Genome–Wide Investigation of the CBL–CIPK Gene Family in Oil Persimmon: Evolution, Function and Expression Analysis during Development and Stress
Abstract
1. Introduction
2. Results
2.1. Identification of CBL–CIPK Genes in Oil Persimmon
2.2. Phylogenetic Analysis of the CBL–CIPK Gene Family
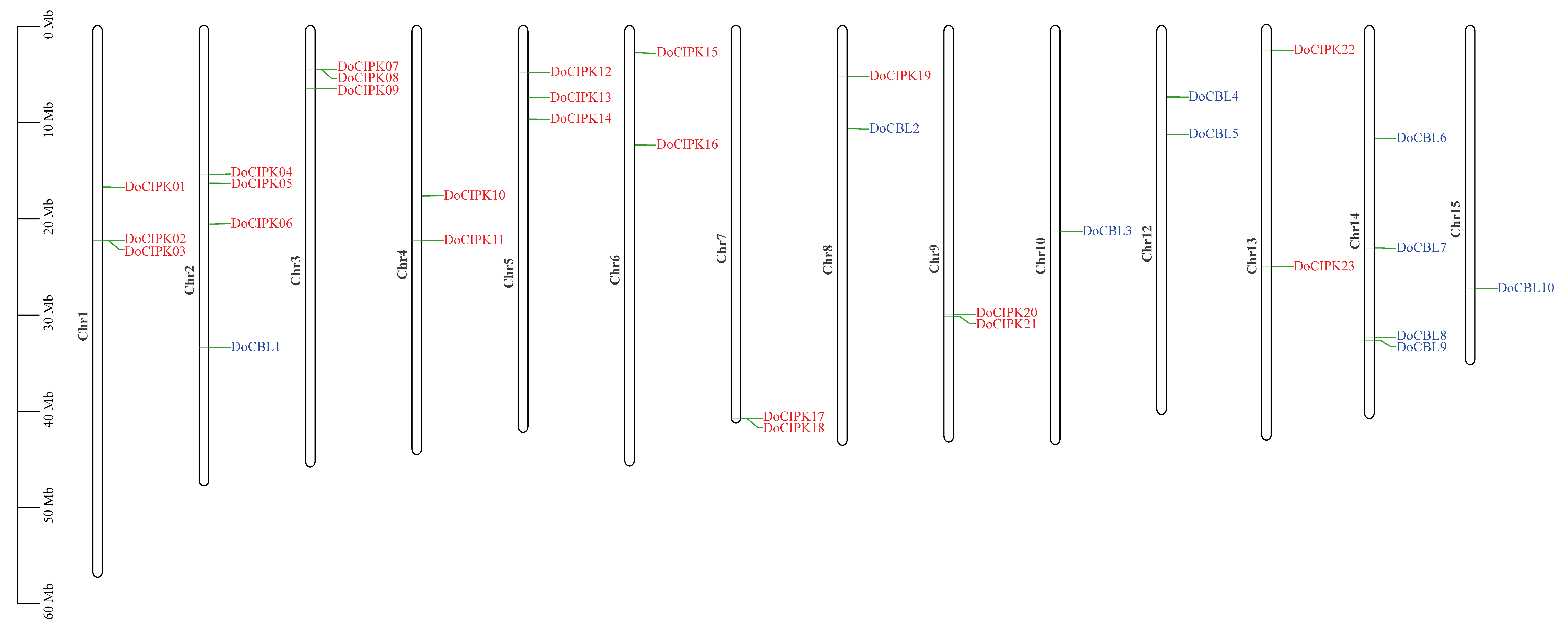
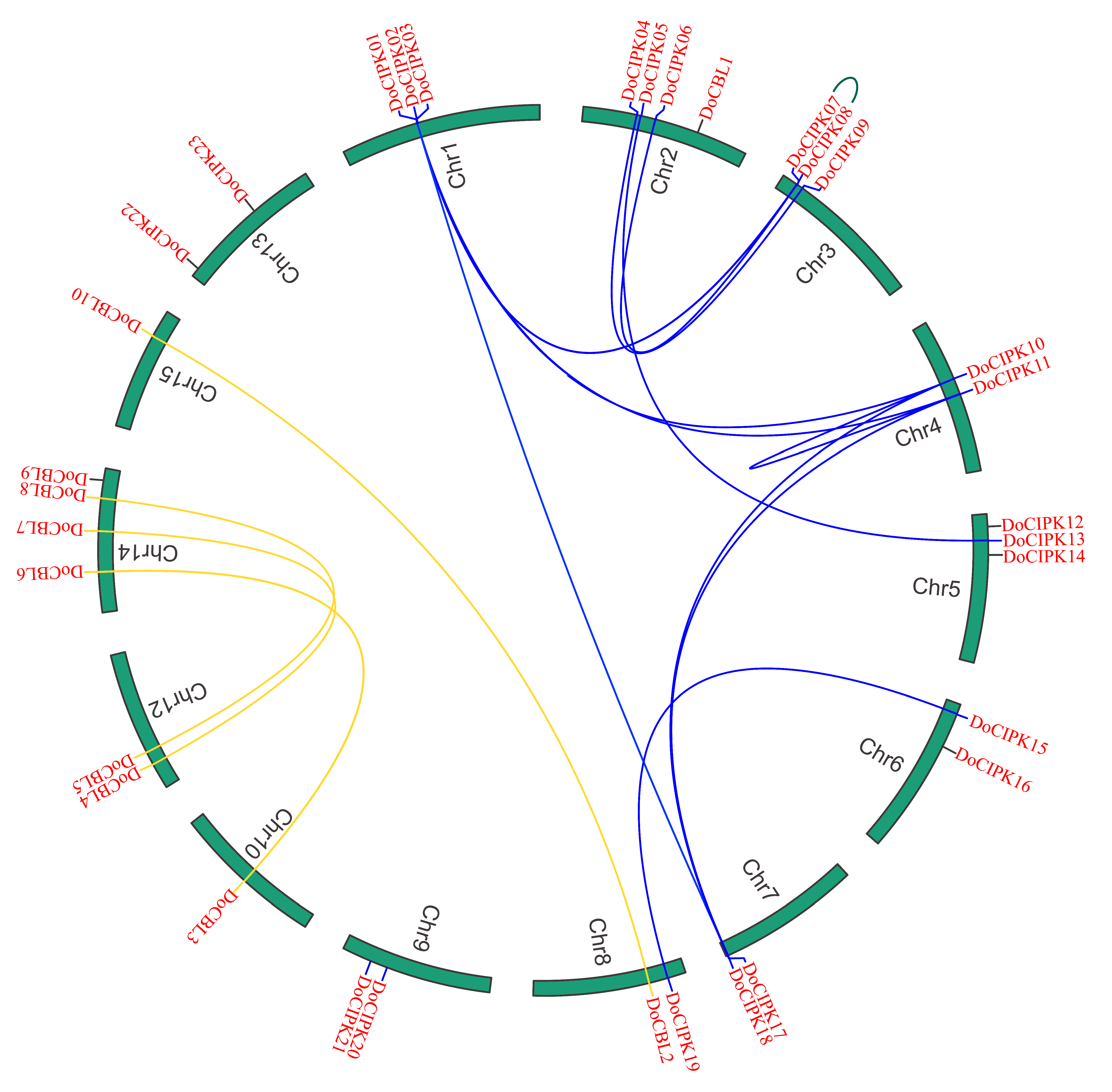
2.3. Chromosomal Location and Gene Duplication
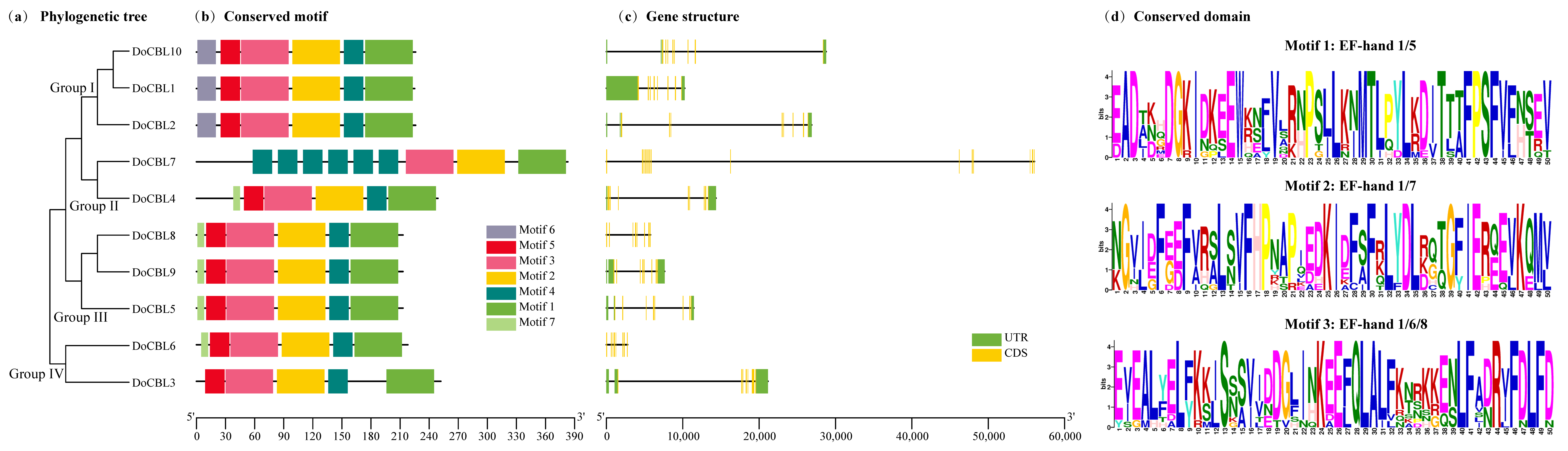
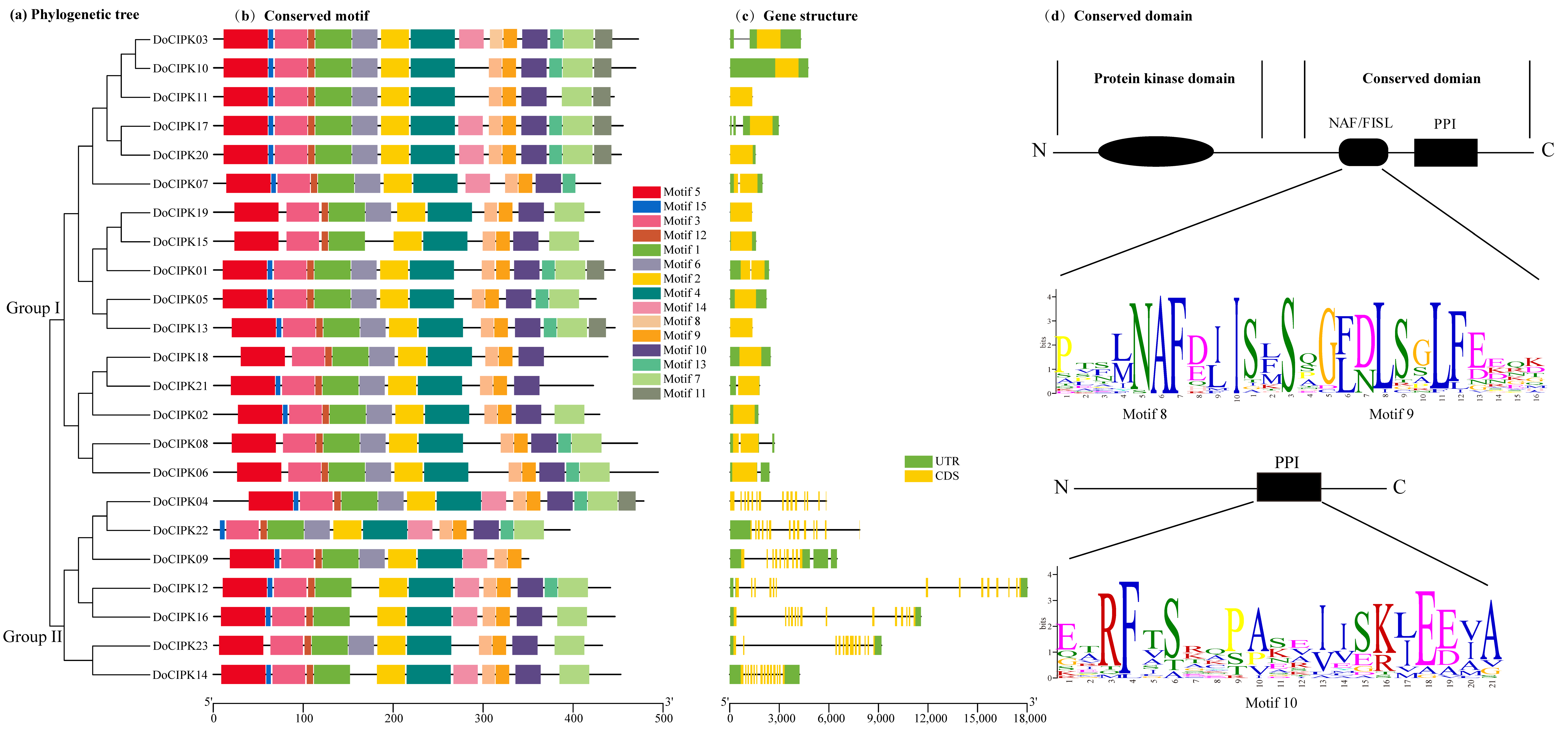
2.4. Conserved Motifs and Gene Structure Analysis
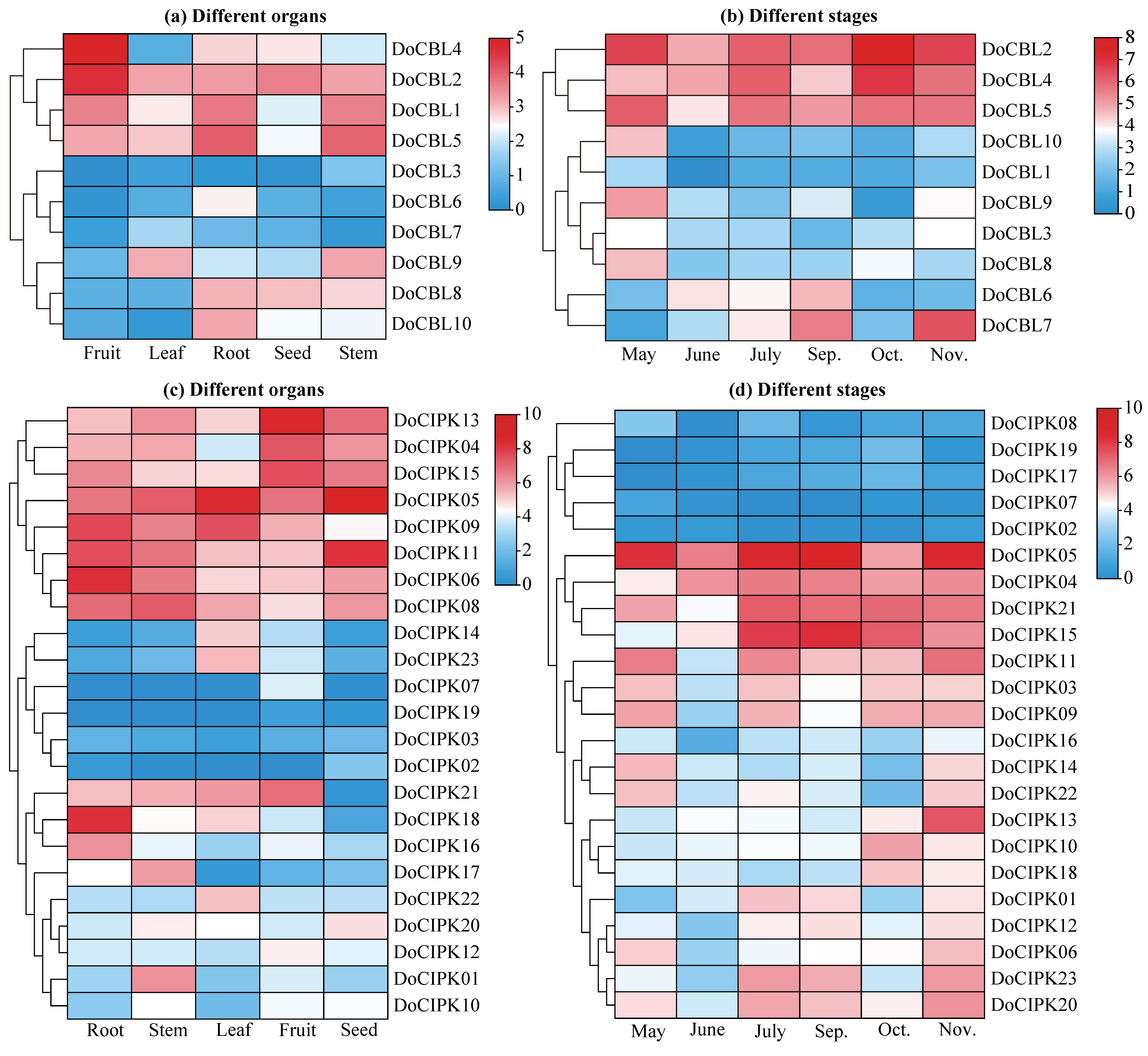
2.5. Expression Patterns of DoCBL and DoCIPK Genes by Transcriptome
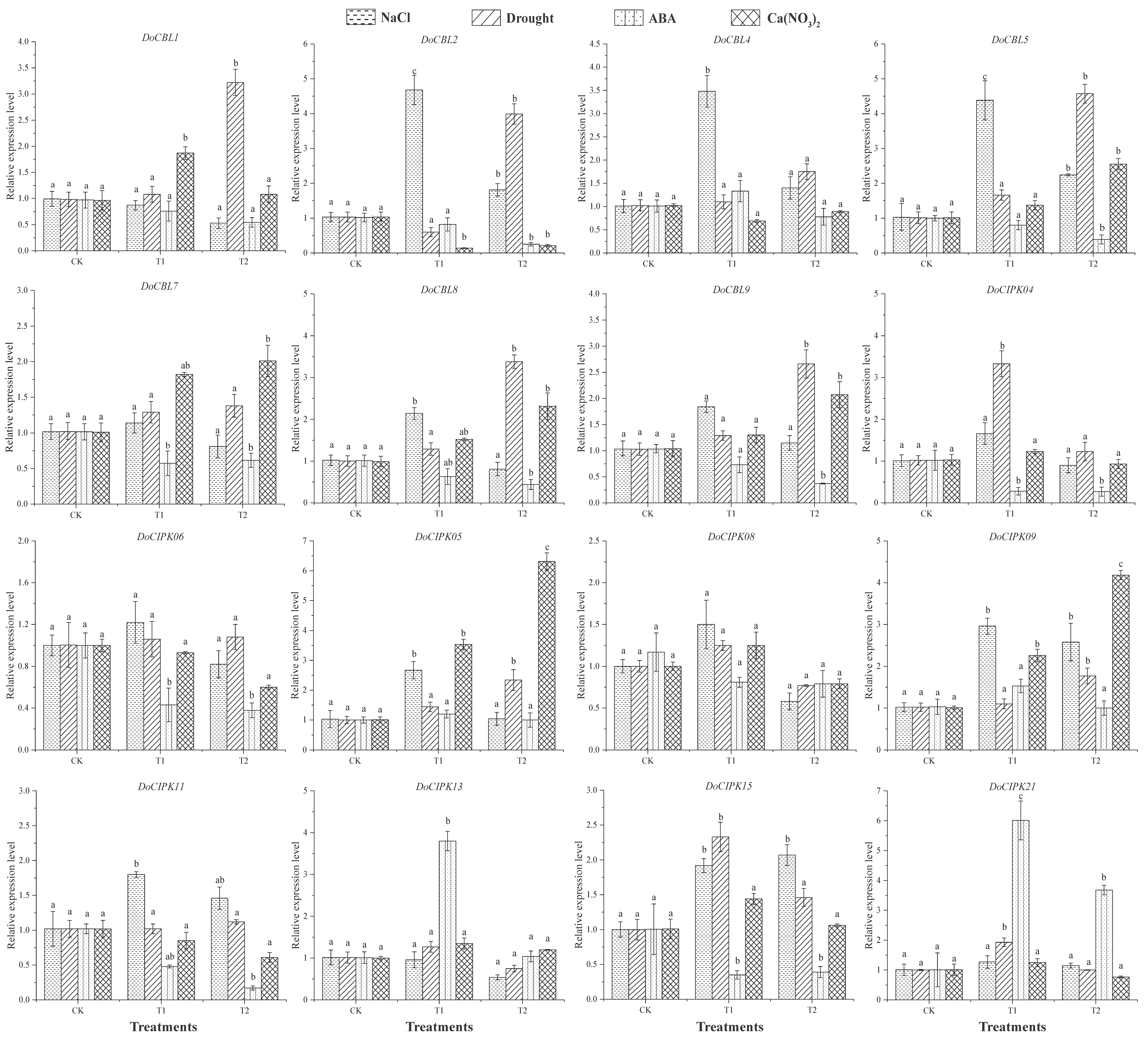
2.6. Expression Levels of DoCBL and DoCIPK Genes under Different Stresses
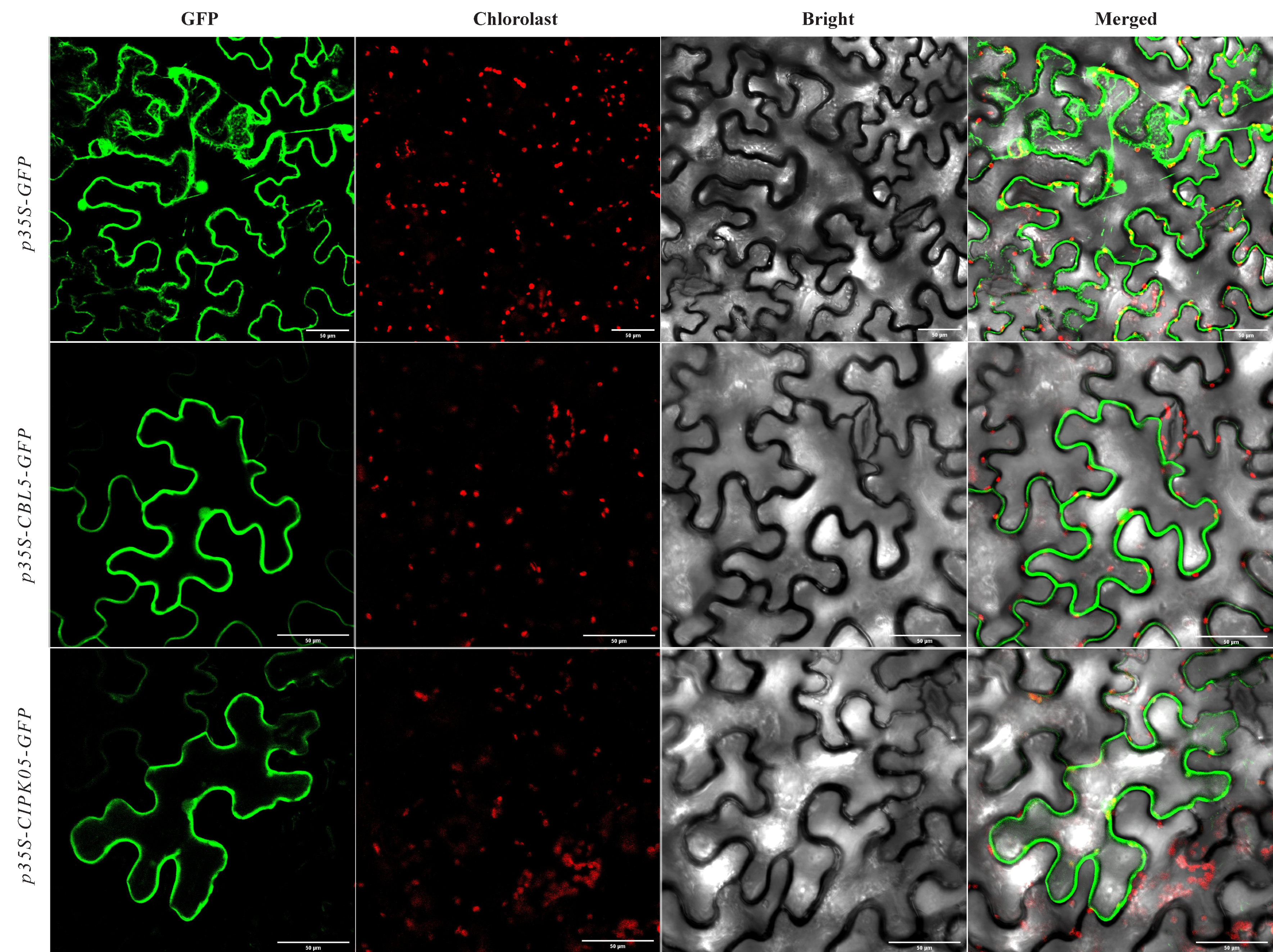
2.7. Gene Cloning and Subcellular Localization of DoCBL5 and DoCIPK05
2.8. The Interaction Network of DoCBL5 and DoCIPK05
3. Discussion
4. Materials and Methods
4.1. Identification of CBL–CIPK Gene Family in Oil Persimmon
4.2. Phylogenetic Analysis of CBL–CIPK Gene Family
4.3. Chromosomal Location, Gene Duplication, Gene Structure and Conserved Motifs
4.4. Plant Materials and Growth Conditions
4.5. Expression Patterns of DoCBL and DoCIPK Genes
4.6. Gene Cloning and Subcellular Localization
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Riveras, E.; Alvarez, J.M.; Vidal, E.A.; Oses, C.; Vega, A.; Gutiérrez, R.A. The calcium ion Is a second messenger in the nitrate signaling pathway of Arabidopsis. Plant Physiol. 2015, 169, 1397–1404. [Google Scholar] [CrossRef] [PubMed]
- Sheng, L. The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 2009, 14, 37–42. [Google Scholar]
- Petit-Glatron, M.F.; Grajcar, L.; Munz, A.; Chambert, R. The contribution of the cell wall to a transmembrane calcium gradient could play a key role in Bacillus subtilis protein secretion. Mol. Microbiol. 2010, 9, 1097–1106. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Shankhdhar, D.; Shankhdhar, S.C. The role of calcium in plant signal transduction under macronutrient deficiency stress. In Plant Macronutrient Use Efficiency; Sharma, A., Shankhdhar, D., Shankhdhar, S.C., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 181–196. [Google Scholar]
