1. Introduction
About 34 viruses are currently known to infect stone fruit species [
1], but the number is increasing constantly due to new virus discovery via high-throughput sequencing (HTS) technology [
2,
3]. However, every new virus found in a specific host species does not necessarily mean that these viruses are of economic importance for agricultural production, and most of them are not biologically characterized [
4]. Only a small number of viruses hold the status of economic importance in stone fruits; above all are the plum pox virus (PPV), prune dwarf virus (PDV) and prunus necrotic ringspot virus (PNRSV), which have a worldwide distribution [
5].
The list of economically important virus diseases in stone fruits is clearly dominated by sharka, whose etiological agent is PPV (genus
Potyvirus, family
Potyviridae). PPV naturally infects cultivated, wild and ornamental plants of the genus
Prunus [
6]. According to Candresse et al. [
7], its impact emanates from the severity of the damage it causes and, as a consequence, it has a quarantine status, which makes it a limiting factor for the transport of germplasm and propagating material. First detected in Bulgaria, PPV quickly spread throughout Europe and is now present in most Western, Central and East European countries in varying degrees [
7]. After three decades, the virus has continued to spread and has now been detected in South and North America, Africa and Asia [
8]. One of the features of this virus is its high serological, molecular and biological variability, which is reflected in the existence of 10 currently defined strains: PPV-D, PPV-M, PPV-REC, PPV-EA, PPV-C, PPV-T, PPV-W, PPV-CR, PPV-CV and PPV-An [
9,
10]. These strains differ in their biological and epidemiological characteristics, including severity, efficiency of aphid transmission, as well as the type of symptoms [
11]. Moreover, the PPV strains differ in their geographical distribution and can express host preferences, therefore being limited to specific regions. Geographically, the most common and important strains are PPV-M, D and REC [
12], the latter being a natural recombinant of the M and D strains [
13]. Strains C, EA, CR and CV infect cherry trees and are limited to specific geographic regions.
PNRSV and PDV, together with apple mosaic virus (ApMV), belong to the genus
Ilarvirus, family
Bromoviridae, and are often found in mixed infections [
14]. These viruses alone or in combination can severely affect fruit trees, both in terms of fruit yield and tree growth [
15]. Single or mixed infections can lead to a wide range of symptoms depending on plant species and climatic conditions [
16].
In addition to PPV, PNRSV and PDV, stone fruit viruses that are often found in orchards around the world and included in certification schemes are: apple chlorotic leafspot virus (ACLSV; family Betaflexviridae, genus Trichovirus), ApMV, myrobalan latent ringspot virus (MLRSV; family Secoviridae, genus Nepovirus) and plum bark necrosis stem pitting-associated virus (PBNSPaV; family Closteroviridae, genus Ampelovirus). In general, viruses are a constant companion of stone fruit production in all regions of the world but with differences in the present species, strains, severity and economic importance for a specific region. Therefore, the occurrence and distribution patterns of different viruses can vary between countries.
In Bosnia and Herzegovina (B&H), plum represents a valuable fruit crop with a centuries-old tradition of production. Plums are produced both for fresh consumption and processing, which entails the production of jam and brandy [
17,
18]. At a certain period during the last century, former Yugoslavia (which among others used to include Bosnia and Herzegovina, Croatia and Serbia) was the biggest producer of plums in the world [
19]. At that time, the production in Bosnia and Herzegovina was mainly based on the traditional cultivar ‘Požegača’ (synonyms: ‘Bistrica’, ‘Mađarica’, ‘Hauszwetsche’), which is highly susceptible to PPV [
20]. The rapid spread of PPV throughout all parts of former Yugoslavia has led to a significant decrease in plum production [
21], consequently leading to the introduction of the cultivar ‘Stanley’ and regional cultivars obtained through the Čačak breeding program [
19]. Cultivars developed through this breeding program, such as ‘Čačanska lepotica’, still represent a significant share of today’s plum production in Bosnia and Herzegovina. Although the country has favorable climatic conditions for the production of high quality plum fruit [
22], the production is still in stagnation. In addition to reasons such as economic and market instability caused by the recent war and slow economic recovery of the country, a significant limiting factor in the production are plant diseases, among which virus infections are the most prominent.
The rapid spread of PPV in the former Yugoslavia since the first report in 1936 [
23], has led to an increase in scientific interest in plant virology and has produced a number of scientific papers and detailed distribution maps of the virus [
21,
24]. It is worth mentioning that the region of former Yugoslavia is considered the center of origin of the recombinant PPV strain [
25,
26]. Nevertheless, fresh data on virus distributions among plant species in Bosnia and Herzegovina are scarce. The last notable study on the distribution of plum viruses was conducted by Matić et al. [
27] in 2008 and reported an overall virus infection incidence of 36% for all surveyed stone fruit trees (plum, peach, cherry, apricot, as well as myrobalan and blackthorn). During the aforementioned study, an infection incidence of 30% was detected in plum with the following viruses: PPV (19.5%), PDV (4.2%), PNRSV (2.9%) and ACLSV (1.7%). In addition, Matić et al. [
28] provided insights into the distribution patterns of PPV, as well as the occurrence of PPV strains, pointing out PPV-D as the most common strain and providing first isolate sequences of the three major strains (three for the REC strain, one each for the M and D strains) [
29]. Data on virus occurrence in a plant crop and distribution patterns within a country are important for the development of adequate phytosanitary measures but also bring useful epidemiological information.
