qi, Y.; Zhong, G.Y.; Song, G.Q. CRISPR Cas9- and Cas12a-mediated gusA editing in transgenic blueberry. PCTOC 2022, 148, 217–229. [Google Scholar] [CrossRef]
- Alok, A.; Sandhya, D.; Jogam, P.; Rodrigues, V.; Bhati, K.K.; Sharma, H.; Kumar, J. The rise of the CRISPR/Cpf1 system for efficient genome editing in plants. Front. Plant Sci. 2020, 11, 264. [Google Scholar] [CrossRef]
- Bandyopadhyay, A.; Kancharla, N.; Javalkote, V.S.; Dasgupta, S.; Brutnell, T.P. CRISPR-Cas12a (Cpf1): A versatile tool in the plant genome editing tool box for agricultural advancement. Front. Plant Sci. 2020, 11, 584151. [Google Scholar] [CrossRef]
- Ming, M.; Long, H.; Ye, Z.; Pan, C.; Chen, J.; Tian, R.; Sun, C.; Xue, Y.; Zhang, Y.; Li, J.; et al. Highly efficient CRISPR systems for loss-of-function and gain-of-function research in pear calli. Hortic. Res. 2022, 9, uhac148. [Google Scholar] [CrossRef]
- Jia, H.; Wang, Y.; Su, H.; Huang, X.; Wang, N. LbCas12a-D156R efficiently edits LOB1 effector binding elements to generate canker-resistant citrus plants. Cells 2022, 11, 315. [Google Scholar] [CrossRef] [PubMed]
- Pausch, P.; Al-Shayeb, B.; Bisom-Rapp, E.; Tsuchida, C.A.; Li, Z.; Cress, B.F.; Knott, G.J.; Jacobsen, S.E.; Banfield, J.F.; Doudna, J.A. CRISPR-CasF from huge phages is a hypercompact genome editor. Science 2020, 369, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Guo, D.; Cao, Y.; Li, Y.; Ma, R.; Liu, W. Application of CRISPR/CasΦ2 system for genome editing in plants. Int. J. Mol. Sci. 2022, 23, 5755. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Wu, X.; Markel, K.; Malzahn, A.A.; Kundagrami, N.; Sretenovic, S.; Zhang, Y.; Cheng, Y.; Shih, P.M.; Qi, Y. CRISPR–Act3.0 for highly efficient multiplexed gene activation in plants. Nat. Plants 2021, 7, 942–953. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Li, G.; Malzahn, A.A.; Cheng, Y.; Leyson, B.; Sretenovic, S.; Gurel, F.; Coleman, G.D.; Qi, Y. Boosting plant genome editing with a versatile CRISPR-Combo system. Nat. Plants 2022, 8, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Paul, H.M.; Istanto, D.D.; Heldenbrand, J.; Hudson, M.E. CROPSR: An automated platform for complex genome-wide CRISPR gRNA design and validation. BMC Bioinform. 2022, 23, 74. [Google Scholar] [CrossRef]
- Pramanik, D.; Shelake, R.M.; Kim, M.J.; Kim, J.-Y. CRISPR-mediated engineering across the central dogma in plant biology for basic research and crop improvement. Mol. Plant 2021, 14, 127–150. [Google Scholar] [CrossRef]
- Buchholzer, M.; Frommer, W.B. An increasing number of countries regulate genome editing in crops. New Phytol. 2022, 23, 12–15. [Google Scholar] [CrossRef]
- Barrangou, R. CRISPR craziness: A response to the EU Court ruling. CRISPR J. 2018, 1, 251–252. [Google Scholar] [CrossRef]
- Faure, J.D.; Napier, J.A. Europe’s first and last field trial of gene-edited plants? Elife 2018, 7, e42379. [Google Scholar] [CrossRef]
- Metje-Sprink, J.; Sprink, T.; Hartung, F. Genome-edited plants in the field. Curr. Opin. Biotechnol. 2020, 61, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Morineau, C.; Bellec, Y.; Tellier, F.; Gissot, L.; Kelemen, Z.; Nogue, F.; Faure, J.D. Selective gene dosage by CRISPR-Cas9 genome editing in hexaploid Camelina sativa. Plant Biotechnol. J. 2017, 15, 729–739. [Google Scholar] [CrossRef]