- Sanyal, S.K.; Pandey, A.; Pandey, G.K. The CBL–CIPK signaling module in plants: A mechanistic perspective. Physiol. Plant 2015, 155, 89–108. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Mohanta, N.; Mohanta, Y.K.; Parida, P.; Bae, H. Genome-wide identification of calcineurin b-Like (CBL) gene family of plants reveals novel conserved motifs and evolutionary aspects in calcium signaling events. BMC Plant Biol. 2015, 15, 189. [Google Scholar] [CrossRef]
- Batistic, O.; Sorek, N.; Schültke, S.; Kudla, S.Y. Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell 2008, 20, 1346–1362. [Google Scholar] [CrossRef]
- de la Torre, F.; Gutiérrez-Beltrán, E.; Pareja-Jaime, Y.; Chakravarthy, S.; Martin, G.B.; del Pozoa, O. The tomato calcium sensor CBL10 and its interacting protein kinase CIPK6 define a signaling pathway in plant immunity. Plant Cell 2013, 25, 2748–2764. [Google Scholar] [CrossRef]
- Albrecht, V.; Ritz, O.; Linder, S.; Harter, K.; Kudla, J. The NAF domain defines a novel protein–protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 2001, 20, 1051–1063. [Google Scholar] [CrossRef]
- Ohta, M.; Guo, Y.; Halfter, U.; Zhu, J.K. A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. USA 2003, 100, 11771–11776. [Google Scholar] [CrossRef]
- Ji, H.T.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The salt overly sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef]
- Yin, X.; Xia, Y.; Xie, Q.; Cao, Y.; Wang, Z.; Hao, G.; Song, J.; Zhou, Y.; Jiang, X. The protein kinase complex CBL10–CIPK8–SOS1 functions in Arabidopsis to regulate salt tolerance. J. Exp. Bot. 2019, 6, 1801–1814. [Google Scholar] [CrossRef] [PubMed]
- Batistic, O.; Kudla, J. Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 2004, 219, 915–924. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, C.; Tang, R.J.; Xu, H.X.; Luan, S. Calcineurin B-Like proteins CBL4 and CBL10 mediate two independent salt tolerance pathways in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 2421. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Kolukisaoglu, U.; Weinl, S.; Blazevic, D.; Batistic, O.; Kudla, J. Calcium sensors and their Interacting protein kinases: Genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 2004, 134, 43–58. [Google Scholar] [CrossRef]
