Genome-Wide Identification and Expression Analysis of the KCS Gene Family in Yellow Horn Reveal Their Putative Function on Abiotic Stress Responses and Wax Accumulation
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide Identification of KCS Genes in Yellow Horn
2.2. Physicochemical Properties, Subcellular Localization and Transmembrane Alpha Helix
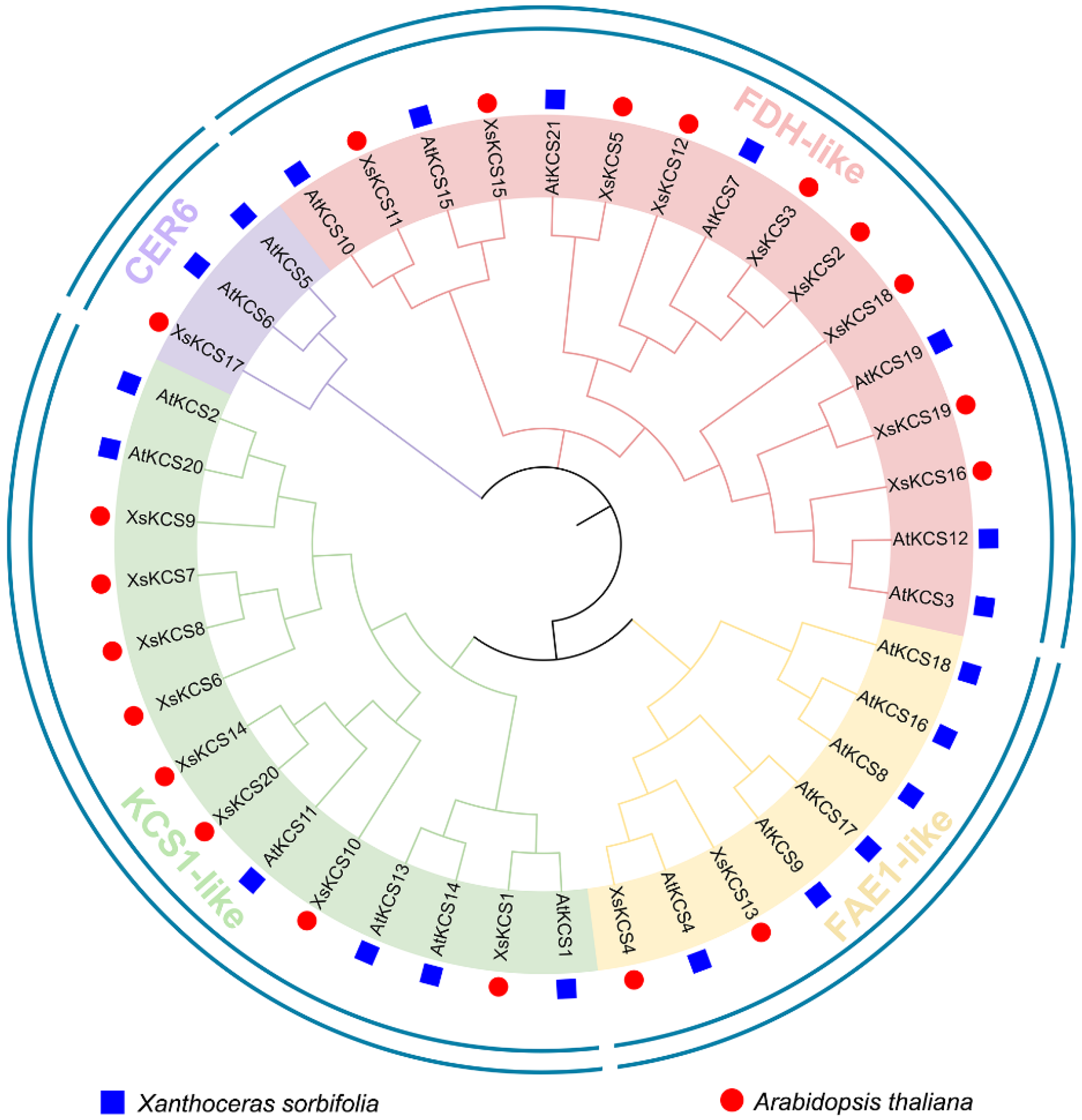
2.3. Phylogenetic Analyses of XsKCS Proteins
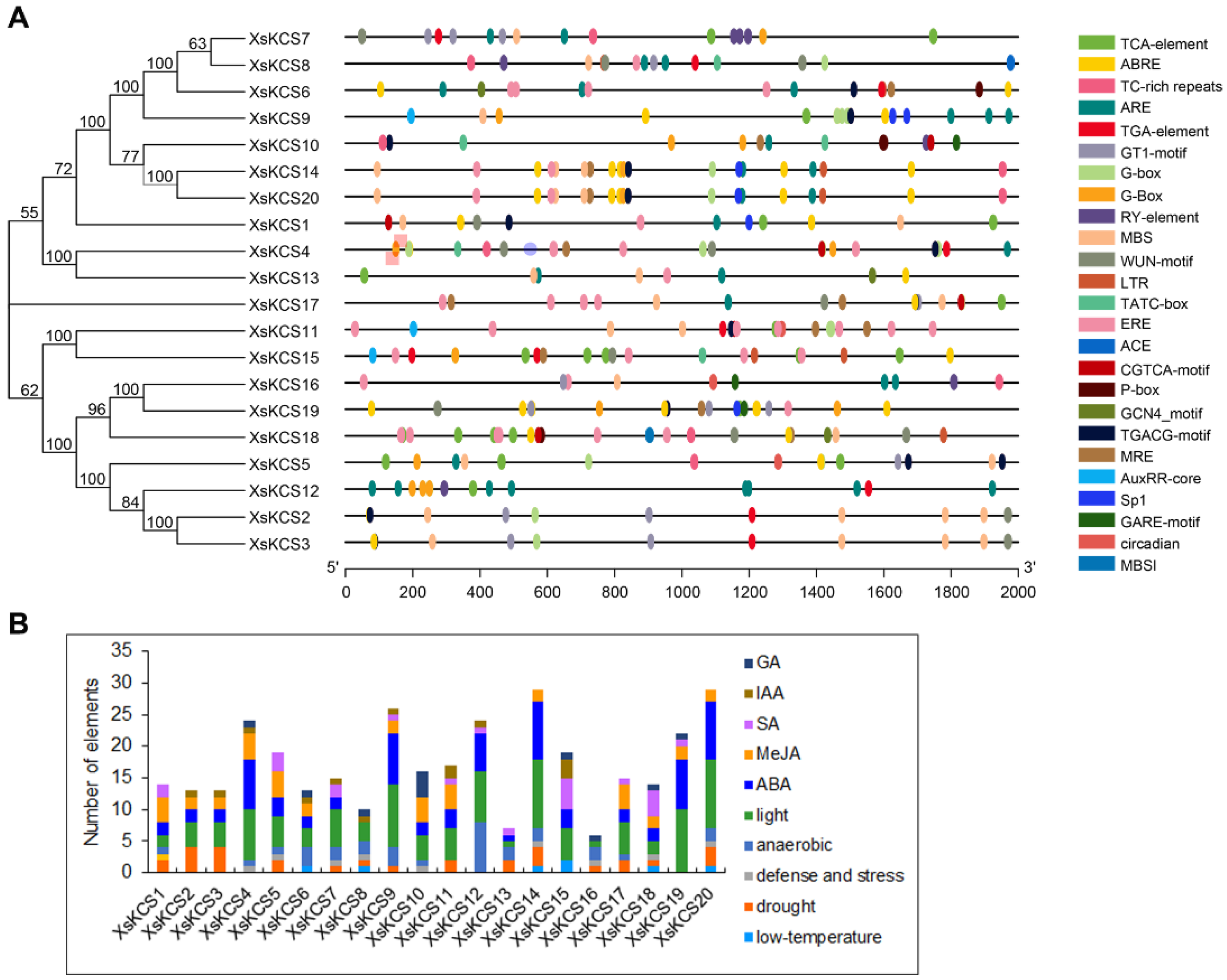
2.4. Gene Structure and Motif Analysis of XsKCS Genes
2.5. Cis-Element Prediction for Promoters
2.6. Chromosome Localization and Synteny Analysis
2.7. Plant Materials and Treatments
2.8. Quantitative Real-Time (qRT)-PCR Analysis
2.9. RNA Sequencing and RNA-Seq Data Analysis
3. Results
3.1. Identification of KCS Genes in Yellow Horn, Physicochemical Properties and Subcellular Localization
3.2. Phylogenetic Analysis of the XsKCS Family
3.3. Conserved Motif and Gene Structure Analysis of XsKCS Genes
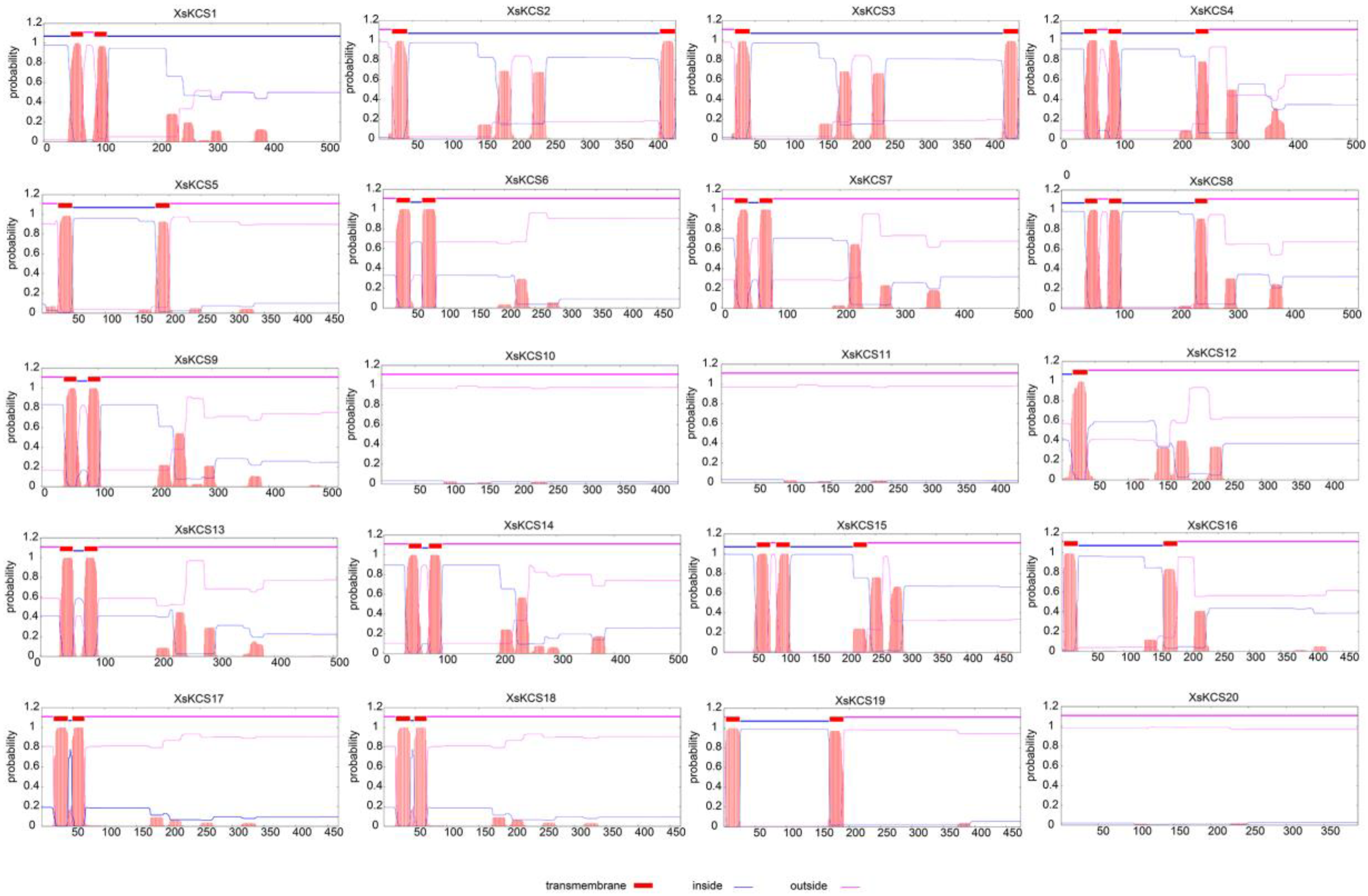
3.4. Transmembrane Alpha Helix of XsKCS Proteins
3.5. Chromosomal Localization and Gene Duplication Analysis of XsKCS Gene Family
3.6. Analysis of Cis-Acting Regulatory Elements of the XsKCS Promoter
3.7. Expression Profiles of XsKCS Genes in Different Tissues of Yellow Horn
3.8. Expression Profile of XsKCS Genes under Abiotic Stress
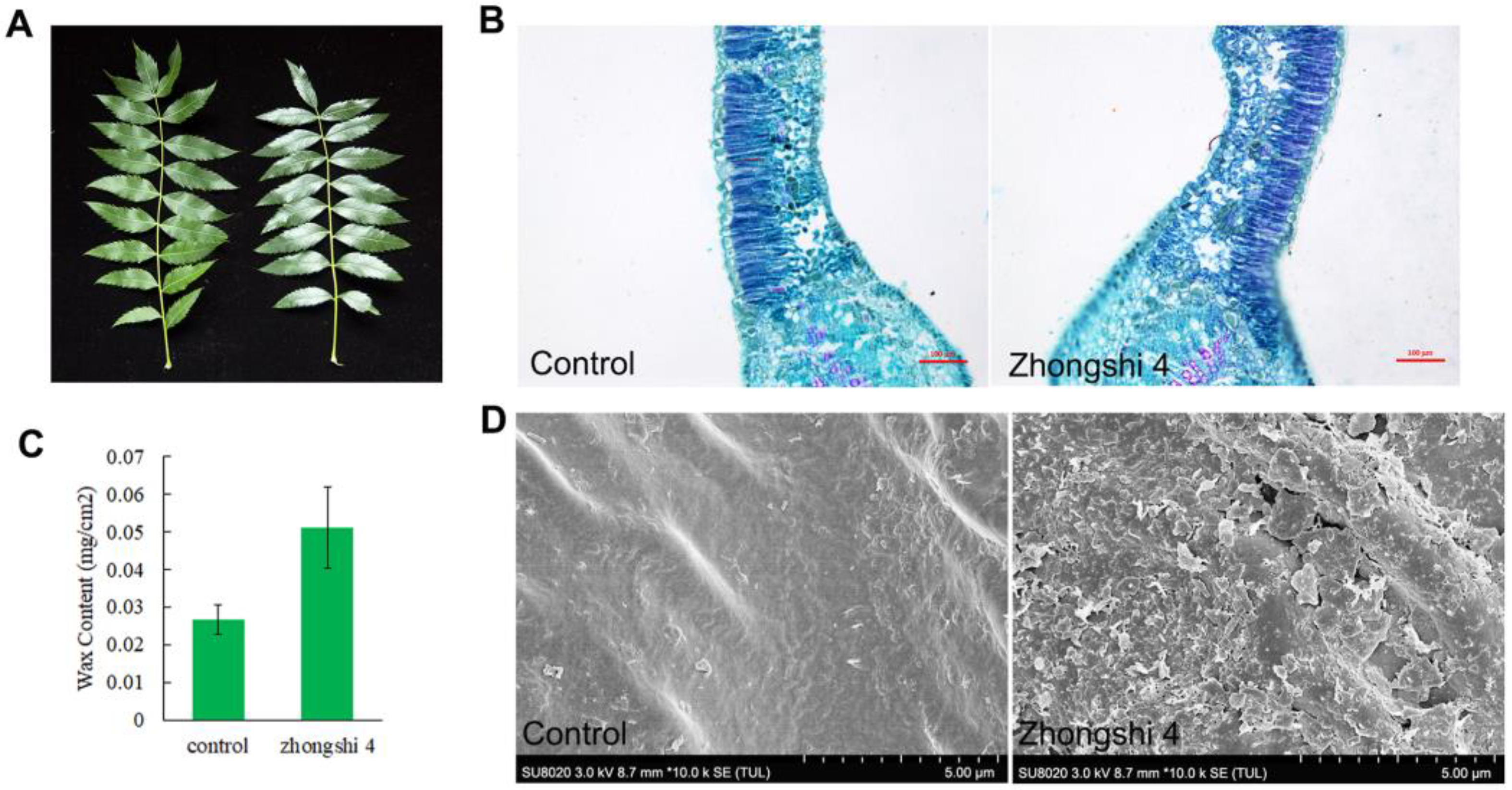
3.9. Transcriptome Analysis of the Expression Patterns of XsKCS Genes in High-Wax Yellow Horn
