Characterization and Expression Analysis of the UDP Glycosyltransferase Family in Pomegranate (Punica granatum L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Identification of Pomegranate UGT Gene Family
2.3. Phylogenetic Analysis of PgUGTs
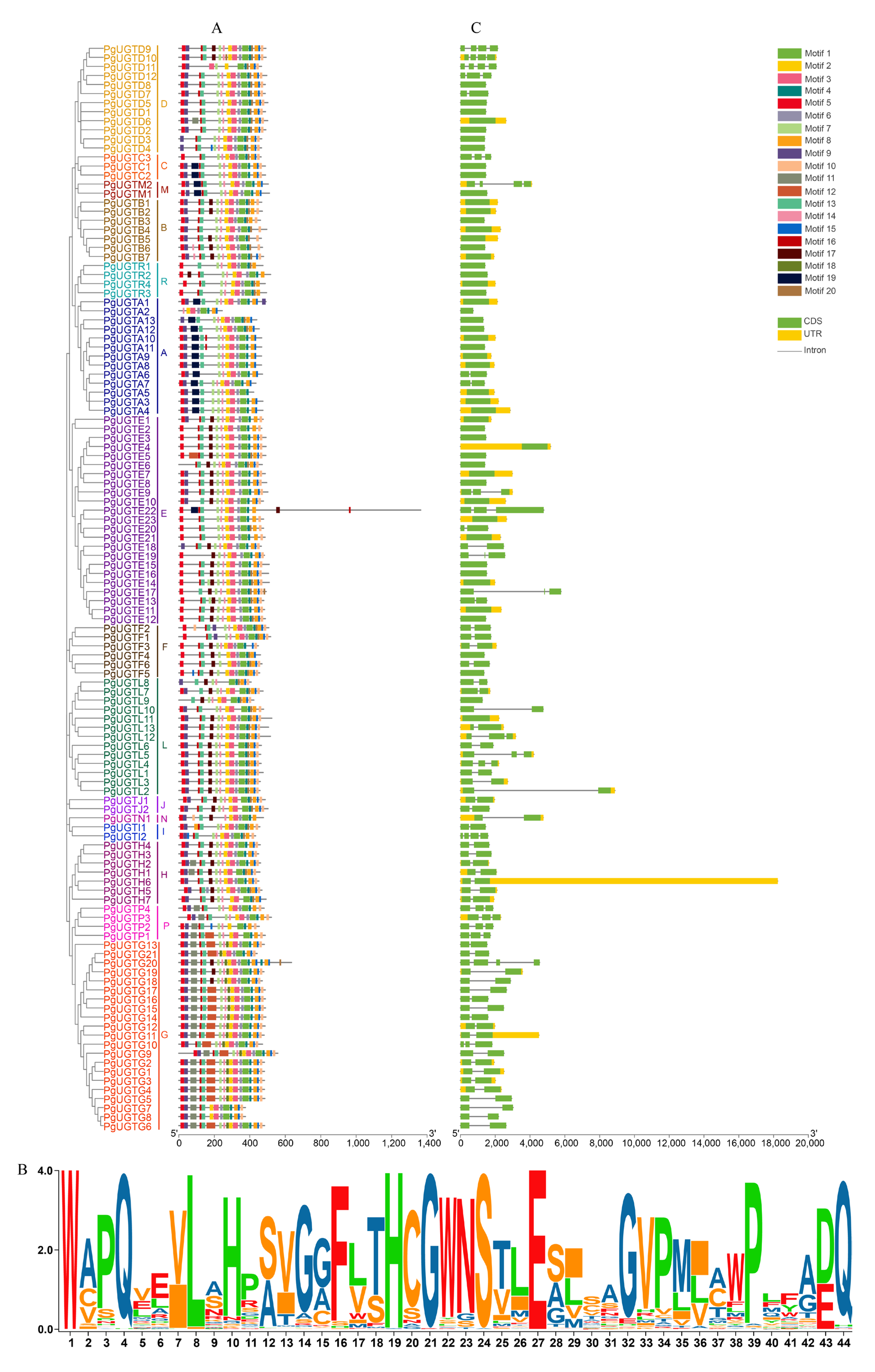
2.4. Gene Structure and Conservative Motif Analysis of PgUGTs
2.5. Chromosomal Locations and Gene Duplication of PgUGTs
2.6. Cis-Regulatory Element Analysis of PgUGTs
2.7. Expression Pattern Analysis of PgUGTs
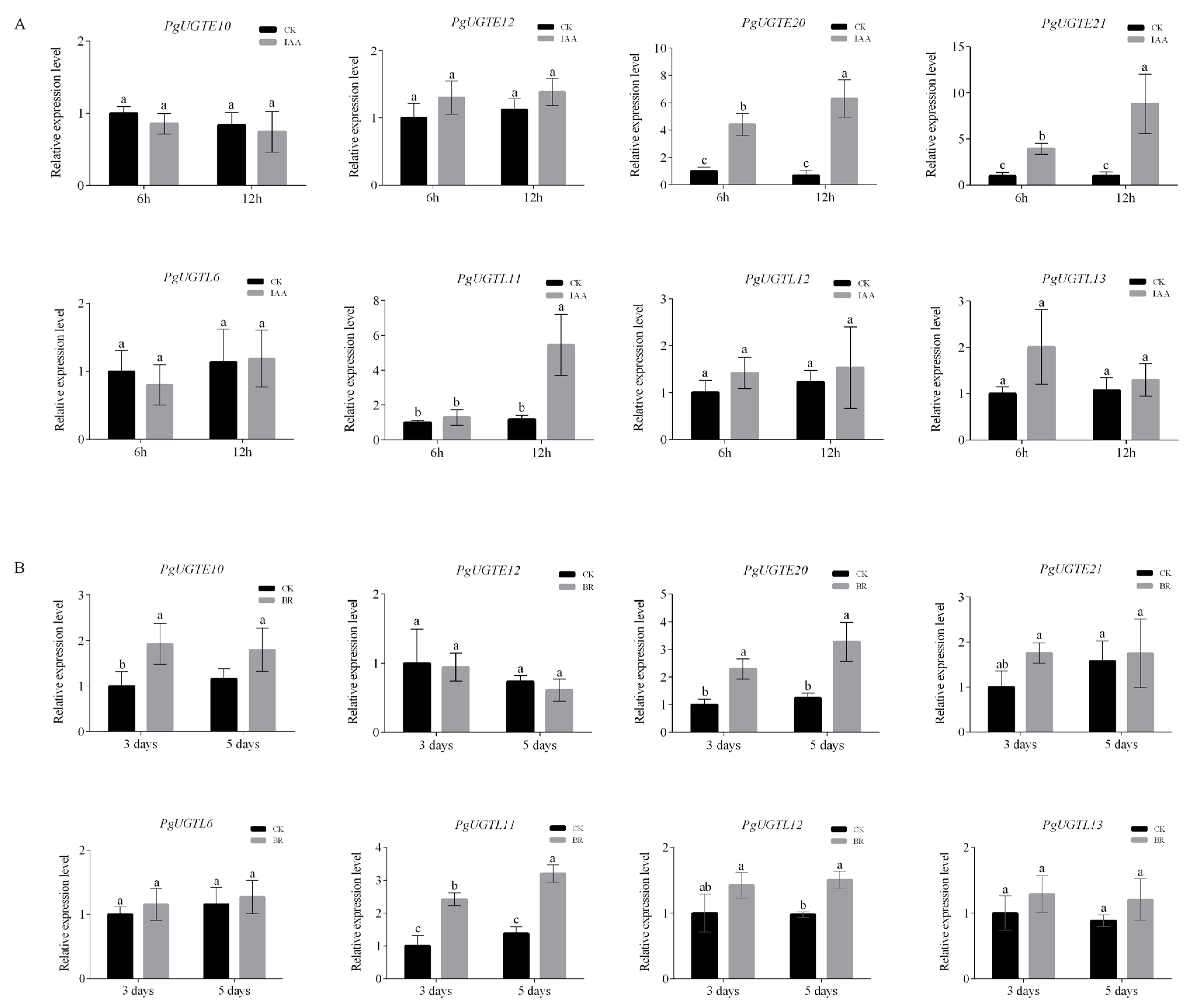
2.8. Quantitative Real-Time PCR (qRT-PCR) Analysis
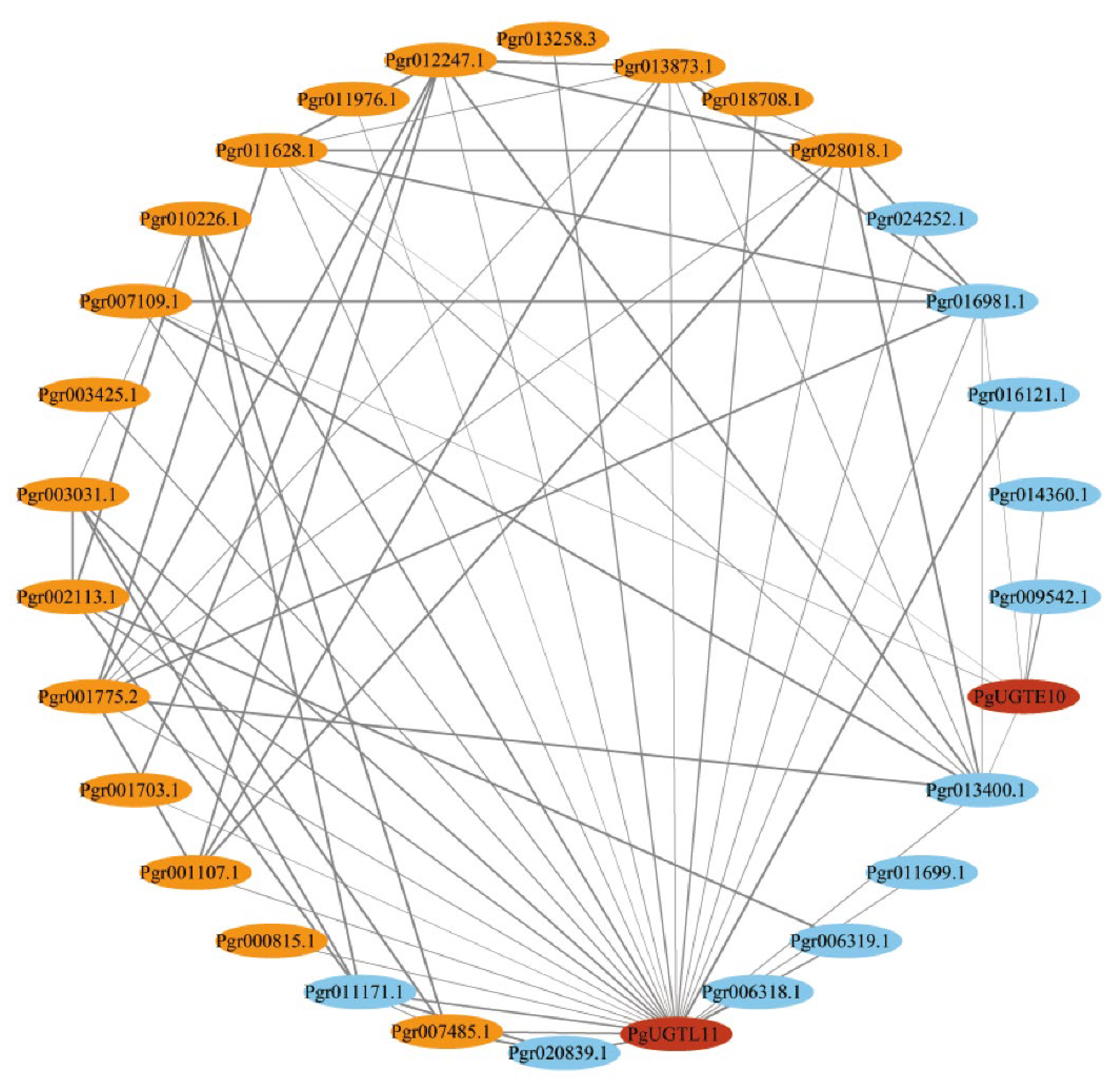
2.9. Co-Expression Analysis
2.10. Subcellular Localization of PgUGTL11 and PgUGTE10
3. Results
3.1. Identification and Phylogenetic Analysis of PgUGTs
3.2. Conserved Motifs and Gene Structure Analysis of PgUGTs
3.3. Gene Duplication and Synteny Analysis of PgUGTs
3.4. Expression Profiles of PgUGTs Based on RNA-seq Data
3.5. CRE Identification and Relative Expression of PgUGTs Responding to Exogenous Hormones
3.6. Co-Expression Analysis of Candidate PgUGTs
3.7. Subcellular Localization of PgUGTL11 and PgUGTE10
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Niu, J.; Cao, D.; Li, H.; Xue, H.; Chen, L.; Liu, B.; Cao, S. Quantitative proteomics of pomegranate varieties with contrasting seed hardness during seed development stages. Tree Genet. Genomes 2018, 14, 14. [Google Scholar] [CrossRef]
- Heber, D. Multitargeted therapy of cancer by ellagitannins. Cancer Lett. 2008, 269, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Huang, Y.; Tian, S.; Li, G.; Cao, S. Relationship of seed hardness development and microstructure of seed coat cell in soft seed pomegranate. Acta Hortic. Sin. 2017, 44, 1174–1180. [Google Scholar] [CrossRef]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.J.; Miao, Y.C.; Zhang, K.W. Sequestration and transport of lignin monomeric precursors. Molecules 2011, 16, 710–727. [Google Scholar] [CrossRef]
- Dima, O.; Morreel, K.; Vanholme, B.; Kim, H.; Ralph, J.; Boerjan, W. Small glycosylated lignin oligomers are stored in Arabidopsis leaf vacuoles. Plant Cell 2015, 27, 695–710. [Google Scholar] [CrossRef]
- Luo, M.; Dennis, E.S.; Berger, F.; Peacock, W.J.; Chaudhury, A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 17531–17536. [Google Scholar] [CrossRef]
- Garcia, D.; Saingery, V.; Chambrier, P.; Mayer, U.; Jurgens, G.; Berger, F. Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 2003, 131, 1661–1670. [Google Scholar] [CrossRef]
- Figueiredo, D.D.; Batista, R.A.; Roszakt, P.J.; Hennig, L.; Kohler, C. Auxin production in the endosperm drives seed coat development in Arabidopsis. Elife 2016, 5, e20542. [Google Scholar] [CrossRef]
