Evaluation of Reference Genes Suitable for Gene Expression during Root Enlargement in Cherry Radish Based on Transcriptomic Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Extraction of Total RNA and Synthesis of cDNA
2.3. Screening of Candidate RGs and Primer Design
2.4. Specificity Detection of Candidate RGs Primers
2.5. Analysis of RT-qPCR and Amplification Efficiency
2.6. Data Analysis
3. Results
3.1. Analysis of RNA Quality, Primer Specificity, and Amplification Efficiency
3.2. Ct Value Analysis of Candidate RGs
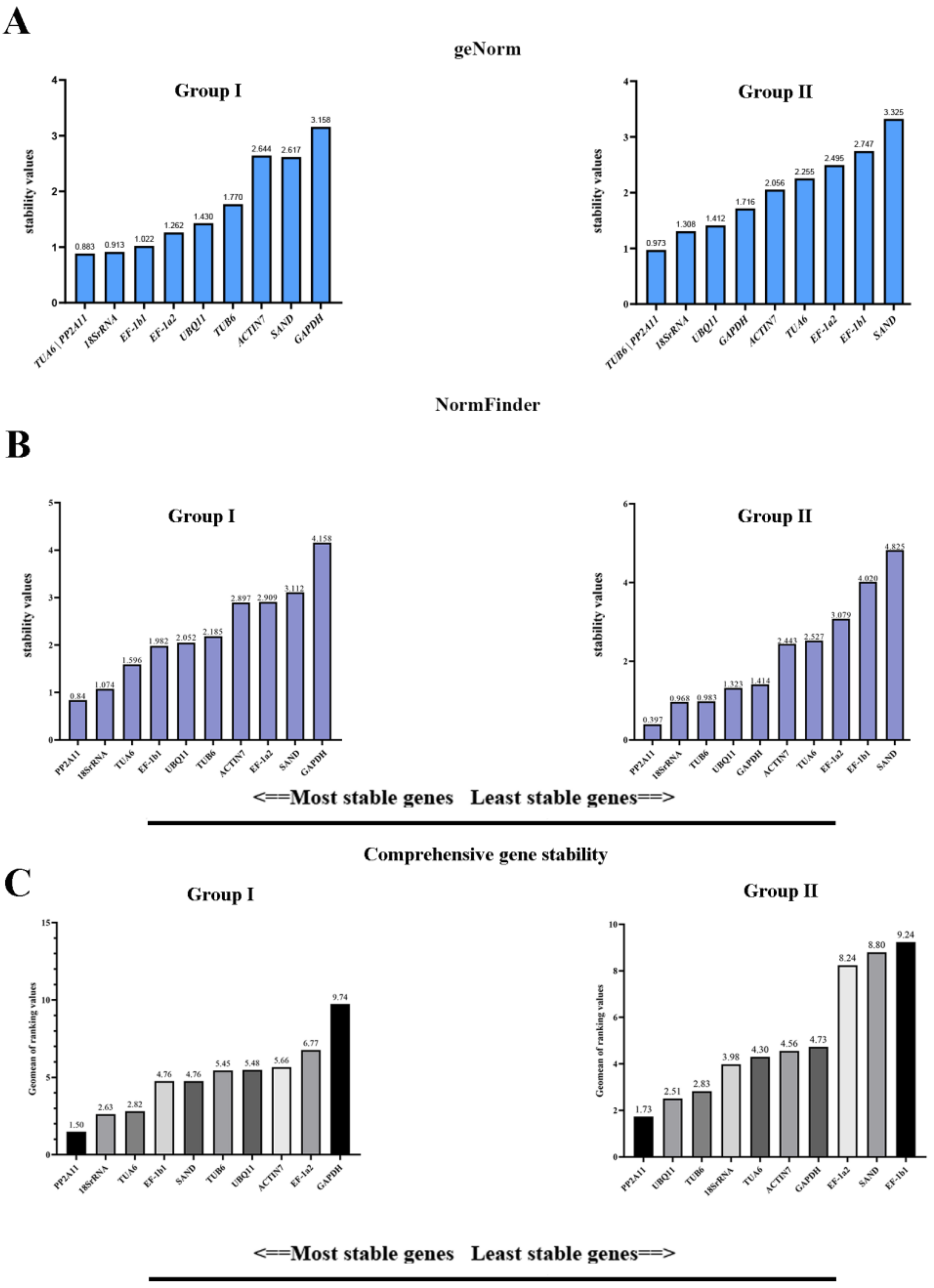
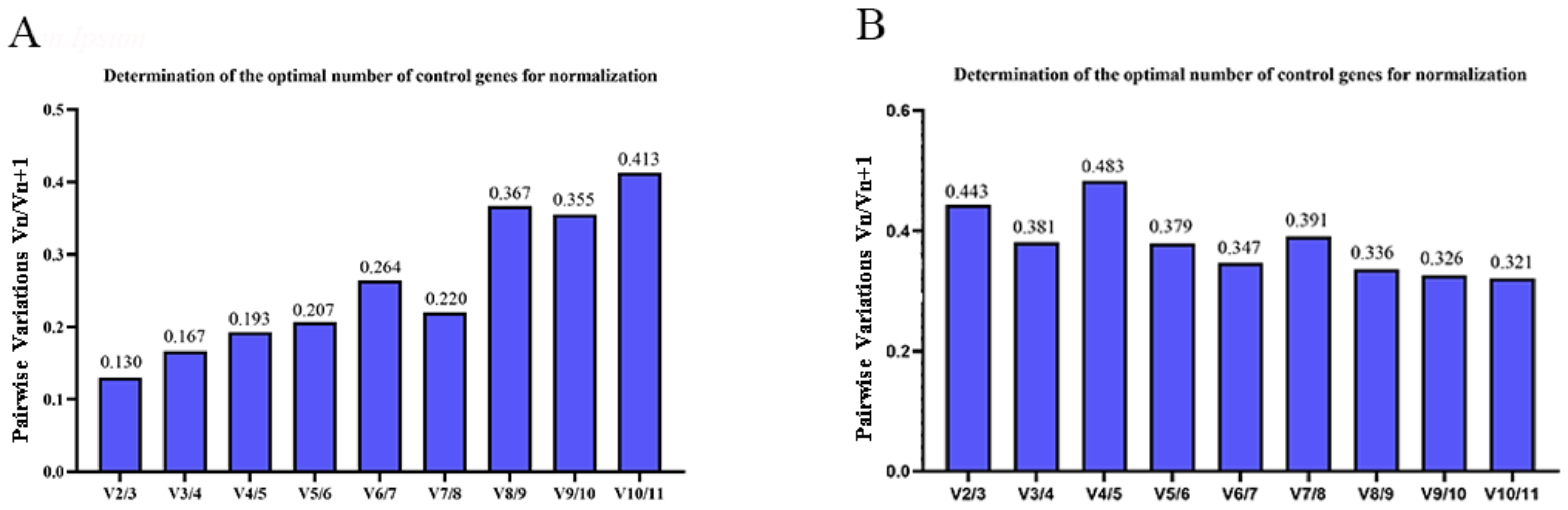
3.3. Stability Analysis by geNorm, NormFinder, and BestKeeper
3.4. Comprehensive Analysis of Candidate RGs
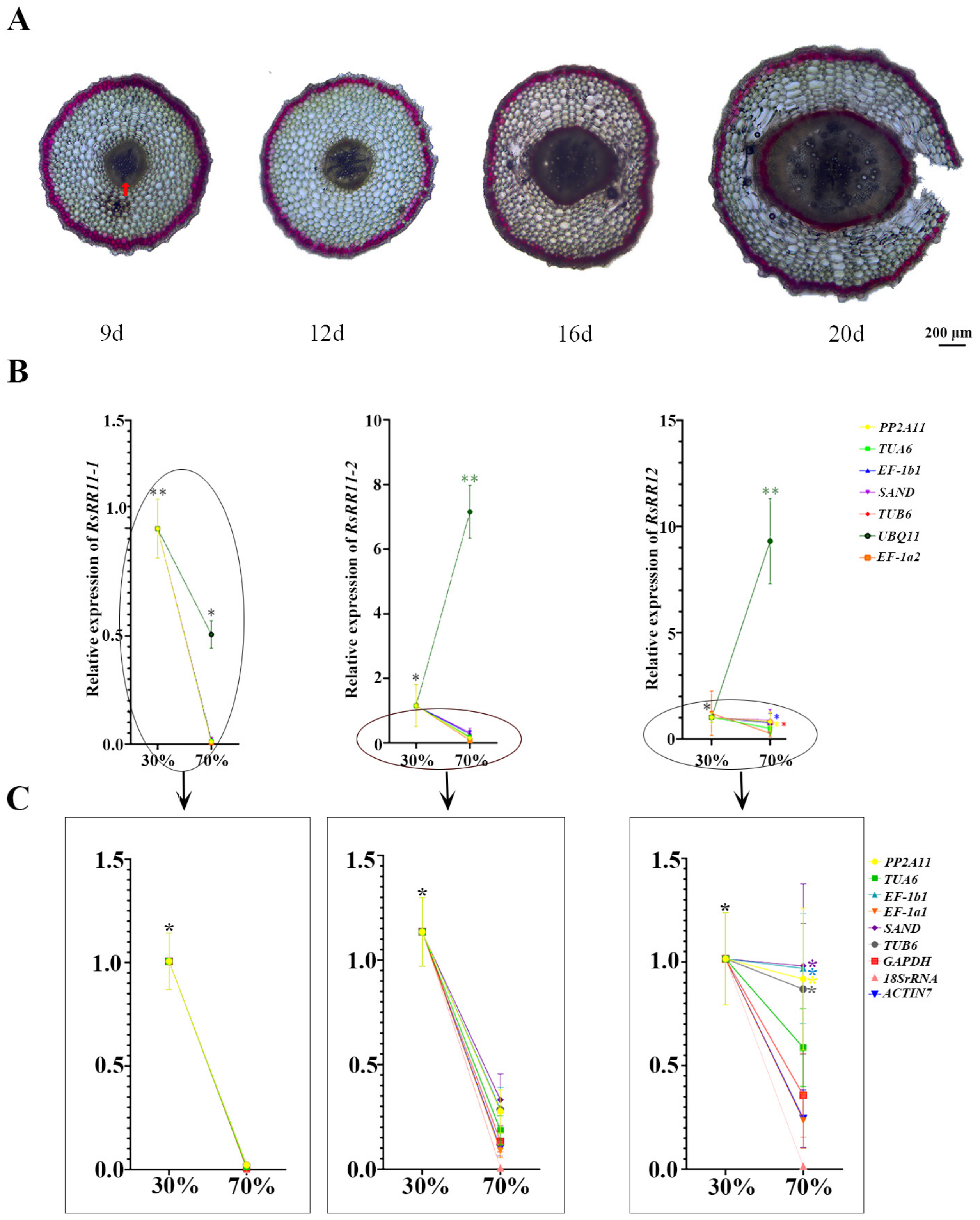
3.5. Validation of RGs Expression Canonicalization of B-Type RRs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, Y.; Zhang, S.; Zhang, H.; Li, F.; Li, G.; Fan, C.; Sun, R.; Zhang, S. QTL mapping and candidate gene identification of swollen root formation in Turnip. Int. J. Mol. Sci. 2021, 22, 653. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Shou, S. Physiological Research on the Constitution and Succulention in Raphanus sativus L.’s Succulent Root. North. Hortic. 2006, 1, 17–19. [Google Scholar]
- Yu, R.; Wang, J.; Xu, L.; Wang, Y.; Wang, R.; Zhu, X.; Sun, X.; Luo, X.; Xie, Y.; Everlyne, M. Transcriptome profiling of taproot reveals complex regulatory networks during taproot thickening in radish (Raphanus sativus L.). Front. Plant Sci. 2016, 7, 1210. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Paschold, A.; Marcon, C.; Liu, S.; Tai, H.; Nestler, J.; Yeh, C.-T.; Opitz, N.; Lanz, C.; Schnable, P.S. The Aux/IAA gene rum1 involved in seminal and lateral root formation controls vascular patterning in maize (Zea mays L.) primary roots. J. Exp. Bot. 2014, 65, 4919–4930. [Google Scholar] [CrossRef]
- Bustos-Sanmamed, P.; Mao, G.; Deng, Y.; Elouet, M.; Khan, G.A.; Bazin, J.; Turner, M.; Subramanian, S.; Yu, O.; Crespi, M. Overexpression of miR160 affects root growth and nitrogen-fixing nodule number in Medicago truncatula. Funct. Plant Biol. 2013, 40, 1208–1220. [Google Scholar] [CrossRef]