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- De Bigault Du Granrut, A.; Cacas, J.L. How Very-Long-Chain Fatty Acids Could Signal Stressful Conditions in Plants? Front. Plant Sci. 2016, 7, 1490. [Google Scholar] [CrossRef] [PubMed]
- Joubès, J.; Raffaele, S.; Bourdenx, B.; Garcia, C.; Laroche-Traineau, J.; Moreau, P.; Domergue, F.; Lessire, R. The VLCFA elongase gene family in Arabidopsis thaliana: Phylogenetic analysis, 3D modelling and expression profiling. Plant Mol. Biol. 2008, 67, 547–566. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Guo, Y.J. Progress in the study on genes encoding enzymes involved in biosynthesis of very long chain fatty acids and cuticular wax in plants. Hereditas 2008, 30, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.-S.; Zhang, Y.-M.; Sun, X.-Q.; Li, M.-M.; Hang, Y.-Y.; Xue, J.-Y. Evolution of the KCS gene family in plants: The history of gene duplication, sub/neofunctionalization and redundancy. Mol. Genet. Genom. 2016, 291, 739–752. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Mei, W.; Wan, H.; Xu, R.; Cheng, Y. Comprehensive analysis of KCS gene family in Citrinae reveals the involvement of CsKCS2 and CsKCS11 in fruit cuticular wax synthesis at ripening. Plant Sci. 2021, 310, 110972. [Google Scholar] [CrossRef]
- Lian, X.-Y.; Wang, X.; Gao, H.-N.; Jiang, H.; Mao, K.; You, C.-X.; Li, Y.-Y.; Hao, Y.-J. Genome wide analysis and functional identification of MdKCS genes in apple. Plant Physiol. Biochem. 2021, 151, 299–312. [Google Scholar] [CrossRef]
- Xiao, G.-H.; Wang, K.; Huang, G.; Zhu, Y.-X. Genome-scale analysis of the cotton KCS gene family revealed a binary mode of action for gibberellin A regulated fiber growth. J. Integr. Plant Biol. 2016, 58, 577–589. [Google Scholar] [CrossRef]