- Cuéllar, T.; Azeem, F.; Andrianteranagna, M.; Pascaud, F.; Verdeil, J.-L.; Sentenac, H.; Zimmermann, S.; Gaillard, I. Potassium transport in developing fleshy fruits: The grapevine inward K+ channel VvK1.2 is activated by CIPK-CBL complexes and induced in ripening berry flesh cells. Plant J. 2013, 73, 1006–1018. [Google Scholar] [CrossRef] [PubMed]
- Kanwar, P.; Sanyal, S.K.; Tokas, I.; Yadav, A.K.; Pandey, A.; Kapoor, S.; Pandey, G.K. Comprehensive structural, interaction and expression analysis of CBL and CIPK complement during abiotic stresses and development in rice. Cell Calcium 2014, 56, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.J.; Manik, S.M.N.; Shi, S.J.; Chao, J.T.; Jin, Y.R.; Wang, Q.; Liu, H.B. Mechanisms and physiological roles of the CBL-CIPK networking system in Arabidopsis thaliana. Genes 2016, 7, 62. [Google Scholar] [CrossRef]
- Xiang, Y.; Huang, Y.M.; Xiong, L.Z. Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 2007, 144, 1416. [Google Scholar] [CrossRef]
- Tao, S.; Yan, W.; Meng, W.; Li, T.; Yi, Z.; Wang, X.; Wei, S.; He, G.; Yang, G. Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 265. [Google Scholar]
- Niu, L.L.; Dong, B.Y.; Song, Z.H.; Dong, M.; Fu, Y.J. Genome-wide identification and characterization of CIPK family and analysis responses to various stresses in apple (Malus domestica). Int. J. Mol. Sci. 2018, 19, 2131. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Jing, L.; Hui, L.; Li, X.; Yang, Q.; Cheng, Z.M.; Chang, Y. Characterization of CIPK family in asian pear (Pyrus bretschneideri Rehd) and co-expression analysis related to salt and osmotic stress responses. Front. Plant Sci. 2016, 7, 1361. [Google Scholar] [CrossRef] [PubMed]
- Duangjai, S.; Wallnfer, B.; Samuel, R.; Munzinger, J.; Chase, M.W. Generic delimitation and relationships in Ebenaceae sensu lato: Evidence from six plastid DNA regions. Am. J. Bot. 2006, 93, 1808–1827. [Google Scholar] [CrossRef] [PubMed]