Taking into consideration the significant amount of time that has passed since the study conducted by Matić et al. [
27,
28,
29], it is likely that the situation in the field has drastically changed. It is important to note that the previously mentioned virological studies conducted on plum viruses in Bosnia and Herzegovina were limited to sequencing of the purified PCR product, obtained through the amplification of short viral sequences. As stated above, the novel HTS approach presents a much more powerful tool for obtaining information on genetic diversity. As detailed by Maclot et al. [
30], HTS enables characterizations of almost all viruses in a sample without
a priori knowledge of which viruses may be present, therefore prompting new virus discoveries. Additionally, HTS enables the reconstruction of full genome sequences. HTS has been successfully applied in the study of fruit tree viruses in the past decade, leading to new virus discoveries, new strain detections and natural plant host findings. For stone fruit viruses, the benefits of HTS are best seen in the case of little cherry virus 1 (LChV-1). LChV-1 was primarily only associated with cherry trees, but with the use of HTS technologies has now been detected in a wider host range and in different countries around the world [
31,
32,
33,
34]. Therefore, the present study relied on HTS to obtain the full genome sequence of the most prevalent PPV strain.
The objective of this study was to fill the information gap by providing new insights into the occurrence and distribution of seven stone fruit viruses, to evaluate infection incidence in international and traditional plum cultivars, to identify the existing PPV strains and to investigate the genetic diversity of PPV isolates.
2. Materials and Methods
The research consisted of three phases. The first phase involved on-site symptom evaluation combined with serological assays and was aimed at obtaining biological data (symptom cards) and assessment of infection incidence and distribution patterns. The second phase focused on molecular detection and strain typing of PPV, whereas the third phase aimed at obtaining a full genome sequence of the most common PPV strain.
2.1. Field Inspections and Symptom Evaluations
The initial screening survey included 15 orchards in the northwest, central and northeast regions of Bosnia and Herzegovina, representing the main plum-producing regions of the country. Field monitoring with visual symptom evaluations were carried out in all surveyed orchards.
2.2. Plant Material and Serological Assay
A total of 292 symptomatic and 176 asymptomatic leaves were collected in spring (May–June). Each sample represented an individual tree and consisted of 15–20 fully developed leaves collected from all four sides of the canopy (east–west–north–south orientation). An overview of the number of collected samples from each orchard is given in
Supplementary Table S1. The samples were taken from all parts of surveyed orchards. The sample-set consisted mainly of the traditional plum cultivar ‘Požegača’, its synonyms and the main commercial plum cultivars such as ‘Čačanska lepotica’, ‘Stanley’, ‘Čačanska rodna’ and ‘Čačanska rana’. In addition, a number of local cultivars (‘Trnovača’, ‘Savka’ and ‘Kaorka’) and individual trees of introduced cultivars (‘Čačanski šećer’ and ‘Reine-Claude’) that exhibited virus-like symptoms were sampled. All samples were kept frozen at −20 °C prior to testing.
In addition to PPV, PDV and PNRSV as the viruses of the greatest economic importance, screening for apple chlorotic ringspot virus (ACLSV), apple mosaic virus (ApMV) and myrobalan latent ringspot virus (MLRSV) was also conducted in this study because they are listed in the EPPO’s regulation scheme for
Prunus planting material. Plum bark necrosis stem pitting-associated virus (PBNSPaV) was also included in the study as it was reported from neighboring Serbia [
35].
A total of 468 samples were tested for the presence of PPV, MLRSV, PDV and PBNSPaV by DAS-ELISA [
36], ApMV and PNRSV by DASI-ELISA [
37], and ACLSV by cocktail-ELISA. PPV, ApMV, PNRSV, MLRSV and PBNSPaV were screened using commercially available ELISA kits from Agritest (Valenzano, Italy), whereas ACLSV and PDV were tested using kits from Loewe Biochemica (Sauerlach, Germany). All tests were performed according to the manufacturers’ instructions. Samples displaying absorbance values at 405 nm at least three times higher than the mean value of the negative controls were considered positive. Absorbance was measured 1 h after application of the substrate buffer using the Biotek EL800 spectrophotometer (Biotek, VT, USA).