- Immanen, J.; Nieminen, K.; Smolander, O.P.; Kojima, M.; Serra, J.A.; Koskinen, P.; Zhang, J.; Elo, A.; Mahonen, A.P.; Street, N.; et al. Cytokinin and auxin display distinct but interconnected distribution and signaling profiles to stimulate cambial activity. Curr. Biol. 2016, 26, 1990–1997. [Google Scholar] [CrossRef]
- Xu, C.Z.; Shen, Y.; He, F.; Fu, X.K.; Yu, H.; Lu, W.X.; Li, Y.L.; Li, C.F.; Fan, D.; Wang, H.C.; et al. Auxin-mediated Aux/IAA-ARF-HB signaling cascade regulates secondary xylem development in Populus. New Phytol. 2019, 222, 752–767. [Google Scholar] [CrossRef]
- Uggla, C.; Mellerowicz, E.J.; Sundberg, B. Indole-3-acetic acid controls cambial growth in Scots pine by positional signaling. Plant Physiol. 1998, 117, 113–121. [Google Scholar] [CrossRef]
- Korasick, D.A.; Enders, T.A.; Strader, L.C. Auxin biosynthesis and storage forms. J. Exp. Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef]
- Jin, H.; Do, J.; Shin, S.J.; Choi, J.W.; Choi, Y.I.; Kim, W.; Kwon, M. Exogenously applied 24-epi brassinolide reduces lignification and alters cell wall carbohydrate biosynthesis in the secondary xylem of Liriodendron tulipifera. Phytochemistry 2014, 101, 40–51. [Google Scholar] [CrossRef]
- Ge, Y.H.; Li, C.Y.; Tang, R.X.; Sun, R.H.; Li, J.R. Effects of postharvest brassinolide dipping on quality parameters and antioxidant activity in peach fruit. In Proceedings of the 3rd International Symposium on Postharvest Pathology—Using Science to Increase Food Availability, Bari, Italy, 7–11 June 2015; pp. 377–384. [Google Scholar] [CrossRef]
- Sanchez-Rodriguez, C.; Rubio-Somoza, I.; Sibout, R.; Persson, S. Phytohormones and the cell wall in Arabidopsis during seedling growth. Trends Plant Sci. 2010, 15, 291–301. [Google Scholar] [CrossRef]
- Ibanes, M.; Fabregas, N.; Chory, J.; Cano-Delgado, A.I. Brassinosteroid signaling and auxin transport are required to establish the periodic pattern of Arabidopsis shoot vascular bundles. Proc. Natl. Acad. Sci. USA 2009, 106, 13630–13635. [Google Scholar] [CrossRef]
- Oda, Y.; Mimura, T.; Hasezawa, S. Regulation of secondary cell wall development by cortical microtubules during tracheary element differentiation in Arabidopsis cell suspensions. Plant Physiol. 2005, 137, 1027–1036. [Google Scholar] [CrossRef]
- Shen, H.; Mazarei, M.; Hisano, H.; Escamilla-Trevino, L.; Fu, C.X.; Pu, Y.Q.; Rudis, M.R.; Tang, Y.H.; Xiao, X.R.; Jackson, L.; et al. A genomics approach to deciphering lignin biosynthesis in switchgrass. Plant Cell 2013, 25, 4342–4361. [Google Scholar] [CrossRef]
- Ostrowski, M.; Jakubowska, A. UDP-Glycosyltransferases of Plant Hormones. Postep. Biol. Komorki 2013, 40, 141–160. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Hanada, K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011, 66, 182–193. [Google Scholar] [CrossRef]