- Woolf, A.B.; Ben-Arie, R. Persimmon (Diospyros kaki L.). In Postharvest Biology and Technology of Tropical and Subtropical Fruits; Yahia, E.M., Ed.; Woodhead Publishing: Cambridge, UK, 2011; Volume 4, pp. 166–193, 194e. [Google Scholar]
- Suo, Y.; Sun, P.; Cheng, H.; Han, W.; Fu, J. A high-quality chromosomal genome assembly of Diospyros oleifera Cheng. GigaScience 2020, 9, giz164. [Google Scholar] [CrossRef]
- Zhu, Q.G.; Xu, Y.; Yang, Y.; Guan, C.F.; Zhang, Q.Y.; Huang, J.W.; Grierson, D.; Chen, K.S.; Gong, B.C.; Yin, X.R. The persimmon (Diospyros oleifera Cheng) genome provides new insights into the inheritance of astringency and ancestral evolution. Hortic. Res. 2019, 6, 138. [Google Scholar] [CrossRef]
- Yue, X.; Liu, J.; Dong, C.; Cheng, Z.M. The CBL and CIPK gene family in grapevine (Vitis vinifera): Genome-wide analysis and expression profiles in response to various abiotic stresses. Front. Plant Sci. 2017, 8, 978. [Google Scholar]
- Tang, R.J.; Wang, C.; Li, K.; Luan, S. The CBL–CIPK calcium signaling network: Unified paradigm from 20 years of discoveries. Trends Plant Sci. 2020, 25, 604–617. [Google Scholar] [CrossRef]
- Kleist, T.J.; Spencley, A.L.; Luan, S. Comparative phylogenomics of the CBL-CIPK calcium-decoding network in the moss Physcomitrella, Arabidopsis, and other green lineages. Front. Plant Sci. 2014, 5, 187. [Google Scholar] [CrossRef]
- Aslam, M.; Fakher, B.; Jakada, B.H.; Zhao, L.; Cao, S.; Cheng, Y.; Qin, Y. Genome-wide identification and expression profiling of CBL-CIPK gene family in pineapple (Ananas comosus) and the role of AcCBL1 in abiotic and biotic stress response. Biomolecules 2019, 9, 293. [Google Scholar] [CrossRef]
- Chen, P.H.; Yang, J.; Mei, Q.L.; Liu, H.Y.; Cheng, Y.P.; Ma, F.W.; Mao, K. Genome-wide analysis of the apple CBL family reveals that Mdcbl10.1 functions positively in modulating apple salt tolerance. Int. J. Mol. Sci. 2021, 22, 12430. [Google Scholar] [CrossRef]
- Linder, M.E.; Deschenes, R.J. Palmitoylation: Policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 2007, 8, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Smotrys, J.E.; Linder, M.E. Palmitoylation of intracellular signaling proteins: Regulation and function. Annu. Rev. Biochem. 2004, 73, 559–587. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Wang, Q.L.; Chen, Q.; Xiang, N.; Yang, Y.Q.; Yang, Y.P. Genome-wide identification and functional analysis of the calcineurin b-like protein and calcineurin b-like protein-interacting protein kinase gene families in turnip (Brassica rapa var. rapa). Front. Plant Sci. 2017, 8, 1191. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.H.; Xia, X.L.; Yin, W.L.; Zhang, H.C. Comparative genomic analysis of CIPK gene family in Arabidopsis and Populus. Plant Growth Regul. 2007, 52, 101–110. [Google Scholar] [CrossRef]
- Li, R.F.; Zhang, J.W.; Wei, J.H.; Wang, H.Z.; Wang, Y.Z.; Ma, R.C. Functions and mechanisms of the CBL–CIPK signaling system in plant response to abiotic stress. Prog. Nat. Sci. 2009, 19, 667–676. [Google Scholar] [CrossRef]
- Yu, J.; Wang, J.; Lin, W.; Li, S.G.; Li, H.; Zhou, J.; Ni, P.X.; Dong, W.; Hu, S.N.; Zeng, C.Q.; et al. The Genomes of Oryza sativa: A history of duplications. PLoS Biol. 2005, 3, e38. [Google Scholar] [CrossRef]
- Schlicker, A.; Domingues, F.S.; Rahnenführer, J.; Lengauer, T. A new measure for functional similarity of gene products based on Gene Ontology. BMC Bioinform. 2006, 7, 302. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Q.Q.; Zhou, L.; Ren, F.; Li, D.D.; Li, X.B. Arabidopsis CBL-interacting protein kinase (CIPK6) is involved in plant response to salt/osmotic stress and ABA. Mol. Biol. Rep. 2013, 40, 4759–4767. [Google Scholar] [CrossRef]
- Xu, J.; Li, H.D.; Chen, L.Q.; Wang, Y.; Liu, L.; He, L.; Wu, W. A protein kinase, interacting with two calcineurin b-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 2006, 125, 1347–1360. [Google Scholar] [CrossRef]
- Chen, L.; Ren, F.; Zhou, L.; Wang, Q.Q.; Zhong, H.; Li, X.B. The Brassica napus Calcineurin B-Like 1/CBL-interacting protein kinase 6 (CBL1/CIPK6) component is involved in the plant response to abiotic stress and ABA signalling. J. Exp. Bot. 2012, 63, 6211–6222. [Google Scholar] [CrossRef]
- Batistic, O.; Kudla, J. Plant calcineurin B-like proteins and their interacting protein kinases. Biochim. Biophys. Acta 2009, 1793, 985–992. [Google Scholar] [CrossRef]