2.3. Plant Material and Molecular Assay
The molecular study focused exclusively on three plum cultivars (‘Stanley’, ‘Čačanska lepotica’, ‘Požegača’) that dominate plum production in the sampled orchards in B&H. The selected orchards included both the predominant cultivars and the main geographical locations of plum production in the country. Field visits and sampling were conducted from 2017 to 2018 at two orchards in the northwest and three orchards in the northeast part of the country. A total of 45 leaf samples were collected representing three main cultivars. The dataset consisted of 12 samples collected from trees that previously tested positive for PPV by ELISA (‘Požegača’) and 33 samples collected without any prior testing (‘Stanley’ and ‘Č. lepotica’). The sample consisted of 15–20 fully developed leaves that were taken from all sides of the canopy representing one tree in the orchard.
Total RNA was extracted from leaf tissue using the RNeasy Plant mini kit (Qiagen, Germantown, MD, USA) after grinding samples in liquid nitrogen to fine powder. RNA quality and quantity was measured using a UV/VIS spectrophotometer NanoDrop 1000 (ThermoFischer, Waltham, MA, USA). RT-PCR was carried out using the One-Step RT-PCR kit (Qiagen) in 10 µL of reaction volume containing: 100 ng total RNA as a template, 0.4 µL enzyme mix, 2 µL reaction buffer (5×), 2 µL Q-solution, 3 µL RNase-free water, 0.4 µL dNTP mix and 0.6 µL of each primer (10 µM). The P1/P5 primer combination [
38] was used to amplify a 243 bp fragment of the C-terminal CP gene, ensuring the detection of PPV. Strain-specific primers were used to differentiate isolates belonging to the most common strains reported from B&H and surrounding countries: D, M and REC [
39]. Reverse transcription was carried out at 50 °C for 30 min followed by an initial activation step at 95 °C for 15 min in accordance with the manufacturer’s instruction. The following thermal cycling scheme was used in 40 repeats: denaturation at 94 °C for 15 s, annealing at 52 °C for 1 min and elongation at 72 °C for 45 s. A final elongation step at 72 °C for 10 min was performed. RT-PCR results were verified by electrophoresis in a 1.5% TBE agarose gel. The size of amplified products was determined using a siZer-100 DNA marker (iNtROn, Seongnam, South Korea).
2.4. Total RNA Extraction and HTS
A sample of the cultivar ‘Požegača’ with confirmed PPV-D infection from an orchard in the northwest region with high infection incidence was selected for HTS. Total RNA was extracted using the Spectrum Total Plant RNA Kit (ThermoFischer) followed by quantification and quality control using the NanoDrop 1000 (ThermoFischer). For ribosomal RNA depletion, the Ribo-Zero Plant Kit (Illumina, San Diego, CA, USA) was used and the library was prepared using the TrueSeq Standard Total RNA Library Prep Kit (Illumina). Sequencing was performed on the Illumina NextSeq 500 platform with a sequencing length of 2 × 150 nucleotides (nt) (University of Liege, Gembloux, Belgium). Data were analysed using Geneious software, version 10.1.2. (Biomatters, Auckland, New Zealand). Duplicate reads were first eliminated and reads were further paired and merged. De novo assembly was performed using the SPAdes algorithm as a plugin in Geneious. De novo contigs were annotated using the TBLASTX module of Geneious for homology with a local database of virus and viroid genomes downloaded from the RefSeq database of NCBI.
2.5. Cloning and Sequencing
Sample O7/68 (cv. ‘Požegača’), collected in the same orchard from the northwest region as O7/80 showed an infection with PPV-REC. To obtain further molecular data, this isolate was gel-purified using a NucleoSpin PCR Clean-Up Kit (Macherey-Nagel, Düren, Germany) and subjected to quality control. The purified PCR products were ligated into the pJet 1.2 vector using the Clonejet PCR Cloning Kit (ThermoFischer) according to manufacturer’s instructions. Plasmid DNA extraction was performed using the NucleoSpin Plasmid DNA Purification Kit (Macherey-Nagel). Selected clones were subjected to Sanger sequencing (StarSeq, Mainz, Germany).
2.6. Phylogenetic Analysis
Sequence similarities of the obtained isolates were confirmed using the BlastN program in GenBank. Phylogenetic analysis was conducted on the whole genome and partial genome sequence. The corresponding region of the previously generated sequences from B&H was extracted from the whole genome and included in additional phylogenetic analysis to obtain information about their sequence similarities.
Sequence alignments were performed using the MUSCLE algorithm in MEGA ver. 7.0.26 [
40]. Sequence analysis also included isolates representing each of the 10 PPV strains, which allowed observation of diversity between strains at the nucleotide and amino acid levels. Phylogenetic trees were constructed from nucleotide sequence alignments using maximum likelihood (ML) under the GTR+G+I model.