- Paquette, S.; Møller, B.L.; Bak, S. On the origin of family 1 plant glycosyltransferases. Phytochemistry 2003, 62, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.; Li, Y.; Lim, E.-K.; Bowles, D.J. Higher plant glycosyltransferases. Genome Biol. 2001, 2, reviews3004.1. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Yao, S.; Dai, X.; Yin, Q.; Liu, Y.; Jiang, X.; Wu, Y.; Qian, Y.; Pang, Y.; Gao, L.; et al. Identification of UDP-glycosyltransferases involved in the biosynthesis of astringent taste compounds in tea (Camellia sinensis). J. Exp. Bot. 2016, 67, 2285–2297. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, P.; Wang, Y.; Dong, R.; Yu, H.; Hou, B. Genome-wide identification and phylogenetic analysis of family-1 UDP glycosyltransferases in maize (Zea mays). Planta 2014, 239, 1265–1279. [Google Scholar] [CrossRef]
- Caputi, L.; Malnoy, M.; Goremykin, V.; Nikiforova, S.; Martens, S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012, 69, 1030–1042. [Google Scholar] [CrossRef]
- Lim, E.-K.; Li, Y.; Parr, A.; Jackson, R.; Ashford, D.A.; Bowles, D.J. Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in Arabidopsis. J. Biol. Chem. 2001, 276, 4344–4349. [Google Scholar] [CrossRef]
- Lim, E.-K.; Jackson, R.G.; Bowles, D.J. Identification and characterisation of Arabidopsis glycosyltransferases capable of glucosylating coniferyl aldehyde and sinapyl aldehyde. FEBS Lett. 2005, 579, 2802–2806. [Google Scholar] [CrossRef]
- Lanot, A.; Hodge, D.; Jackson, R.G.; George, G.L.; Elias, L.; Lim, E.K.; Vaistij, F.E.; Bowles, D.J. The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis thaliana. Plant J. 2006, 48, 286–295. [Google Scholar] [CrossRef]
- Lin, J.S.; Huang, X.X.; Li, Q.; Cao, Y.; Bao, Y.; Meng, X.F.; Li, Y.J.; Fu, C.; Hou, B.K. UDP-glycosyltransferase 72B1 catalyzes the glucose conjugation of monolignols and is essential for the normal cell wall lignification in Arabidopsis thaliana. Plant J. 2016, 88, 26–42. [Google Scholar] [CrossRef]
- Wang, H.; Feng, X.; Zhang, Y.; Wei, D.; Zhang, Y.; Jin, Q.; Cai, Y. PbUGT72AJ2-mediated glycosylation plays an important role in lignin formation and stone cell development in pears (Pyrus bretschneideri). Int. J. Mol. Sci. 2022, 23, 7893. [Google Scholar] [CrossRef]
- Szerszen, J.B.; Szczyglowski, K.; Bandurski, R.S. iaglu, a gene from Zea mays involved in conjugation of growth hormone indole-3-acetic acid. Science 1994, 265, 1699–1701. [Google Scholar] [CrossRef]
- Jackson, R.G.; Lim, E.-K.; Li, Y.; Kowalczyk, M.; Sandberg, G.; Hoggett, J.; Ashford, D.A.; Bowles, D.J. Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J. Biol. Chem. 2001, 276, 4350–4356. [Google Scholar] [CrossRef]