- Lan, W.Z.; Lee, S.C.; Che, Y.F.; Jiang, Y.Q.; Sheng, L. Mechanistic analysis of AKT1 regulation by the CBL-CIPK-PP2CA interactions. Mol. Plant 2011, 4, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Weinl, S.; Kudla, J. The CBL–CIPK Ca2+-decoding signaling network: Function and perspectives. New Phytol. 2009, 184, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.K.; Liu, C.Y.; Huang, X.B.; Zhang, H.Y.; Yuan, Z.H. Land-plant phylogenomic and pomegranate transcriptomic analyses reveal an evolutionary scenario of CYP75 genes subsequent to whole genome duplications. J. Plant Biol. 2019, 62, 48–60. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Hartmann, A.; Tesch, D.; Nothwang, H.G.; Binindaemonds, O.R.P. Evolution of the cation chloride cotransporter family: Ancient origins, gene-losses, and subfunctionalization through duplication. Mol. Biol. Evol. 2014, 31, 434–447. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Zhang, C.F.; Zhang, T.K.; Luebert, F.; Xiang, Y.Z.; Huang, C.-H.; Hu, Y.; Rees, M.; Frohlich, M.W.; Qi, J.; Weigend, M.; et al. Asterid phylogenomics/phylotranscriptomics uncover morphological evolutionary histories and support phylogenetic placement for numerous whole genome duplications. Mol. Biol. Evol. 2020, 37, 3188–3210. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; Debarry, J.D.; Tan, X.F.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.S.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.T.; Deng, Y.Q.; Fan, H.; Sun, Q.J.; Sui, N.; Wang, B.S. Effects of NaCl stress on the growth and photosynthetic characteristics of Ulmus pumila L. seedlings in sand culture. Photosynthetica 2014, 52, 313–320. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Qiu, Y.; Na, R.; Meerja, F.; Lu, Q.S.; Yang, C.; Tian, L. A Golden Gate and Gateway double-compatible vector system for high throughput functional analysis of genes. Plant Sci. 2018, 271, 117–126. [Google Scholar] [CrossRef] [PubMed]


| Gene ID | Name | Length | Mw * (kDa) | pI * | Arabidopsis Orthologs | EF—Hands | Cys Amino Acid Location | Gly Amino Acid Location |
|---|---|---|---|---|---|---|---|---|
| EVM0026572.3 | DoCBL1 | 225 | 25.8 | 5.46 | AT4G26570.1 | 3 | C (4, 18) | G (7) |
| EVM0017523.1 | DoCBL2 | 226 | 26.05 | 4.93 | AT4G26570.1 | 3 | C (4, 12, 18) | G (7) |