- Costaglioli, P.; Joubès, J.; Garcia, C.; Stef, M.; Arveiler, B.; Lessire, R.; Garbay, B. Profiling candidate genes involved in wax biosynthesis in Arabidopsis thaliana by microarray analysis. Biochim. et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2005, 1734, 247–258. [Google Scholar] [CrossRef]
- James, D.W., Jr.; Lim, E.; Keller, J.; Plooy, I.; Ralston, E.; Dooner, H.K. Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. Plant Cell 1995, 7, 309–319. [Google Scholar] [CrossRef]
- Pruitt, R.E.; Vielle-Calzada, J.-P.; Ploense, S.E.; Grossniklaus, U.; Lolle, S.J. FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc. Natl. Acad. Sci. USA 2000, 97, 1311–1316. [Google Scholar] [CrossRef]
- Todd, J.; Post-Beittenmiller, D.; Jaworski, J. KCS1encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis inArabidopsis thaliana. Plant J. 1999, 17, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Fiebig, A.; Mayfield, J.A.; Miley, N.L.; Chau, S.; Fischer, R.L.; Preuss, D. Alterations in CER6, a Gene Identical to CUT1, Differentially Affect Long-Chain Lipid Content on the Surface of Pollen and Stems. Plant Cell 2000, 12, 2001–2008. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Suh, M.C. Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species. Plant Cell Rep. 2015, 34, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Fan, S.; Bi, Q.; Wang, S.; Hu, X.; Chen, M.; Wang, L. Seed morphology, oil content and fatty acid composition variability assessment in yellow horn (Xanthoceras sorbifolium Bunge) germplasm for optimum biodiesel production. Ind. Crops Prod. 2017, 97, 425–430. [Google Scholar] [CrossRef]