- Jackson, R.G.; Kowalczyk, M.; Li, Y.; Higgins, G.; Ross, J.; Sandberg, G.; Bowles, D.J. Over-expression of an Arabidopsis gene encoding a glucosyltransferase of indole-3-acetic acid: Phenotypic characterisation of transgenic lines. Plant J. 2002, 32, 573–583. [Google Scholar] [CrossRef]
- Tognetti, V.B.; Van Aken, O.; Morreel, K.; Vandenbroucke, K.; van de Cotte, B.; De Clercq, I.; Chiwocha, S.; Fenske, R.; Prinsen, E.; Boerjan, W.; et al. Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 2010, 22, 2660–2679. [Google Scholar] [CrossRef]
- Jin, S.-H.; Ma, X.-M.; Han, P.; Wang, B.; Sun, Y.-G.; Zhang, G.-Z.; Li, Y.-J.; Hou, B.-K. UGT74D1 is a novel auxin glycosyltransferase from Arabidopsis thaliana. PLoS ONE 2013, 8, e61705. [Google Scholar] [CrossRef]
- Poppenberger, B.; Fujioka, S.; Soeno, K.; George, G.L.; Vaistij, F.E.; Hiranuma, S.; Seto, H.; Takatsuto, S.; Adam, G.; Yoshida, S. The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc. Natl. Acad. Sci. USA 2005, 102, 15253–15258. [Google Scholar] [CrossRef]
- Husar, S.; Berthiller, F.; Fujioka, S.; Rozhon, W.; Khan, M.; Kalaivanan, F.; Elias, L.; Higgins, G.S.; Li, Y.; Schuhmacher, R. Overexpression of the UGT73C6 alters brassinosteroid glucoside formation in Arabidopsis thaliana. BMC Plant Biol. 2011, 11, 51. [Google Scholar] [CrossRef]
- Ono, N.N.; Qin, X.; Wilson, A.E.; Li, G.; Tian, L. Two UGT84 family glycosyltransferases catalyze a critical reaction of hydrolyzable tannin biosynthesis in pomegranate (Punica granatum). PLoS ONE 2016, 11, e0156319. [Google Scholar] [CrossRef]
- Wilson, A.E.; Wu, S.; Tian, L. PgUGT95B2 preferentially metabolizes flavones/flavonols and has evolved independently from flavone/flavonol UGTs identified in Arabidopsis thaliana. Phytochemistry 2019, 157, 184–193. [Google Scholar] [CrossRef]
- Shi, M.; Liu, X.; Zhang, H.; He, Z.; Yang, H.; Chen, J.; Feng, J.; Yang, W.; Jiang, Y.; Yao, J.-L. The IAA-and ABA-responsive transcription factor CgMYB58 upregulates lignin biosynthesis and triggers juice sac granulation in pummelo. Hortic. Res. 2020, 7, 139. [Google Scholar] [CrossRef]
- Soeno, K.; Fujioka, S.; Hiranuma, S.; Seto, H.; Yoshida, S. Metabolic conversion of castasterone and brassinolide into their glucosides in higher plants. J. Plant Growth Regul. 2006, 25, 195–202. [Google Scholar] [CrossRef]