| EVM0005359.2 | DoCBL3 | 252 | 29.13 | 5.02 | AT5G24270.1 | 3 | / | G (2) |
| EVM0012501.1 | DoCBL4 | 249 | 28.72 | 4.82 | AT4G26570.1 | 3 | C (7, 9) | / |
| EVM0016230.1 | DoCBL5 | 213 | 24.4 | 4.74 | AT4G17615.2 | 3 | C (3) | G (2) |
| EVM0025671.1 | DoCBL6 | 218 | 24.75 | 5.04 | AT1G64480.1 | 3 | C (20, 40) | G (7) |
| EVM0028457.1 | DoCBL7 | 383 | 44.49 | 9.1 | AT4G26570.1 | 3 | C (32, 46) | / |
| EVM0015273.1 | DoCBL8 | 213 | 24.4 | 4.74 | AT4G17615.1 | 3 | C (3) | G (2) |
| EVM0011958.1 | DoCBL9 | 213 | 24.4 | 4.74 | AT4G17615.2 | 3 | C (3) | G (2) |
| EVM0006583.2 | DoCBL10 | 226 | 26.34 | 4.89 | AT4G26570.1 | 3 | C (4, 12, 18) | / |
| Gene ID | Name | Length | Mw * (kDa) | pI * | Arabidopsis Orthologs | Domains | ||
| EVM0026935.1 | DoCIPK01 | 446 | 50.31 | 7.98 | AT5G10930.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0017998.1 | DoCIPK02 | 429 | 48.22 | 8.48 | AT2G30360.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0029929.1 | DoCIPK03 | 472 | 53.17 | 8.57 | AT5G58380.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0019494.1 | DoCIPK04 | 478 | 53.27 | 8.35 | AT1G30270.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0027553.1 | DoCIPK05 | 425 | 47.57 | 9.2 | AT4G30960.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0031579.3 | DoCIPK06 | 494 | 55.22 | 7.64 | AT4G18700.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0007664.1 | DoCIPK07 | 430 | 47.93 | 9.31 | AT5G45820.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0005656.1 | DoCIPK08 | 471 | 53.09 | 8.84 | AT4G18700.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0030600.1 | DoCIPK09 | 350 | 39.43 | 8.96 | AT1G30270.1 | Ser–Thr kinases, NAF | ||
| EVM0006423.1 | DoCIPK10 | 469 | 53.51 | 9.15 | AT5G58380.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0006143.1 | DoCIPK11 | 445 | 50.09 | 8.63 | AT5G01810.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0019559.1 | DoCIPK12 | 441 | 49.61 | 8.23 | AT5G35410.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0031872.1 | DoCIPK13 | 446 | 49.72 | 8.98 | AT4G30960.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0005984.1 | DoCIPK14 | 453 | 51.28 | 5.77 | AT5G57630.