- Taylor, D.C.; Guo, Y.; Katavic, V.; Mietkiewska, E.; Francis, T.; Bettger, W. New Seed Oils for Improved Human and Animal Health: Genetic Manipulation of the Brassicaceae for Oils Enriched in Nervonic Acid. Modif. Seed Compos. Promot. Health Nutr. 2015, 51, 219–232. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, X.J.; Wang, M.K.; Bi, Q.X.; Cui, Y.F.; Wang, L.B. Transcriptome and physiological analyses provide insights into the leaf epicuticular wax accumulation mechanism in yellowhorn. Hortic. Res. 2021, 8, 134. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Cui, D.; Li, H.; Bi, Q.; Hu, H.; Cui, T.; Liu, X.; Wang, L. New Xanthoceras sorbifolium Cultivars ‘Zhongshi 4’ and ‘Zhongshi 9’. Acta Hortic. Sin. 2020, 47, 3115–3116. [Google Scholar] [CrossRef]
- Ghanevati, M.; Jaworski, J.G. Engineering and mechanistic studies of the Arabidopsis FAE1 beta-ketoacyl-CoA synthase, FAE1 KCS. Eur. J. Biochem. 2002, 269, 3531–3539. [Google Scholar] [CrossRef]
- Lewandowska, M.; Keyl, A.; Feussner, I. Wax biosynthesis in response to danger: Its regulation upon abiotic and biotic stress. New Phytol. 2020, 227, 698–713. [Google Scholar] [CrossRef]
- Rui, C.; Chen, X.; Xu, N.; Wang, J.; Zhang, H.; Li, S.; Huang, H.; Fan, Y.; Zhang, Y.; Lu, X.; et al. Identification and Structure Analysis of KCS Family Genes Suggest Their Reponding to Regulate Fiber Development in Long-Staple Cotton Under Salt-Alkaline Stress. Front. Genet. 2022, 13, 812449. [Google Scholar] [CrossRef]