- Qin, G.; Xu, C.; Ming, R.; Tang, H.; Guyot, R.; Kramer, E.M.; Hu, Y.; Yi, X.; Qi, Y.; Xu, X. The pomegranate (Punica granatum L.) genome and the genomics of punicalagin biosynthesis. Plant J. 2017, 91, 1108–1128. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, D302–D305. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2010, 39, D225–D229. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Baldauf, S.; Lim, E.K.; Bowles, D.J. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 4338–4343. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Hoang, M.; Wu, C.-N.; Lin, C.-F.; Nguyen, H.T.T.; Chiou, M.-T.; Lin, C.-N. Genetic characterization of feline panleukopenia virus from dogs in Vietnam reveals a unique Thr101 mutation in VP2. PeerJ 2020, 8, e9752. [Google Scholar] [CrossRef]
- Zhang, J. Evolution by gene duplication: An update. Trends Ecol. Evol. 2003, 18, 292–298. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Zhao, X.-Q.; Wang, J.; Wong, G.K.-S.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Qin, G.; Liu, C.; Li, J.; Qi, Y.; Gao, Z.; Zhang, X.; Yi, X.; Pan, H.; Ming, R.; Xu, Y. Diversity of metabolite accumulation patterns in inner and outer seed coats of pomegranate: Exploring their relationship with genetic mechanisms of seed coat development. Hortic. Res. 2020, 7, 10. [Google Scholar] [CrossRef]
- Kwiatkowska, M.; Norman, G.; Parker, D. PRISM 4.0: Verification of probabilistic real-time systems. In Computer Aided Verification. CAV 2011. Lecture Notes in Computer Science; Gopalakrishnan, G., Qadeer, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 585–591. [Google Scholar]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef]
- Zhou, K.; Hu, L.; Li, P.; Gong, X.; Ma, F. Genome-wide identification of glycosyltransferases converting phloretin to phloridzin in Malus species. Plant Sci. 2017, 265, 131–145. [Google Scholar] [CrossRef]
- Gachon, C.M.; Langlois-Meurinne, M.; Saindrenan, P. Plant secondary metabolism glycosyltransferases: The emerging functional analysis. Trends Plant Sci. 2005, 10, 542–549. [Google Scholar] [CrossRef]
- Lim, E.-K.; Doucet, C.J.; Li, Y.; Elias, L.; Worrall, D.; Spencer, S.P.; Ross, J.; Bowles, D.J. The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J. Biol. Chem. 2002, 277, 586–592. [Google Scholar] [CrossRef]
- Tanaka, K.; Hayashi, K.; Natsume, M.; Kamiya, Y.; Sakakibara, H.; Kawaide, H.; Kasahara, H. UGT74D1 catalyzes the glucosylation of 2-Oxindole-3-Acetic Acid in the auxin metabolic pathway in Arabidopsis. Plant Cell Physiol. 2014, 55, 218–228. [Google Scholar] [CrossRef]
- Aoi, Y.; Hira, H.; Hayakawa, Y.; Liu, H.Q.; Fukui, K.; Dai, X.H.; Tanaka, K.; Hayashi, K.I.; Zhao, Y.D.; Kasahara, H. UDP-glucosyltransferase UGT84B1 regulates the levels of indole-3-acetic acid and phenylacetic acid in Arabidopsis. Biochem. Biophys. Res. Commun. 2020, 532, 244–250. [Google Scholar] [CrossRef]