- Neequaye, M.; Stavnstrup, S.; Harwood, W.; Lawrenson, T.; Hundleby, P.; Irwin, J.; Troncoso-Rey, P.; Saha, S.; Traka, M.H.; Mithen, R.; et al. CRISPR-Cas9-mediated gene editing of MYB28 genes impair glucoraphanin accumulation of Brassica oleracea in the field. CRISPR J. 2021, 4, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Mallapaty, S. China’s approval of gene-edited crops energizes researchers. Nature 2022, 602, 559–560. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef]
- Li, S.; Lin, D.; Zhang, Y.; Deng, M.; Chen, Y.; Lv, B.; Li, B.; Lei, Y.; Wang, Y.; Zhao, L.; et al. Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 2022, 602, 455–460. [Google Scholar] [CrossRef]

| Plant | Targeted Gene | Trait | References |
|---|---|---|---|
| Apple Malus domestica | DIPM-1, 2, 4 | Fire blight disease resistance | [20,45] |
| MdDIPM4 | |||
| MdPDS | Photobleaching, albinism | [25,42,46] | |
| Md/PcPDS | |||
| CNGC2 | Resistance to Botryosphaeria dothidea | [47] | |
| Banana Musa Whilliams cv. Cavendish | PDS | Photobleaching, albinism, dwarfing | [48] |
| Musa acuminata | PDS | Photobleaching, albinism | [40,49] |
| Musa balbisiana | BSOLV | Banana streak virus resistance | [23] |
| Musa acuminata ‘Gros Michel’ | MaGA20ox2 | Semi-dwarf phenotype | [26] |
| Musa acuminata | MaACO1 | Fruit ripening delay, extended shelf life | [27] |
| Musa spp. | LCYε | Sixfold enhancement of β-carotene content in fruits | [50] |
| DMR6 | Banana Xanthomonas wilt resistance | [51] | |
| Blueberry Vaccinum corymbosum | CEN | Dwarfism, lack of precocious flowering | [52] |
| Cacao Theobroma cacao | TcNPR3 | Resistance to Ph. tropicalis | [53] |
| Carrizo citrange Citrus sinensis L. Osb. x Poncirus trifoliata L. Raf. | CsALS | Herbicide resistance | [29] |
| Coffee Coffea canephora | CcPDS | Photobleaching, albinism | [54] |
| Grapefruit Citrus paradisi | CsLOB1 | Citrus canker resistance | [55,56,57,58] |
| Grapes Vitis vinifera L. | IdnDH | Failure of tartaric acid biosynthesis | [59,60] |
| PDS | Photobleaching, albinism | [31,32,61] | |
| WRKY52 | Botrytis cinerea resistance | [62] | |
| MLO-7 | Resistance to powdery mildew | [20] | |
| VvMLO3, VvMLO4 | Resistance to powdery mildew | [63] | |
| VvPR4b | Sensitivity to downey mildew | [64] | |
| VvCCD8 | Highly branched phenotype | [65] | |
| Kiwifruit Actinidia chinensis | AcPDS | Photobleaching, albinism | [66] |
| AcCen4, AcCen | Compact growth, terminal flowering | [67] | |
| AcCen4, SyGl | Rapid flowering | [28] | |
| Kumquat Fortunella hindsii | FhPDS | Photobleaching, albinism | [24] |
| FhCCD4b | No mutant phenotype | ||
| FhDUO1 | |||
| FhNZZ | Leaf curling, longer pedicel length | [68] | |
| Melon Cucumis melo | CmPDS | Photobleaching, albinism | [69] |
| CmNAC-NOR, | Shelf life | [70,71] | |
| CTR1-like, ROS1 | |||
| Orange Citrus sinensis Wanjincheng | CsWRKY22 | Delayed citrus canker symptoms | [72] |
| Papaya Carica papaya L. | CpPDS | Photobleaching, albinism | [73] |
| Pear Pyrus communis L. | MdTFL1, Pc TFL1 | Early flowering | [25] |
| Pyrus bretschneideri | Md/PcALS | Herbicide resistance | [42] |
| PbPAT14 | Dwarf yellowing phenotype | [74] | |
| Pomegranate Punica granatum L. | PgUGT84A23, | Change of phenolic metabolites | [75] |
| PgUGT84A24 | |||
| Red raspberry Rubus idaeus L. | F3′H | No mutant phenotype | [76] |