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0022207.1 | DoCIPK15 | 422 | 46.34 | 9.45 | AT3G23000.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0020525.1 | DoCIPK16 | 446 | 50.79 | 6.88 | AT4G24400.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0016074.1 | DoCIPK17 | 455 | 51.25 | 8.82 | AT5G58380.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0026193.1 | DoCIPK18 | 438 | 49.49 | 8.63 | AT5G01820.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0014392.1 | DoCIPK19 | 429 | 47.64 | 8.43 | AT3G23000.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0019419.1 | DoCIPK20 | 453 | 51.09 | 9.42 | AT5G58380.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0016829.1 | DoCIPK21 | 422 | 47.83 | 8.38 | AT2G30360.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0009367.1 | DoCIPK22 | 396 | 44.68 | 6.69 | AT1G30270.1 | Ser–Thr kinases, NAF, PPI | ||
| EVM0000183.1 | DoCIPK23 | 432 | 48.63 | 6.35 | AT3G17510.1 | Ser–Thr kinases, NAF, PPI | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Wang, Y.; Yao, J.; Yang, X.; Wu, K.; Teng, G.; Gong, B.; Xu, Y. Genome–Wide Investigation of the CBL–CIPK Gene Family in Oil Persimmon: Evolution, Function and Expression Analysis during Development and Stress. Horticulturae 2023, 9, 30. https://doi.org/10.3390/horticulturae9010030
Liu C, Wang Y, Yao J, Yang X, Wu K, Teng G, Gong B, Xu Y. Genome–Wide Investigation of the CBL–CIPK Gene Family in Oil Persimmon: Evolution, Function and Expression Analysis during Development and Stress. Horticulturae. 2023; 9(1):30. https://doi.org/10.3390/horticulturae9010030
Chicago/Turabian StyleLiu, Cuiyu, Yanpeng Wang, Jin Yao, Xu Yang, Kaiyun Wu, Guoxin Teng, Bangchu Gong, and Yang Xu. 2023. "Genome–Wide Investigation of the CBL–CIPK Gene Family in Oil Persimmon: Evolution, Function and Expression Analysis during Development and Stress" Horticulturae 9, no. 1: 30. https://doi.org/10.3390/horticulturae9010030
APA StyleLiu, C., Wang, Y., Yao, J., Yang, X., Wu, K., Teng, G., Gong, B., & Xu, Y. (2023). Genome–Wide Investigation of the CBL–CIPK Gene Family in Oil Persimmon: Evolution, Function and Expression Analysis during Development and Stress. Horticulturae, 9(1), 30. https://doi.org/10.3390/horticulturae9010030