- Ullmannova, V.; Haskovec, C. The use of housekeeping genes (HKG) as an internal control for the detection of gene expression by quantitative real-time RT-PCR. Folia Biol.-Praha- 2003, 49, 211–216. [Google Scholar]
- Meng, Y.; Li, N.; Tian, J.; Gao, J.; Zhang, C. Identification and validation of reference genes for gene expression studies in postharvest rose flower (Rosa hybrida). Sci. Hortic. 2013, 158, 16–21. [Google Scholar] [CrossRef]
- Frericks, M.; Esser, C. A toolbox of novel murine house-keeping genes identified by meta-analysis of large scale gene expression profiles. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2008, 1779, 830–837. [Google Scholar] [CrossRef]
- Kudo, T.; Sasaki, Y.; Terashima, S.; Matsuda-Imai, N.; Takano, T.; Saito, M.; Kanno, M.; Ozaki, S.; Suwabe, K.; Suzuki, G.; et al. Identification of reference genes for quantitative expression analysis using large-scale RNA-seq data of Arabidopsis thaliana and model crop plants. Genes Genet. Syst. 2016, 91, 111–125. [Google Scholar] [CrossRef]
- Jain, M.; Nijhawan, A.; Tyagi, A.K.; Khurana, J.P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006, 345, 646–651. [Google Scholar] [CrossRef]
- Škiljaica, A.; Jagić, M.; Vuk, T.; Leljak Levanić, D.; Bauer, N.; Markulin, L. Evaluation of reference genes for RT-qPCR gene expression analysis in Arabidopsis thaliana exposed to elevated temperatures. Plant Biol. 2022, 24, 367–379. [Google Scholar] [CrossRef]
- Kim, B.-R.; Nam, H.-Y.; Kim, S.-U.; Kim, S.-I.; Chang, Y.-J. Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnol. Lett. 2003, 25, 1869–1872. [Google Scholar] [CrossRef]
- Jain, N.; Vergish, S.; Khurana, J.P. Validation of house-keeping genes for normalization of gene expression data during diurnal/circadian studies in rice by RT-qPCR. Sci. Rep. 2018, 8, 3203. [Google Scholar] [CrossRef]
- Yuanyuan, X.; Xianwen, Z.; Yiqin, G.; Liang, X.; Yan, W.; Liwang, L. Evaluation of reference genes for gene expression studies in radish (Raphanus sativus L.) using quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2012, 424, 398–403. [Google Scholar]
