Genome-Wide Identification and Expression Analysis of Eggplant DIR Gene Family in Response to Biotic and Abiotic Stresses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Chromosomal Mapping of DIR Family Genes in Eggplant
2.2. Gene Structure Analysis and Phylogenetic Tree Construction of DIR Family Genes in Eggplant
2.3. Gene Duplication and Synteny Analysis of DIR Family Genes
2.4. RNA-Seq Reanalysis of Eggplant Transcriptome Sequencing Data
2.5. Organ-Specific Expression Analysis of DIR Family Genes in Eggplant
2.6. The Expression Profiles Analysis of Eggplant DIR Family Genes under Stresses
3. Results
3.1. Genome-Wide Identification of DIR Family Genes in Eggplant
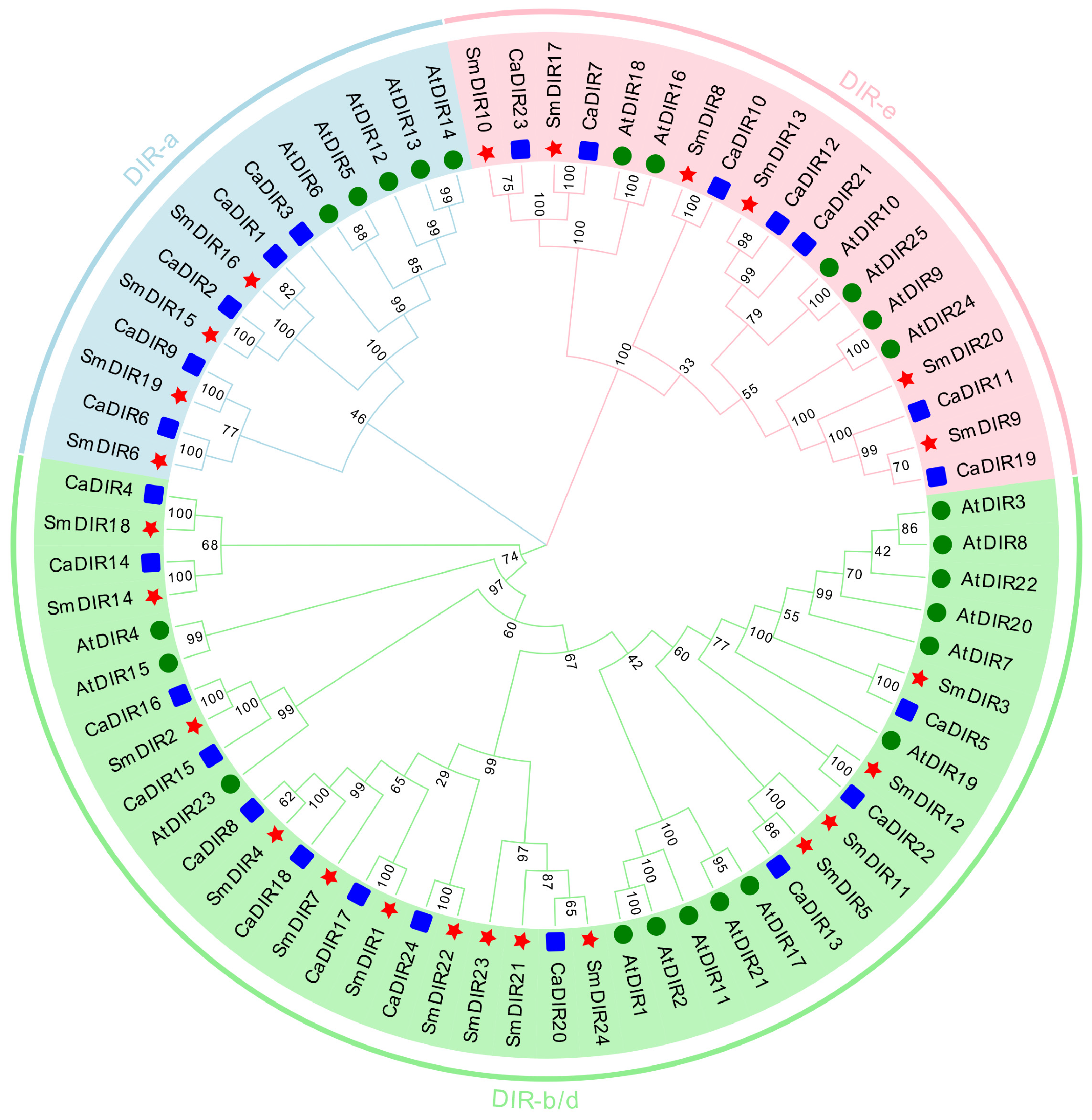