Visualization of phylogenetic trees was performed using FigTree ver.1.4.3 (
http://tree.bio.ed.ac.uk/software/Figtree/, accessed 1 May 2022). Sequence identity matrixes for amino acids and nucleotides were generated in BioEDIT ver. 7.0.5.3 [
41].
3. Results
3.1. Field Inspections and Symptom Evaluation
The most common symptoms observed in plum orchards were discoloration in the form of chlorotic ring patterns on the leaves and vein banding (
Figure 1A,B). These manifestations of symptoms were subsequently determined by ELISA to be indicative of the presence of PPV in the examined trees. Through strain typing using RT-PCR, the aforementioned symptoms of irregular shapes and semicircles with chlorotic discoloration were associated with infections with the PPV-D strain, while the classic ring patterns were observed in trees infected with PPV-REC (
Figure 1C–E).
Differences in symptom intensity were observed between traditional and international cultivars, with ‘Požegača’ and local cultivars showing a stronger symptomatic response (
Figure 1F). Among international cultivars, a strong symptomatic response to PPV was observed in ‘Čačanska lepotica’ and ‘Stanley’ in the form of rings, semicircles and lesions.
Although PDV and PNRSV were detected, they could not be linked to any specific symptoms. Mixed infections of PPV+PDV, later detected by ELISA, could not be associated with any observed symptoms
3.2. Results of Serological Assays
Out of 468 samples, 243 tested positive to at least one virus included in the investigation, leading to an overall infection incidence of 51.9%. Specifically, we detected infections with PPV (48.7%), PDV (2.99%), PNRSV (0.21%) and a mixed infection with PPV+PDV (eight samples, 1.71%). Other combinations of mixed infections were not detected by ELISA. Infections with ApMV, ACLSV, MLRSV and PBNSPaV were not detected in this study.
Of the 292 symptomatic samples, 78.7% tested positive, the remaining 21.3% showed absorbance below the ELISA cut-off value and were considered negative. Among the asymptomatic samples, 8.52% tested positive (PPV and PDV detected)
PPV was detected in all surveyed regions, while PDV and PNRSV were detected only in the northwestern part of B&H. The northwestern region had an infection incidence of 50.4%, with the following frequencies: PPV 46.9%, PDV 3.13% and PNRSV 0.24%. A mixed infection with PPV+PDV was detected (1.71%). Infection incidence was higher in traditional plum cultivars (57.3%) than in international cultivars (36.1%). An overview of the results for the individual locations can be found in
Supplementary Table S1.
3.3. Results of Molecular Assays
Of the 45 samples, 36 tested positive for the presence of PPV using the universal P1/P5 primer. To determine the presence of PPV-D, PPV-REC and PPV-M, the positive samples were subjected to strain typing by RT-PCR. The most frequently detected strain was PPV-D, which was found in 12 samples (33.3%), followed by PPV-REC, which was detected in six samples (16.7%). A mixed infection with two strains (PPV-D+PPV-REC) was found in three samples (8.33%) of cv. ‘Požegača’, all from the same orchard in the northwestern region of Bosnia and Herzegovina. RT-PCR confirmed the presence of PPV in all 12 ELISA-positive samples. Of the randomly selected symptomatic samples, seven proved to be negative. Strain PPV-M was not detected in this study.
Sixteen samples of the cv. ‘Požegača’ tested positive for the presence of PPV of which nine samples were infected with PPV-D and four samples were positive for PPV-REC. All samples of this cultivar originated from the orchards in the northwest region. Three samples proved to carry a mixed infection of PPV-D+PPV-REC. In the cultivar ‘Stanley’, from the northeast region, PPV was found in nine out of ten tested samples. In three samples the PPV-D strain could be detected, and in two samples it was PPV-REC. Out of 16 tested samples of the cultivar ‘Čačanska lepotica’, 11 were positive for PPV. For this cultivar, only one sample could be associated with a strain, namely PPV-D. Interestingly, 21 out of the 36 PPV-positive samples could not be assigned to any of the strains studied (D, M or REC). A detailed overview of the results is given in
Supplementary Table S2.
Regarding the regional distribution, PPV-D and PPV-REC were found in all regions studied, with a higher frequency in the northwestern part (52.6%) than in the northeastern region (23.1%). However, it is worth noting that 70% of positive samples (14/20) from the northeastern region could not be associated with any of the strains analyzed. In addition, co-infections with PPV-D+PPV-REC were detected in the same orchard (three samples).
3.4. Sequencing Results
Because of the higher prevalence of occurrence, PPV-D was selected for genome sequencing using HTS. After quantity and quality controls, sample PPV_O7/80 originating from an orchard in the northwestern region was sequenced. A total of 2.5 million reads of 2 × 150 nt were generated.
A 9.743 nts sequence belonging to the PPV-D strain and encompassing the entire genome was obtained by HTS and de novo assembly (isolate PPV_O7/80; accession number in GenBank: MW412433).