- Duan, M.; Wang, J.; Zhang, X.; Yang, H.; Wang, H.; Qiu, Y.; Song, J.; Guo, Y.; Li, X. Identification of optimal reference genes for expression analysis in radish (Raphanus sativus L.) and its relatives based on expression stability. Front. Plant Sci. 2017, 8, 1605. [Google Scholar] [CrossRef]
- Wang, W. Studies on the Physiological and mRNA Differential Display and DNA Methylation during Fleshy Taproot Development in Radish (Raphanus sativus L.). Master’s Thesis, Nanjing Agricultural University, Nanjing, China, 2007. [Google Scholar]
- Jin, Y.; Liu, F.; Huang, W.; Sun, Q.; Huang, X. Identification of reliable reference genes for qRT-PCR in the ephemeral plant Arabidopsis pumila based on full-length transcriptome data. Sci. Rep. 2019, 9, 8408. [Google Scholar] [CrossRef]
- Guo, R.; Li, W.; Wang, X.; Chen, B.; Huang, Z.; Liu, T.; Chen, X.; XuHan, X.; Lai, Z. Effect of photoperiod on the formation of cherry radish root. Sci. Hortic. 2019, 244, 193–199. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Lindbjerg, A.C.; Ledet, J.J.; Falck, Ø.T. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar]
- Zou, J.; Young, S.; Zhu, F.; Gheyas, F.; Skeans, S.; Wan, Y.; Wang, L.; Ding, W.; Billah, M.; McClanahan, T. Microarray profile of differentially expressed genes in a monkey model of allergic asthma. Genome Biol. 2002, 3, research0020.1. [Google Scholar] [CrossRef]
- Chen, Y.; Tan, Z.; Hu, B.; Yang, Z.; Xu, B.; Zhuang, L.; Huang, B. Selection and validation of reference genes for target gene analysis with quantitative RT-PCR in leaves and roots of bermudagrass under four different abiotic stresses. Physiol. Plant. 2015, 155, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef]
- Udvardi, M.K.; Czechowski, T.; Scheible, W.R. Eleven golden rules of quantitative RT-PCR. Plant Cell 2008, 20, 1736–1737. [Google Scholar] [CrossRef] [PubMed]
- Expósito-Rodríguez, M.; Borges, A.A.; Borges-Pérez, A.; Pérez, J.A. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 2008, 8, 131. [Google Scholar] [CrossRef] [PubMed]
- Cruz, F.; Kalaoun, S.; Nobile, P.; Colombo, C.; Almeida, J.; Barros, L.M.G.; Romano, E.; Grossi-de-Sá, M.F.; Vaslin, M.; Alves-Ferreira, M. Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Mol. Breed. 2009, 23, 607–616. [Google Scholar] [CrossRef]