- Tong, T.; Fang, Y.-X.; Zhang, Z.; Zheng, J.; Zhang, X.; Li, J.; Niu, C.; Xue, D.; Zhang, X. Genome-wide identification and expression pattern analysis of the KCS gene family in barley. Plant Growth Regul. 2021, 93, 89–103. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef]
- Koralewski, T.E.; Krutovsky, K.V. Evolution of Exon-Intron Structure and Alternative. PLoS ONE 2011, 6, e18055. [Google Scholar] [CrossRef]
- Heidari, P.; Puresmaeli, F.; Mora-Poblete, F. Genome-Wide Identification, and Molecular Evolution of The Magnesium Transporter (MGT) Gene Family in Citrullus Lanatus and Cucumis Sativus. Agronomy 2022, 12, 2253. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The Evolutionary Fate and Consequences of Duplicate Genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Vision, T.J.; Brown, D.G.; Tanksley, S.D. The Origins of Genomic Duplications in Arabidopsis. Science 2000, 290, 2114–2117. [Google Scholar] [CrossRef] [PubMed]
- Faraji, S.; Heidari, P.; Amouei, H.; Filiz, E.; Abdullah; Poczai, P. Investigation and Computational Analysis of the Sulfotransferase (SOT) Gene Family in Potato (Solanum tuberosum): Insights into Sulfur Adjustment for Proper Development and Stimuli Responses. Plants 2021, 10, 2597. [Google Scholar] [CrossRef] [PubMed]
- Heidari, P.; Abdullah; Faraji, S.; Poczai, P. Magnesium transporter Gene Family: Genome-Wide Identification and Characterization in Theobroma cacao, Corchorus capsularis, and Gossypium hirsutum of Family Malvaceae. Agronomy 2021, 11, 1651. [Google Scholar] [CrossRef]