Because orchard O7 had a high infection incidence and co-existence of PPV-D+PPV-REC, and in order to obtain further molecular information about the REC strain, a partial genome sequence from one isolate (designated PPV_O7/68) was Sanger sequenced after strain-specific amplification and cloning. The sequence obtained corresponded to the (Cter) NIb/(Nter) CP region (622 bp) belonging to the PPV-REC strain, and was deposited in the GenBank database (MW412434).
3.5. Genetic Diversity of PPV Isolates
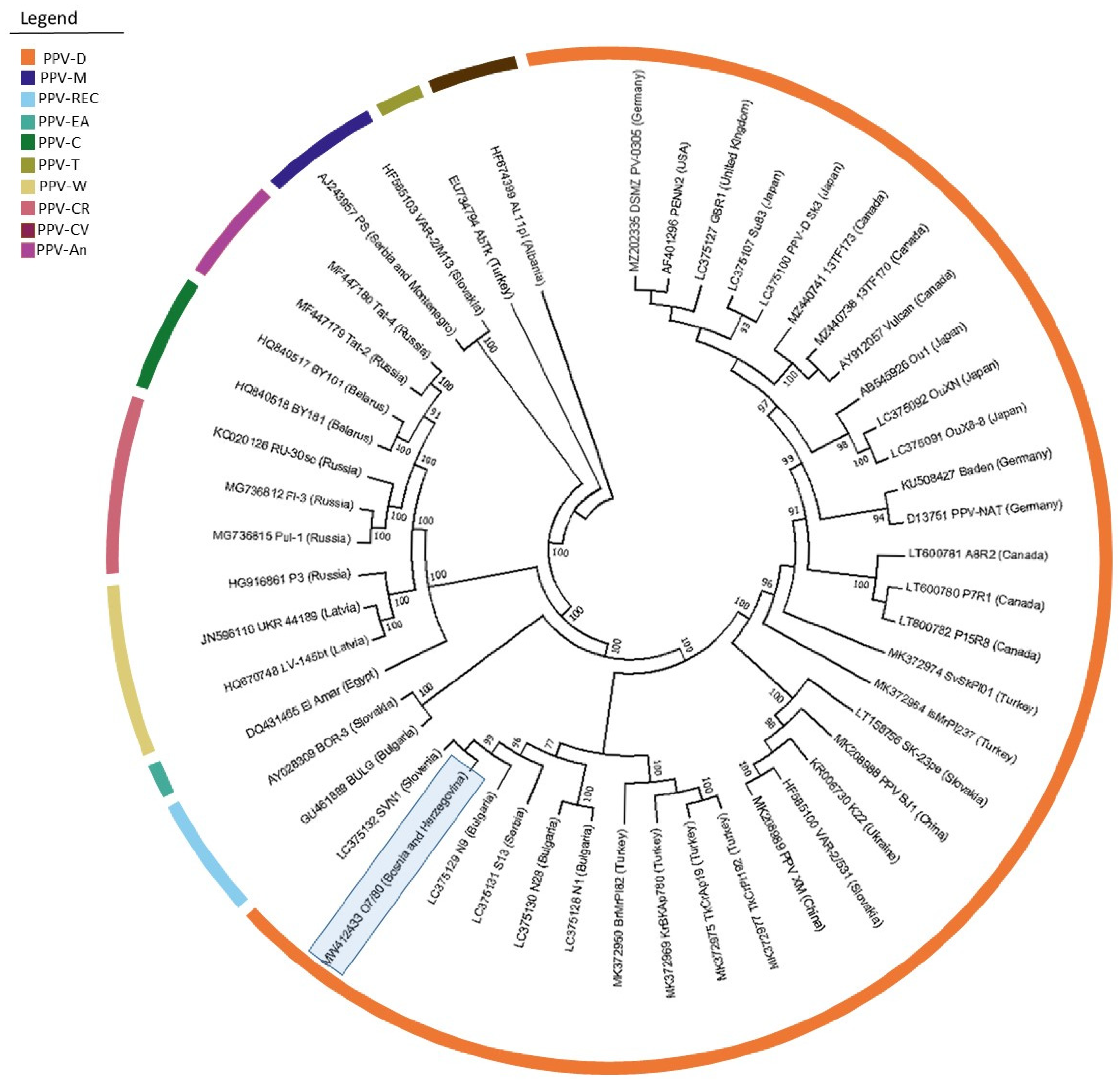
The obtained complete genome sequence (isolate PPV_O7/80; MW412433) was compared with 27 other D strain isolates, as well as with representatives of the individual PPV strains, giving a total of 50 isolates used for comparison (
Supplementary Table S3). The isolate from B&H had the highest nucleotide sequence identity of 97.7% along with isolates S13 (LC375131) from Serbia and isolate IsMrPI237 (MK372964) from Turkey. Nucleotide similarities of 97.6% were found with isolates SVN1 (LC375132) from Slovenia, N9 (LC375129) from Bulgaria and TkCrPI192 (MK372975) from Turkey. However, the amino acid similarity comparison between the analyzed isolates showed the highest identity values of 95.6% with isolates S13 (LC375131), SVN1 (LC375132) and N9 (LC375129).