- Zhang, K.; Li, M.; Cao, S.; Sun, Y.; Long, R.; Kang, J.; Yan, L.; Cui, H. Selection and validation of reference genes for target gene analysis with quantitative real-time PCR in the leaves and roots of Carex rigescens under abiotic stress. Ecotoxicol. Environ. Saf. 2019, 168, 127–137. [Google Scholar] [CrossRef]
- Nieminen, K.; Immanen, J.; Laxell, M.; Kauppinen, L.; Tarkowski, P.; Dolezal, K.; Tähtiharju, S.; Elo, A.; Decourteix, M.; Ljung, K. Cytokinin signaling regulates cambial development in poplar. Proc. Natl. Acad. Sci. USA 2008, 105, 20032–20037. [Google Scholar] [CrossRef]
- Wang, H. Regulation of vascular cambium activity. Plant Sci. 2020, 291, 110322. [Google Scholar] [CrossRef]
- Zhang, F.; May, A.; Irish, V.F. Type-B Arabidopsis response regulators directly activate WUSCHEL. Trends Plant Sci. 2017, 22, 815–817. [Google Scholar] [CrossRef]
- Acheampong, A.K.; Shanks, C.; Cheng, C.-Y.; Schaller, G.E.; Dagdas, Y.; Kieber, J.J. EXO70D isoforms mediate selective autophagic degradation of type-A ARR proteins to regulate cytokinin sensitivity. Proc. Natl. Acad. Sci. USA 2020, 117, 27034–27043. [Google Scholar] [CrossRef]
- Kai, I.; Takafumi, Y.; Akihiro, Y.; Takeshi, M. Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 47–57. [Google Scholar]
- Gancheva, M.S.; Dodueva, I.E.; Lebedeva, M.A.; Tvorogova, V.E.; Tkachenko, A.A.; Lutova, L.A. Identification, expression, and functional analysis of CLE genes in radish (Raphanus sativus L.) storage root. BMC Plant Biol. 2016, 16, 7. [Google Scholar] [CrossRef]
- Fu, Y.; Niu, F.; Jia, H.; Wang, Y.; Guo, B.; Wei, Y. Reference gene selection for real-time quantitative PCR assays in different tissues of Huperzia serrata based on full-length transcriptome sequencing. Plant Direct 2021, 5, e362. [Google Scholar] [CrossRef]
- Small, B.C.; Murdock, C.A.; Bilodeau-Bourgeois, A.L.; Peterson, B.C.; Waldbieser, G.C. Stability of reference genes for real-time PCR analyses in channel catfish (Ictalurus punctatus) tissues under varying physiological conditions. Comp. Biochem. Physiol. B 2008, 151, 296–304. [Google Scholar] [CrossRef]