- Du, Y.; Zhao, Q.; Li, W.; Geng, J.; Li, S.; Yuan, X.; Gu, Y.; Zhong, J.; Zhang, Y.; Du, J. Genome-wide identification of the LBD transcription factor genes in common bean (Phaseolus vulgaris L.) and expression analysis under different abiotic stresses. J. Plant Interact. 2022, 17, 731–743. [Google Scholar] [CrossRef]
- Pinweha, N.; Asvarak, T.; Viboonjun, U.; Narangajavana, J. Involvement of miR160/miR393 and their targets in cassava responses to anthracnose disease. J. Plant Physiol. 2015, 174, 26–35. [Google Scholar] [CrossRef]
- Zeng, M.-H.; Liu, S.-H.; Yang, M.-X.; Zhang, Y.-J.; Liang, J.-Y.; Wan, X.-R.; Liang, H. Characterization of a gene encoding clathrin heavy chain in maize up-regulated by salicylic acid, abscisic acid and high boron supply. Int. J. Mol. Sci. 2013, 14, 15179–15198. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N. Abscisic acid and abiotic stress signaling. Plant Signal. Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Sun, M.; Jiang, X.; Sun, H.; Dang, X.; Cong, H.; Qiao, F. Glycinebetaine biosynthesis in response to osmotic stress depends on jasmonate signaling in watermelon suspension cells. Front. Plant Sci. 2018, 9, 1469. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Pati, P.K. Analysis of cis-acting regulatory elements of Respiratory burst oxidase homolog (Rboh) gene families in Arabidopsis and rice provides clues for their diverse functions. Comput. Biol. Chem. 2016, 62, 104–118. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Pati, P.K.; Pati, A.M.; Nagpal, A.K. In-silico analysis of cis-acting regulatory elements of pathogenesis-related proteins of Arabidopsis thaliana and Oryza sativa. PLoS ONE 2017, 12, e0184523. [Google Scholar] [CrossRef]
- Zhou, J.; Hu, Y.; Li, J.; Yu, Z.; Guo, Q. Genome-wide identification and expression analysis of the plant U-box protein gene family in Phyllostachys edulis. Front. Genet. 2021, 2464. [Google Scholar] [CrossRef]
- Song, C.K.; Gu, L.; Liu, J.Y.; Zhao, S.; Hong, X.T.; Schulenburg, K.; Schwab, W. Functional characterization and substrate promiscuity of UGT71 glycosyltransferases from strawberry (Fragaria x ananassa). Plant Cell Physiol. 2015, 56, 2478–2493. [Google Scholar] [CrossRef]
- Lim, E.K.; Bowles, D.J. A class of plant glycosyltransferases involved in cellular homeostasis. EMBO J. 2004, 23, 2915–2922. [Google Scholar] [CrossRef]
- Bowles, D.; Isayenkova, J.; Lim, E.K.; Poppenberger, B. Glycosyltransferases: Managers of small molecules. Curr. Opin. Plant Biol. 2005, 8, 254–263. [Google Scholar] [CrossRef]
- Bowles, D.; Lim, E.K.; Poppenberger, B.; Vaistij, F.E. Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 2006, 57, 567–597. [Google Scholar] [CrossRef]