Phylogenetic analysis revealed the general time reversible model with gamma distributions and invariant sites (GTR+G+I) as the best model for nucleotide substitutions for the whole genome sequence. The inferred phylogenetic tree placed the PPV_O7/80 isolate in the D strain group and formed a clade with the SVN1 isolate (LC375132) from Slovenia (
Figure 2).
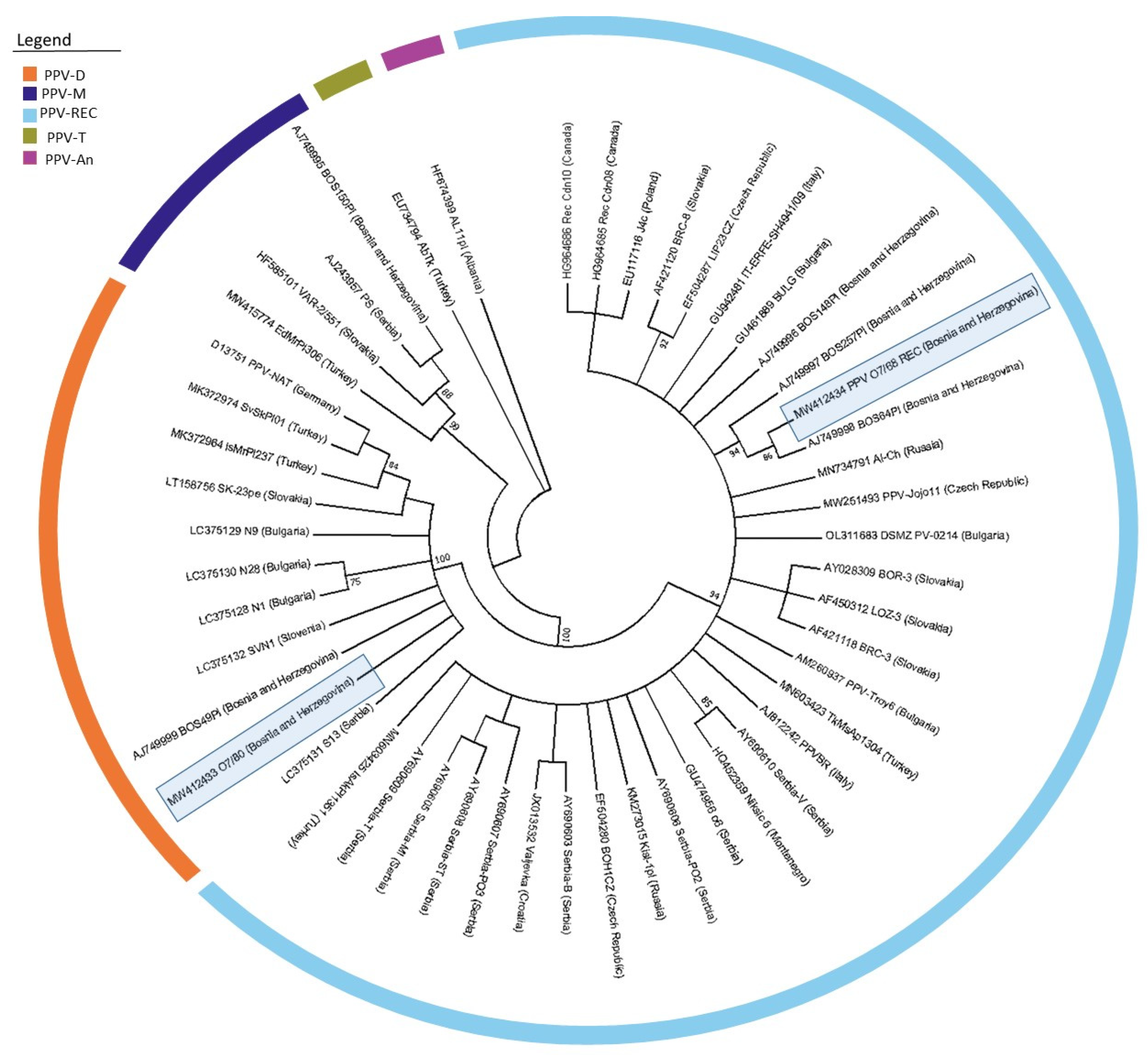
Since no other complete genome sequences from B&H were available in public databases, it was necessary to isolate and analyze the (Cter)NIb/(Nter)CP region of the genome to compare the diversity among the B&H isolates obtained in this study and the five isolates (three isolates of the REC strain, one isolate each of the D and M strains) previously reported. Therefore, this region was extracted from the complete genome sequence of the D strain and analyzed together with the PPV-REC isolate PPV_O7/68 and 48 other isolates (
Supplementary Table S4).