- Chi, C.; Shen, Y.; Yin, L.; Ke, X.; Han, D.; Zuo, Y. Selection and validation of reference genes for gene expression analysis in Vigna angularis using quantitative real-time RT-PCR. PLoS ONE 2016, 11, e0168479. [Google Scholar] [CrossRef]
- Lin, J.; Redies, C. Histological evidence: Housekeeping genes beta-actin and GAPDH are of limited value for normalization of gene expression. Dev. Genes Evol. 2012, 222, 369–376. [Google Scholar] [CrossRef]
- Narsai, R.; Ivanova, A.; Ng, S.; Whelan, J. Defining reference genes in Oryza sativausing organ, development, biotic and abiotic transcriptome datasets. BMC Plant Biol. 2010, 10, 56. [Google Scholar] [CrossRef]
- Liang, W.; Zou, X.; Carballar-Lejarazú, R.; Wu, L.; Sun, W.; Yuan, X.; Wu, S.; Li, P.; Ding, H.; Ni, L.; et al. Selection and evaluation of reference genes for qRT-PCR analysis in Euscaphis konishii Hayata based on transcriptome data. Plant Methods 2018, 14, 42. [Google Scholar] [CrossRef]
- Huang, Z.; Chen, H.; Weng, B.; Liu, B.; Wang, Y.; Liu, P. Screening of reference genes under cadmium stress in Agaricus brasiliensis based on transcriptome sequencing. Acta Microbiol. Sin. 2021, 61, 3458–3470. [Google Scholar]
- Saha, P.; Blumwald, E. Assessing reference genes for accurate transcript normalization using quantitative real-time PCR in pearl millet [ Pennisetum glaucum (L.) R. Br.]. PLoS ONE 2014, 9, e106308. [Google Scholar] [CrossRef]
- Shukla, P.; Reddy, R.A.; Ponnuvel, K.M.; Rohela, G.K.; Shabnam, A.A.; Ghosh, M.K.; Mishra, R.K. Selection of suitable reference genes for quantitative real-time PCR gene expression analysis in Mulberry (Morus alba L.) under different abiotic stresses. Mol. Biol. Rep. 2019, 46, 1809–1817. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Fu, J.; Dai, S.; Sun, Y.; Hong, Y. Reference gene selection for qPCR analysis in cineraria developing flowers. Sci. Hortic. 2013, 153, 64–70. [Google Scholar] [CrossRef]