- Rehman, H.M.; Nawaz, M.A.; Bao, L.; Shah, Z.H.; Lee, J.-M.; Ahmad, M.Q.; Chung, G.; Yang, S.H. Genome-wide analysis of family-1 UDP-glycosyltransferases in soybean confirms their abundance and varied expression during seed development. J. Plant Physiol. 2016, 206, 87–97. [Google Scholar] [CrossRef]
- Cheng, X.; Muhammad, A.; Li, G.; Zhang, J.; Cheng, J.; Qiu, J.; Jiang, T.; Jin, Q.; Cai, Y.; Lin, Y. Family-1 UDP glycosyltransferases in pear (Pyrus bretschneideri): Molecular identification, phylogenomic characterization and expression profiling during stone cell formation. Mol. Biol. Rep. 2019, 46, 2153–2175. [Google Scholar] [CrossRef]
- Wei, Y.; Mu, H.; Xu, G.; Wang, Y.; Li, Y.; Li, S.; Wang, L. Genome-wide analysis and functional characterization of the UDP-glycosyltransferase family in grapes. Horticulturae 2021, 7, 204. [Google Scholar] [CrossRef]
- Levasseur, A.; Pontarotti, P. The role of duplications in the evolution of genomes highlights the need for evolutionary-based approaches in comparative genomics. Biol. Direct 2011, 6, 11. [Google Scholar] [CrossRef]
- Coen, O.; Magnani, E. Seed coat thickness in the evolution of angiosperms. Cell. Mol. Life Sci. 2018, 75, 2509–2518. [Google Scholar] [CrossRef]
- Gardarin, A.; Durr, C.; Mannino, M.R.; Busset, H.; Colbach, N. Seed mortality in the soil is related to seed coat thickness. Seed Sci. Res. 2010, 20, 243–256. [Google Scholar] [CrossRef]
- Kelly, K.M.; Vanstaden, J.; Bell, W.E. Seed coat structure and dormancy. Plant Growth Regul. 1992, 11, 201–209. [Google Scholar] [CrossRef]
- Nooden, L.D.; Blakley, K.A.; Grzybowski, J.M. Control of seed coat thickness and permeability in soybean: A possible adaptation to stress. Plant Physiol. 1985, 79, 543–545. [Google Scholar] [CrossRef]
- Mateo-Bonmati, E.; Casanova-Saez, R.; Simura, J.; Ljung, K. Broadening the roles of UDP-glycosyltransferases in auxin homeostasis and plant development. New Phytol. 2021, 232, 642–654. [Google Scholar] [CrossRef]
- Baldacci-Cresp, F.; Le Roy, J.; Huss, B.; Lion, C.; Creach, A.; Spriet, C.; Duponchel, L.; Biot, C.; Baucher, M.; Hawkins, S.; et al. UDP-glycosyltransferase 72E3 plays a role in lignification of secondary cell walls in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 6094. [Google Scholar] [CrossRef]
- Aloni, R.; Tollier, M.T.; Monties, B. The role of auxin and gibberellin in controlling lignin formation in primary phloem fibers and in xylem of coleus blumei stems. Plant Physiol. 1990, 94, 1743–1747. [Google Scholar] [CrossRef] [PubMed]