The Bosnian-Herzegovinian isolates of the PPV-D strain, PPV_O7/80 (MW412433) and BOS49PI (AJ749999) showed high nucleotide (98.3%) and amino acid (97.4) similarities. In addition, both isolates of the D strain showed high nucleotide (between 96.6 and 97.9%) and amino acid (95.9 to 97.9%) similarities with isolates from Serbia (LC375131) and Slovenia (LC375132), which is also reflected in the positioning of these isolates on the inferred phylogenetic tree (
Figure 3).
The three sequences of previously reported PPV-REC isolates from B&H (AJ749996–AJ749998) have nucleotide similarities from 97.7 to 99.5% and amino acid similarities from 98.4 to 100% with the isolate PPV_O7/68 (MW412434) obtained in our study. In contrast to the PPV-D isolates, the PPV-REC isolates from Bosnia and Herzegovina did not express high similarities with any of the Serbian isolates. The inter-strain diversity between the D and REC isolates from B&H was 15.8%. On the generated phylogenetic tree, all PPV-REC isolates from B&H were clustered together (
Figure 3).
4. Discussion
An overall infection incidence of 51.9%, determined by ELISA among the collected samples, indicates a poor phytosanitary status of plum orchards in all studied regions in Bosnia and Herzegovina. Symptoms associated with virus infections were observed in various forms in all studied orchards: bands and irregular shapes, as well as vein banding and ring patterns. Sharka-associated symptoms in the form of ring patterns or semicircles, as described by Wang et al. [
42], were predominant. PNRSV and PDV, two viruses belonging to the genus
Ilarvirus, have a wide range of symptoms and are often found in co-infections, which can lead to stronger symptomatic reactions. PNRSV infection can be asymptomatic, and if symptom expressions manifest, they range from necrotic rings and mosaics to overall yield reductions [
43,
44]. PDV infections in plum are mostly associated with growth reduction and leaf deformations [
45]. This wide range of symptoms makes field monitoring and evaluations for these viruses difficult. In our study, none of the observed symptoms could be linked to PDV and PNRSV, probably also due to external factors influencing symptom expressions. No symptoms associated with ApMV, ACLSV, MLRSV and PBNSPaV were observed during orchard monitoring.
In this study, 21.3% of the samples that exhibited virus-like symptoms tested negative by ELISA. This is most likely related to the low virus titer in the plants or some other biotic or abiotic factors, especially nutrient deficiency, that could cause similar symptoms [
38]. However, the presence of viruses not included in this study is also a possibility. Among the collected asymptomatic samples, 8.52% tested positive. Symptom evaluation is not a simple endeavor, as several factors can influence expression in plants, including poor phytosanitary status or environmental conditions [
46,
47] that can temporally cover virus symptoms.
PPV, the causal agent of sharka disease in plums, had the largest share in the infection incidence with as much as 49% and was found in all geographical regions surveyed and in the majority of commonly grown plum cultivars. This is a significant increase in PPV infection incidence compared to the results of Matić et al. [
27], who reported a PPV infection incidence of 19.5% in plum. This clearly shows deterioration in the phytosanitary status of plum orchards in B&H during the last decade. Nevertheless, both studies revealed similar patterns of PPV distribution in all major plum-producing regions of B&H. Similarly high levels of PPV infection were also found in neighboring Croatia (51.4%) [
48] and Serbia (55.7%) [
49].
PDV and PNRSV were detected by serological assays with infection incidences of 2.99% and 0.21%, respectively. In comparison, previous research [
27] showed that among sampled plants, PDV infection incidence was 4.2%, while PNRSV was present in 2.9% of tested plants.
Co-infections of PPV+PDV were detected by ELISA but only in eight samples (1.71%). The mixed infection could not be associated with any specific symptoms observed on the trees. Mixed infections in stone fruits (plum, apricot, peach, sweet and sour cherry), although in lower frequency (5.62%), have previously been reported by Matić et al. [
27], who found PNRSV+PDV to be the most common mixed infection in all studied orchards in Bosnia and Herzegovina. In this study, no co-infections with PNRSV and PDV were detected. Overall, it should be noted that these two viruses were not found together in a sampled orchard. Although all samples were tested for the presence of ApMV, ACLSV, MLRSV and PBNSPaV by ELISA, no infections with any of these viruses were found.