- DeLong, A. Switching the flip: Protein phosphatase roles in signaling pathways. Curr. Opin. Plant Biol. 2006, 9, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.H.; Agarwal, S.; Rai, A.; Memaya, M.B.; Mehrotra, S.; Mehrotra, R. Co-expression network analysis of protein phosphatase 2A (PP2A) genes with stress-responsive genes in Arabidopsis thaliana reveals 13 key regulators. Sci. Rep. 2020, 10, 21480. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, Y.; Fang, H.; Shi, H.; Chen, K.; Zhang, Z.; Tan, X. Selection of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in Brassica napus under various stress conditions. Mol. Genet. Genom. 2014, 289, 1023–1035. [Google Scholar] [CrossRef]
- Tang, F.; Chu, L.; Shu, W.; He, X.; Wang, L.; Lu, M. Selection and validation of reference genes for quantitative expression analysis of miRNAs and mRNAs in Poplar. Plant Methods 2019, 15, 35. [Google Scholar] [CrossRef]
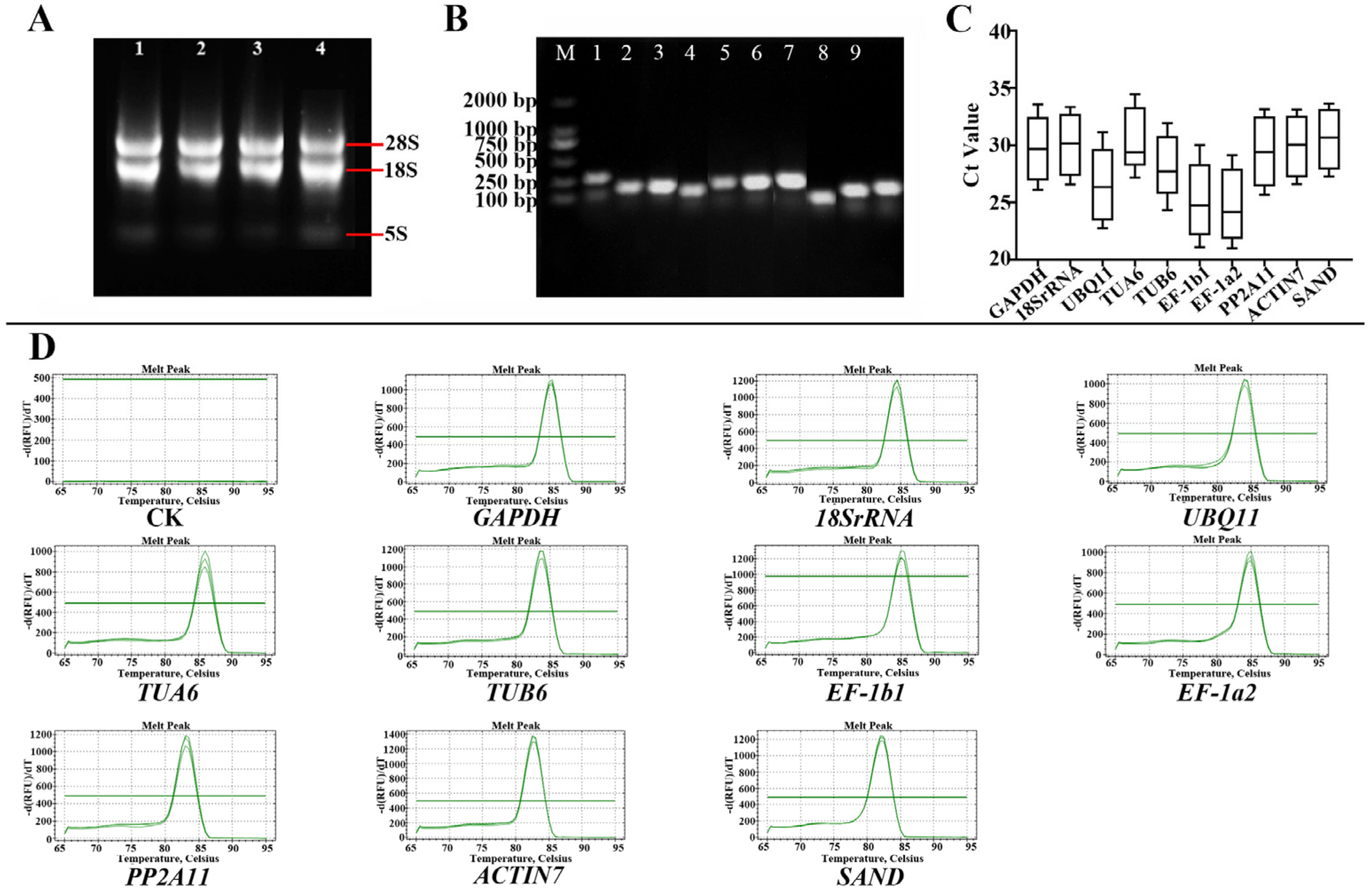
| Primer Number | Gene ID | Gene Annotation | FPKM Value | Gene Description | Primer Sequences (5′-3′) | Tm (℃) | Length of Product (bp) |
|---|---|---|---|---|---|---|---|
| 1 | RSG46678 | GAPDH | 5.37 | Glyceraldehyde-3-phosphate dehydrogenase | F: TTGCCGTCTCCAGAATCCCT | 61.7 | 285 |
| R: CGTGCCAACACCTGAGGAAGT | 62.3 | ||||||
| 2 | RSG01140 | 18SrRNA | 29.56 | 18S ribosomal RNA | F: GACTCAATCGTCCAGAAAGCAG | 59.1 | 186 |
| R: CAAGCGGTTAGAAAGGGGAG | 59.2 | ||||||
| 3 | RSG23795 | UBQ11 | 134.49 | Ubiquitin 11 | F: CATCTCGTTCTCAGGCTTCG | 58.2 | 195 |
| R: CAATGTTCTACCGTCCTCAAGC | 59.1 | ||||||
| 4 | RSG21557 | TUA6 | 397.2 | Tubulin alpha 6 | F: GGTATCCAGGTCGGAAATGC | 59.2 | 193 |
| R: CGTCGATCACAGTGGGCTCT | 60.6 | ||||||
| 5 | RSG05570 | TUB6 | 374.36 | Tubulin beta 6 | F:ACTTCGTTTTCGGGCAATCT | 59 | 258 |
| R: CCTTAGGTGATGGGAAGACAGAG | 59.7 | ||||||
| 6 | RSG42380 | EF-1b1 | 633.74 | Elongation factor 1-beta | F: ATCACTGTCTTTGCTGCTCTTGC | 61.4 | 260 |
| R: TTCTTCTCCTCCTCGGTCTCC | 59.8 | ||||||
| 7 | RSG33614 | EF-1a2 | 524.19 | Elongation factor 1-alpha | F: AAGATGGATGCTACTACCCCTAAG | 58.5 | 297 |
| R: CACTGGCACCGTTCCAATAC | 58.6 | ||||||
| 8 | RSG21210 | PP2A11 | 94.92 | Threonine-protein phosphatase 2A | F: GCTTCCTGGGCTGATTTCGT | 61.6 | 159 |
| R: AGCTTTCTTTGTGCCTTGGTC | 58.8 | ||||||
| 9 | RSG39901 | ACTIN7 | 24.38 | Actin protein | F: CTGAGGACGAACTTGCTTACGA | 59.9 | 225 |
| R: CAGTGTTCTCCAAGAGTTGCCTAT | 59.7 | ||||||