| Strawberry Fragaria vesca | FveARF8 | Dwarfism | [77] |
| FveTAA1 | |||
| FvPDS | Photobleaching, albinism | [78] | |
| FvMYB10, | Changes in anthocyanin synthesis | ||
| FvCHS, | |||
| FvUF3GT, | |||
| FvLDOX | Photobleaching, albinism | ||
| PDS | White berries | [41] | |
| RAP | [79] | ||
| FveSEP3 | Alteration in flowers, abnormal berries | [80] | |
| Strawberry Fragaria vesca, F. x ananassa | FaTM6 | Abnormal petals, anthers, pollen grains and berries | [81] |
| PDS | Photobleaching, albinism | [41] | |
| Sweet orange Citrus sinensis | PDS | Photobleaching, albinism | [39,82,83] |
| CsLOB1 | Citrus canker resistance | [22] | |
| Walnut Juglans regia | JrPDS | Photobleaching, albinism | [84] |
| JrWOX11 | Reduced adventitious root formation and vegetative growth | [85] | |
| Watermelon Citrullus lanatus | ClPDS | Photobleaching, albinism | [86] |
| ClALS | Herbicide resistance | [87] | |
| Clpsk1 | Resistance to Fusarium oxysporum f. sp. niveum | [88] | |
| GlBG1 | Decreased seed size and promoted seed germination | [89] | |
| ClCOMT1 | Decreased melatonin content | [90] |
| Plant | Targeted Gene | Trait | References |
|---|---|---|---|
| Chrysanthemum moriflorium | CpYGFP | Fluorescence | [91] |
| Dendrobium officinale | C3H, C4H, 4CL, CCR, IRX | No mutant phenotype | [92] |
| Japanese gentians Gentiana scabra x G. triflora | Gt5GT, Gt3′GT, Gt5/3′AT | Flower color change | [93] |
| GST1 | Flower color change | [94] | |
| EPH1 | Flower longevity | [95] | |
| Japanese morning glory Ipomoea nil | DFR-B | Flower color change | [96] |
| CCD4 | Flower color change | [97] | |
| EPH1 | Flower longevity | [98] | |
| Lilium longiflorum, | LpPDS | Photobleaching, albinism | [99] |
| L. pumilum | |||
| Petunia Petunia hybrida | PDS | Photobleaching, albinism | [100] |
| NR | Deficiency in nitrate assimilation | [101] | |
| Flower longevity | |||
| ACO1 | Absence of corolla tube venation | [102] | |
| AN4 | Self-incompatibility | [103] | |
| P. inflata | PiSSK1 | [104] | |
| Phalaenopsis equestris | MADS8, MADS36, MADS44 | Long juvenile period | [105] |
| Poinsettia Euphorbia pulcherrima | F3′H | Change of the bract color from red to reddish orange | [106] |
| Torenia fournieri | TfRAD1 | Abnormal shape and color of flowers | [107] |
| Pale blue flowers | |||
| F3H | [108] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rukavtsova, E.B.; Zakharchenko, N.S.; Lebedev, V.G.; Shestibratov, K.A. CRISPR-Cas Genome Editing for Horticultural Crops Improvement: Advantages and Prospects. Horticulturae 2023, 9, 38. https://doi.org/10.3390/horticulturae9010038
Rukavtsova EB, Zakharchenko NS, Lebedev VG, Shestibratov KA. CRISPR-Cas Genome Editing for Horticultural Crops Improvement: Advantages and Prospects. Horticulturae. 2023; 9(1):38. https://doi.org/10.3390/horticulturae9010038
Chicago/Turabian StyleRukavtsova, Elena B., Natalia S. Zakharchenko, Vadim G. Lebedev, and Konstantin A. Shestibratov. 2023. "CRISPR-Cas Genome Editing for Horticultural Crops Improvement: Advantages and Prospects" Horticulturae 9, no. 1: 38. https://doi.org/10.3390/horticulturae9010038
APA StyleRukavtsova, E. B., Zakharchenko, N. S., Lebedev, V. G., & Shestibratov, K. A. (2023). CRISPR-Cas Genome Editing for Horticultural Crops Improvement: Advantages and Prospects. Horticulturae, 9(1), 38. https://doi.org/10.3390/horticulturae9010038