- Salazar, R.; Pollmann, S.; Morales-Quintana, L.; Herrera, R.; Caparros-Ruiz, D.; Ramos, P. In seedlings of Pinus radiata, jasmonic acid and auxin are differentially distributed on opposite sides of tilted stems affecting lignin monomer biosynthesis and composition. Plant Physiol. Biochem. 2019, 135, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, M.; Kegasa, T.; Watanabe, A.; Tamura, M.; Tsutsumi, Y. Expression analysis of transporter genes for screening candidate monolignol transporters using Arabidopsis thaliana cell suspensions during tracheary element differentiation. J. Plant Res. 2018, 131, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.H.; Zhou, Z.J.; Qin, H.J.; Tan, J.H.; Ding, G.J. Exogenous brassinosteroid facilitates xylem development in pinus massoniana seedlings. Int. J. Mol. Sci. 2021, 22, 7615. [Google Scholar] [CrossRef]
- Oh, M.H.; Honey, S.H.; Tax, F.E. The control of cell expansion, cell division, and vascular development by brassinosteroids: A historical perspective. Int. J. Mol. Sci. 2020, 21, 1743. [Google Scholar] [CrossRef]
- Zhang, K.; Sun, Y.; Li, M.N.; Long, R.C. CrUGT87A1, a UDP-sugar glycosyltransferases (UGTs) gene from Carex rigescens, increases salt tolerance by accumulating flavonoids for antioxidation in Arabidopsis thaliana. Plant Physiol. Biochem. 2021, 159, 28–36. [Google Scholar] [CrossRef]
- Wu, B.P.; Cao, X.M.; Liu, H.R.; Zhu, C.Q.; Klee, H.; Zhang, B.; Chen, K.S. UDP-glucosyltransferase PpUGT85A2 controls volatile glycosylation in peach. J. Exp. Bot. 2019, 70, 925–936. [Google Scholar] [CrossRef]
- Zheng, H.; Zhang, C.L.; Wang, Y.T.; Zhou, W.T.; Chen, J.; Yan, X.Y.; Li, Z.Q.; Huang, S.Y.; Li, M.Y.; Tang, Y.; et al. Overexpression of the glucosyltransferase gene BoaUGT74B1 enhances the accumulation of indole glucosinolates in Chinese kale. Sci. Hortic. 2021, 288, 110302. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, G.; Li, J.; Qin, G.; Liu, C.; Liu, X.; Cao, Z.; Jia, B.; Zhang, H. Characterization and Expression Analysis of the UDP Glycosyltransferase Family in Pomegranate (Punica granatum L.). Horticulturae 2023, 9, 119. https://doi.org/10.3390/horticulturae9010119
Li G, Li J, Qin G, Liu C, Liu X, Cao Z, Jia B, Zhang H. Characterization and Expression Analysis of the UDP Glycosyltransferase Family in Pomegranate (Punica granatum L.). Horticulturae. 2023; 9(1):119. https://doi.org/10.3390/horticulturae9010119
Chicago/Turabian StyleLi, Guixiang, Jiyu Li, Gaihua Qin, Chunyan Liu, Xin Liu, Zhen Cao, Botao Jia, and Huping Zhang. 2023. "Characterization and Expression Analysis of the UDP Glycosyltransferase Family in Pomegranate (Punica granatum L.)" Horticulturae 9, no. 1: 119. https://doi.org/10.3390/horticulturae9010119
APA StyleLi, G., Li, J., Qin, G., Liu, C., Liu, X., Cao, Z., Jia, B., & Zhang, H. (2023). Characterization and Expression Analysis of the UDP Glycosyltransferase Family in Pomegranate (Punica granatum L.). Horticulturae, 9(1), 119. https://doi.org/10.3390/horticulturae9010119