| 10 | RSG23793 | SAND | 12.51 | Protein SAND | F: TGAAGGTGGATTGCGTGTTG | 59.9 | 245 |
| R: CATAGAGTTTCTGGTATGCTCGGTA | 60.1 |
| Number | Gene Name | Slope of Standard Curve | Pearson Moment Correlation (r) | r² | Amplification Efficiency (E) |
|---|---|---|---|---|---|
| 1 | GAPDH | −1.86 | −0.999 | 0.998 | 1.220 |
| 2 | 18SrRNA | −1.76 | −0.996 | 0.992 | 1.321 |
| 3 | UBQ11 | −2.09 | −0.993 | 0.986 | 1.030 |
| 4 | TUA6 | −1.74 | −0.965 | 0.932 | 1.340 |
| 5 | TUB6 | −1.77 | −0.984 | 0.968 | 1.306 |
| 6 | EF-1b1 | −2.14 | −0.990 | 0.980 | 1.001 |
| 7 | EF-1a2 | −2.04 | −0.994 | 0.988 | 1.065 |
| 8 | PP2A11 | −1.97 | −0.995 | 0.990 | 1.123 |
| 9 | ACTIN7 | −1.73 | −0.993 | 0.986 | 1.354 |
| 10 | SAND | −1.69 | −0.991 | 0.982 | 1.398 |
| Gene Name | Group Ⅰ | Group Ⅱ | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Correlation Coefficient (r) | Standard Deviation (SD) | Coefficient of Variation (CV) | Gene Stability | Ranking | Correlation Coefficient (r) | Standard Deviation (SD) | Coefficient of Variation (CV) | Gene Stability | Ranking | |
| GAPDH | 0.455 | 3.84 | 11.93 | 3.837 | 9 | 0.925 | 3.41 | 11.64 | 3.408 | 4 |
| 18SrRNA | 0.982 | 3.05 | 9.34 | 3.053 | 4 | 0.968 | 3.57 | 11.89 | 3.57 | 7 |
| UBQ11 | 0.946 | 3.22 | 10.99 | 3.222 | 6 | 0.969 | 3.44 | 13.04 | 3.442 | 5 |
| TUA6 | 0.992 | 3.62 | 10.75 | 3.615 | 7 | 0.815 | 1.99 | 6.46 | 1.994 | 1 |
| TUB6 | 0.832 | 2.93 | 9.20 | 2.934 | 3 | 0.995 | 3.74 | 12.94 | 3.74 | 8 |
| EF-1b1 | 0.963 | 3.69 | 12.48 | 3.69 | 8 | 0.935 | 5.73 | 20.2 | 5.727 | 10 |
| EF-1a2 | 0.990 | 4.42 | 14.71 | 4.422 | 10 | 0.984 | 5.4 | 19.86 | 5.397 | 9 |
| PP2A11 | 0.991 | 3.14 | 9.52 | 3.139 | 5 | 0.980 | 5.23 | 10.78 | 3.232 | 3 |
| ACTIN7 | 0.792 | 0.20 | 0.73 | 0.199 | 2 | 0.828 | 2.71 | 8.12 | 2.712 | 2 |
| SAND | 0.053 | 0.18 | 0.67 | 0.179 | 1 | 0.257 | 3.47 | 11.01 | 3.468 | 6 |
| Sample Groups | Gene Name | Ge Norm | NormFinder | BestKeeper | Delta CT | Geomean of Ranking Values | Recommended Comprehensive Ranking |
|---|---|---|---|---|---|---|---|
| Group Ⅰ | PP2A11 | 1 | 1 | 5 | 1 | 1.50 | 1 |
| 18SrRNA | 3 | 2 | 4 | 2 | 2.63 | 2 | |
| TUA6 | 1 | 3 | 7 | 3 | 2.82 | 3 | |
| EF-1b1 | 4 | 4 | 8 | 4 | 4.76 | 4 | |