- Chen, L.; Hu, W.; Mishra, N.; Wei, J.; Lu, H.; Hou, Y.; Qiu, X.; Yu, S.; Wang, C.; Zhang, H.; et al. AKR2A interacts with KCS1 to improve VLCFAs contents and chilling tolerance of Arabidopsis thaliana. Plant J. 2020, 103, 1575–1589. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Wu, Q.; Yang, L.; Hu, W.; Liu, D.; Liu, Y. Ectopic Expression of CsKCS6 from Navel Orange Promotes the Production of Very-Long-Chain Fatty Acids (VLCFAs) and Increases the Abiotic Stress Tolerance of Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 564656. [Google Scholar] [CrossRef]
- Yang, Z.; Yang, X.; Dong, S.; Ge, Y.; Zhang, X.; Zhao, X.; Han, N. Overexpression of β-Ketoacyl-CoA Synthase from Vitis vinifera L. Improves Salt Tolerance in Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 564385. [Google Scholar] [CrossRef]
- Lian, X.-Y.; Gao, H.-N.; Jiang, H.; Liu, C.; Li, Y.-Y. MdKCS2 increased plant drought resistance by regulating wax biosynthesis. Plant Cell Rep. 2021, 40, 2357–2368. [Google Scholar] [CrossRef]
- Hegebarth, D.; Buschhaus, C.; Joubès, J.; Thoraval, D.; Bird, D.; Jetter, R. Arabidopsis ketoacyl-CoA synthase 16 (KCS16) forms C36 /C38 acyl precursors for leaf trichome and pavement surface wax. Plant Cell Environ. 2017, 40, 1761–1776. [Google Scholar] [CrossRef]
- Huang, H.; Ayaz, A.; Zheng, M.; Yang, X.; Zaman, W.; Zhao, H.; Lü, S. Arabidopsis KCS5 and KCS6 Play Redundant Roles in Wax Synthesis. Int. J. Mol. Sci. 2022, 23, 4450. [Google Scholar] [CrossRef] [PubMed]





| Gene Name | Gene ID | Chromosome | CDS (bp) | Amino Acid (aa) | Molecular Weight (Da) | pI | Subcellular Localization | Group |