Infection with PPV was found in both traditional and international plum cultivars, with a higher infection incidence in traditional (57.3%) than in international cultivars (36.1%). This is probably due to the fact that the highly susceptible traditional cultivar ‘Požegača’ is by far the most commonly grown traditional plum cultivar in B&H. It is important to note that ‘Požegača’ is often found as a marginal or solitary tree in gardens, often in close proximity to commercial orchards. The fact that tolerant cultivars are grown together with traditional, susceptible cultivars might explain the high infection incidence in Čačak selections and the cultivar ‘Stanley’. However, viruses also have the ability to accumulate in tolerant cultivars, sometimes up to the same concentration as in susceptible cultivars [
26]. Thus, tolerant cultivars can act as natural reservoirs of viral inoculum, providing a permanent source of infection that can be transmitted by vectors (e.g., aphids) to highly susceptible cultivars. Because tolerant cultivars do not show strong pathological responses, these virus sources can go unnoticed for many years. Another factor that likely contributed to the decline of plum orchards in B&H is the source of planting material. While virus-free planting material of international cultivars is widely available, traditional cultivars are harder to find and are usually propagated vegetatively from older trees without adequate phytosanitary controls. These trees likely contribute to the further spread of the disease. The unavailability of quality planting material, the existence of trees that act as natural virus reservoirs, combined with the possibility of virus transmissions by aphids, and the lack of adequate phytosanitary regulations and eradication procedures in B&H, have most likely led to the stagnation of plum production.
Molecular strain typing revealed the presence of PPV-D and PPV-REC strains in the sampled areas, with the D strain being more abundant, while the M strain was not detected. The co-infection of D and REC strains found in three samples was not unexpected, considering that mixed populations of PPV strains on a single tree are not uncommon [
50]. It is worth mentioning that the mixed infection PPV-D+PPV-REC is also the most common in Serbia with an incidence of 13% [
49]. Compared to the previous studies by Matić et al. [
28], the data on the prevalence of PPV strains in B&H obtained in our study show a different ratio between strains. However, the two studies agree on the lower presence or, in our case, the complete absence of the M strain. In their study, Matić et al. [
29] identified the M strain in four of sixteen samples examined.
In neighboring countries, somewhat different ratios were observed among PPV strains. In Serbia the most common strain was PPV-REC [
49] and in Croatia it was the PPV-D strain [
48,
51]. In both countries all three PPV strains were reported, with the M strain observed significantly less frequently. The distribution of each PPV strain within geographic regions varies considerably, with the mode of spread differing in time and space among different PPV isolates, host species and environmental conditions [
52]. In general, PPV-REC is widespread in central and southeastern Europe, PPV-M is most common in southern and central Europe and Turkey, while PPV-D has the widest distribution, occurring in Europe, North and South America, and several Asian countries [
9]. In addition, the ratio of occurrence of the different PPV strains varies from country to country. It is also important to note that most of the strain-typed samples (21 of 36) in our study could not be assigned to the three PPV strains mentioned above, raising the question of whether other PPV strains might be common in B&H. Considering the emergence of new strains, such as PPV-CV [
53], the tenth PPV strain identified to date, the standard set of screening primers developed by Šubr et al. [
39] needs to be expanded to include strain-specific primers for the new strains, especially when taking into consideration that global inter-strain diversity of PPV is evolving [
54,
55].
The first complete genome sequence of a PPV-D isolate from B&H was obtained by HTS. Isolate PPV_O7/80 (MW412433) showed a high degree of nucleotide similarity (97.7%) to isolate S13 (LC375131) from Serbia. Considering the history of the two countries, as well as the fact that Čačanska selections originating from Serbia still dominate plum production in B&H, this result was not surprising. It is worth mentioning that as well as the Serbian isolate, no other complete genome sequences from this geographical region are available in GenBank. Isolate PPV_O7/80 also showed high nucleotide identity (97.6%) with isolates N9 (LC375130) from Bulgaria and SVN1 (LC375132) from Slovenia, forming a single clade within the aforementioned Slovenian isolate on the phylogenetic tree. This observation could be linked to introduction events when considering the frequent trade between the northwest part of B&H and Slovenia.
The intra-strain diversity between all analyzed isolates of the PPV-D strain was not higher than 3.7%. The observed inter-strain diversity was up to 23%, therefore clearly separating the representatives of all strains in separate groups (
Figure 2).
To compare isolate PPV_O7/80 (MW412433) with the previously known partial sequences from B&H and surrounding countries, the corresponding (Cter)NIb/(Nter)CP region was excised from the whole genome sequence. As demonstrated by the phylogenetic tree (
Figure 3), our PPV-D isolate was positioned between isolate SVN1 (LC375132) from Slovenia, BOS49PI (AJ 749999) from B&H and isolate S13 from Serbia (LC375131), sharing the amino acid identities of 97.9, 97.4 and 97.4%, respectively. Similarly, the PPV-REC isolate O7/68 (MW412434) obtained in this study showed high nucleotide (97.7 to 99.5%) and amino acid (98.4 to 100%) identity with three previously reported B&H isolates of the same strain. Both cases suggest possible local infection by insect vectors or as a result of the local exchange of planting material within B&H or surrounding countries.