| SAND | 9 | 9 | 1 | 9 | 5.20 | 5 | |
| TUB6 | 7 | 6 | 3 | 7 | 5.45 | 6 | |
| UBQ11 | 6 | 5 | 6 | 5 | 5.48 | 7 | |
| ACTIN7 | 8 | 8 | 2 | 8 | 5.66 | 8 | |
| EF-1a2 | 5 | 7 | 10 | 6 | 6.77 | 9 | |
| GAPDH | 10 | 10 | 9 | 10 | 9.74 | 10 | |
| Group Ⅱ | PP2A11 | 1 | 1 | 3 | 3 | 1.73 | 1 |
| UBQ11 | 4 | 2 | 5 | 1 | 2.51 | 2 | |
| TUB6 | 1 | 4 | 8 | 2 | 2.83 | 3 | |
| 18SrRNA | 3 | 3 | 7 | 4 | 3.98 | 4 | |
| TUA6 | 7 | 7 | 1 | 7 | 4.30 | 5 | |
| ACTIN7 | 6 | 6 | 2 | 6 | 4.56 | 6 | |
| GAPDH | 5 | 5 | 4 | 5 | 4.73 | 7 | |
| EF-1a2 | 8 | 8 | 9 | 8 | 8.24 | 8 | |
| SAND | 10 | 10 | 6 | 10 | 8.80 | 9 | |
| EF-1b1 | 9 | 9 | 10 | 9 | 9.24 | 10 |
| Gene Name | Gene ID | Primer Sequences (5′-3′) | |
|---|---|---|---|
| RsRR11-1 | RSG20107 | F: GGCTTTACCTGAGCAGATTGG | R:GGCTTTGGTGGCGTGAA |
| RsRR11-2 | RSG25754 | F:GAGCGTAAGGACGGGTTTG | R:TGAGGGAGGTCCAGTTCG |
| RsRR12 | RSG31240 | F: AGGGTTACCGATGCCTTTAGA | R:TTGAGCAGGAGGAGGAAGAGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, Y.; Wang, X.; Chen, B.; Zheng, S.; Wang-Pruski, G.; Chen, X.; Guo, R. Evaluation of Reference Genes Suitable for Gene Expression during Root Enlargement in Cherry Radish Based on Transcriptomic Data. Horticulturae 2023, 9, 20. https://doi.org/10.3390/horticulturae9010020
Yao Y, Wang X, Chen B, Zheng S, Wang-Pruski G, Chen X, Guo R. Evaluation of Reference Genes Suitable for Gene Expression during Root Enlargement in Cherry Radish Based on Transcriptomic Data. Horticulturae. 2023; 9(1):20. https://doi.org/10.3390/horticulturae9010020
Chicago/Turabian StyleYao, Yao, Xiaoqian Wang, Bingxing Chen, Shurui Zheng, Gefu Wang-Pruski, Xiaodong Chen, and Rongfang Guo. 2023. "Evaluation of Reference Genes Suitable for Gene Expression during Root Enlargement in Cherry Radish Based on Transcriptomic Data" Horticulturae 9, no. 1: 20. https://doi.org/10.3390/horticulturae9010020
APA StyleYao, Y., Wang, X., Chen, B., Zheng, S., Wang-Pruski, G., Chen, X., & Guo, R. (2023). Evaluation of Reference Genes Suitable for Gene Expression during Root Enlargement in Cherry Radish Based on Transcriptomic Data. Horticulturae, 9(1), 20. https://doi.org/10.3390/horticulturae9010020