|---|---|---|---|---|---|---|---|---|
| XsKCS1 | EVM0006364 | 1 | 1619 | 530 | 59,358.6 | 8.97 | Plasma membrane | KCS1-Like |
| XsKCS2 | EVM0014797 | 2 | 1329 | 425 | 48,631.29 | 8.82 | Plasma membrane | FDH-Like |
| XsKCS3 | EVM0011267 | 2 | 1360 | 445 | 50,066.11 | 9.03 | Plasma membrane | FDH-Like |
| XsKCS4 | EVM0005491 | 2 | 1570 | 514 | 57,801.97 | 9.33 | Plasma membrane | FAE1-Like |
| XsKCS5 | EVM0003350 | 3 | 1403 | 467 | 52,603.33 | 8.86 | Chloroplast | FDH-Like |
| XsKCS6 | EVM0016990 | 3 | 1488 | 487 | 54,849.80 | 9.21 | Plasma membrane | KCS1-Like |
| XsKCS7 | EVM0000575 | 3 | 1537 | 503 | 56,848.36 | 9.43 | Plasma membrane | KCS1-Like |
| XsKCS8 | EVM0020590 | 3 | 1595 | 522 | 59,056.75 | 9.06 | Plasma membrane | KCS1-Like |
| XsKCS9 | EVM0001427 | 3 | 1595 | 522 | 58,892.60 | 9.17 | Plasma membrane | KCS1-Like |
| XsKCS10 | EVM0007196 | 3 | 1329 | 435 | 49,385.24 | 9.39 | Cytoplasm | KCS1-Like |
| XsKCS11 | EVM0002301 | 3 | 1622 | 531 | 60,633.96 | 9.30 | Plasma membrane | FDH-Like |
| XsKCS12 | EVM0020935 | 3 | 1363 | 446 | 50,486.58 | 8.96 | Plasma membrane | FDH-Like |
| XsKCS13 | EVM0022839 | 4 | 1549 | 507 | 56,911.05 | 9.07 | Plasma membrane | FAE1-Like |
| XsKCS14 | EVM0017033 | 6 | 1573 | 515 | 58,158.87 | 9.15 | Endoplasmic reticulum | KCS1-Like |
| XsKCS15 | EVM0009614 | 6 | 1494 | 489 | 55,098.87 | 9.26 | Plasma membrane | FDH-Like |
| XsKCS16 | EVM0019392 | 7 | 1451 | 439 | 50,080.94 | 9.06 | Plasma membrane | FDH-Like |
| XsKCS17 | EVM0010766 | 8 | 1427 | 467 | 52,987.12 | 9.26 | Plasma membrane | CER6 |
| XsKCS18 | EVM0002351 | 12 | 1515 | 496 | 55,996.34 | 9.06 | Endoplasmic reticulum | FDH-Like |
| XsKCS19 | EVM0000167 | 15 | 1445 | 473 | 53,607.15 | 8.89 | Chloroplast | FDH-Like |
| XsKCS20 | EVM0007764 | ctg214 | 1207 | 395 | 44,305.31 | 8.95 | Cytoplasm | KCS1-Like |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Zhao, Z.; Yang, Y.; Xu, H.; Bi, Q.; Wang, L. Genome-Wide Identification and Expression Analysis of the KCS Gene Family in Yellow Horn Reveal Their Putative Function on Abiotic Stress Responses and Wax Accumulation. Horticulturae 2023, 9, 25. https://doi.org/10.3390/horticulturae9010025
Liu X, Zhao Z, Yang Y, Xu H, Bi Q, Wang L. Genome-Wide Identification and Expression Analysis of the KCS Gene Family in Yellow Horn Reveal Their Putative Function on Abiotic Stress Responses and Wax Accumulation. Horticulturae. 2023; 9(1):25. https://doi.org/10.3390/horticulturae9010025
Chicago/Turabian StyleLiu, Xiaojuan, Ziquan Zhao, Yingying Yang, Huihui Xu, Quanxin Bi, and Libing Wang. 2023. "Genome-Wide Identification and Expression Analysis of the KCS Gene Family in Yellow Horn Reveal Their Putative Function on Abiotic Stress Responses and Wax Accumulation" Horticulturae 9, no. 1: 25. https://doi.org/10.3390/horticulturae9010025
APA StyleLiu, X., Zhao, Z., Yang, Y., Xu, H., Bi, Q., & Wang, L. (2023). Genome-Wide Identification and Expression Analysis of the KCS Gene Family in Yellow Horn Reveal Their Putative Function on Abiotic Stress Responses and Wax Accumulation. Horticulturae, 9(1), 25. https://doi.org/10.3390/horticulturae9010025