3.2. Phylogenetic Analysis of DIR Family Genes in Eggplant, Pepper and Arabidopsis
3.3. Gene Structure and Conserved Motifs Analyses of Eggplant DIR Genes
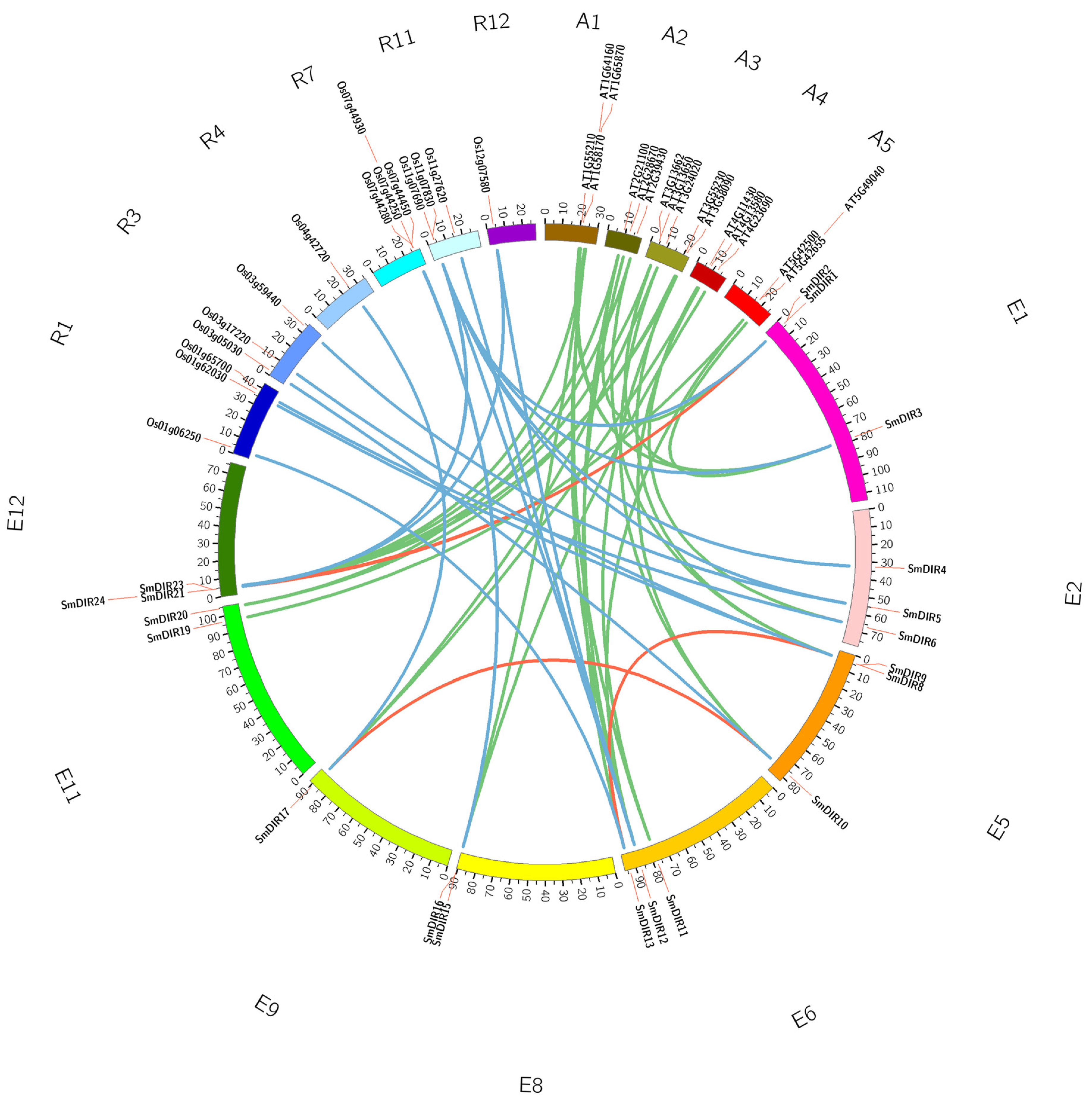
3.4. The Synteny Analysis of DIR Genes among Eggplant, Arabidopsis and Rice
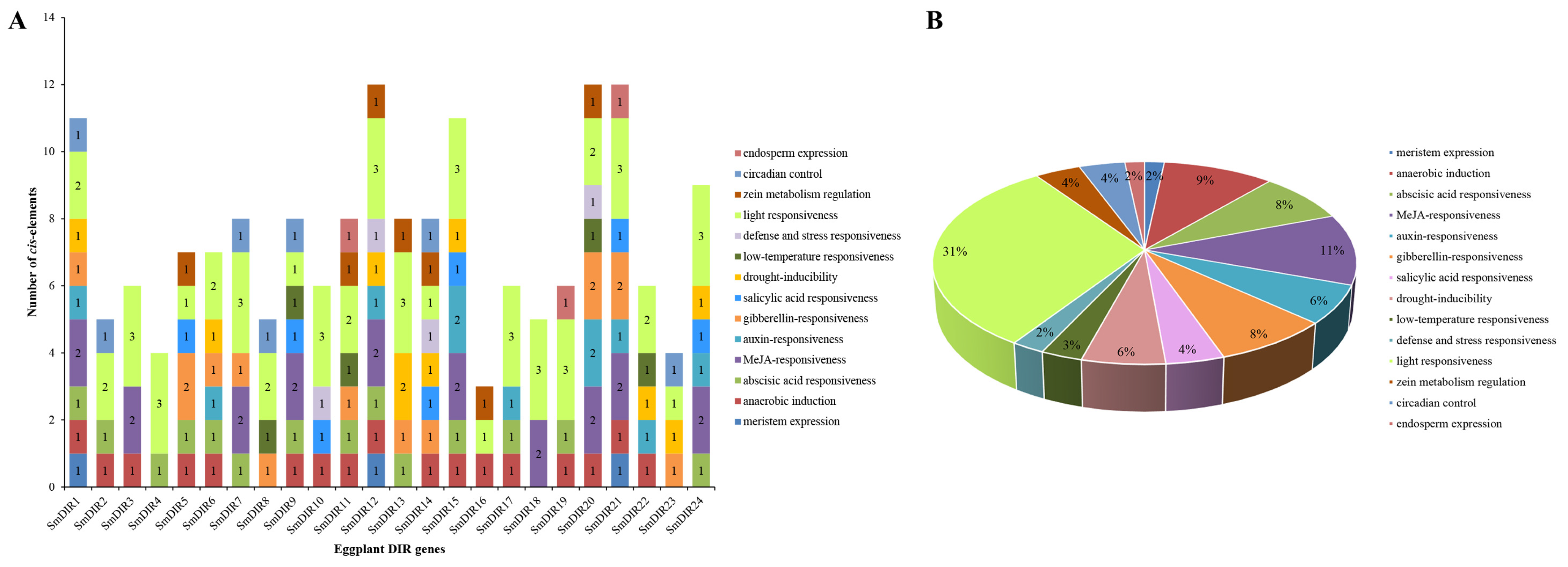
3.5. The Cis-Acting Elements Analysis of Eggplant DIR Genes
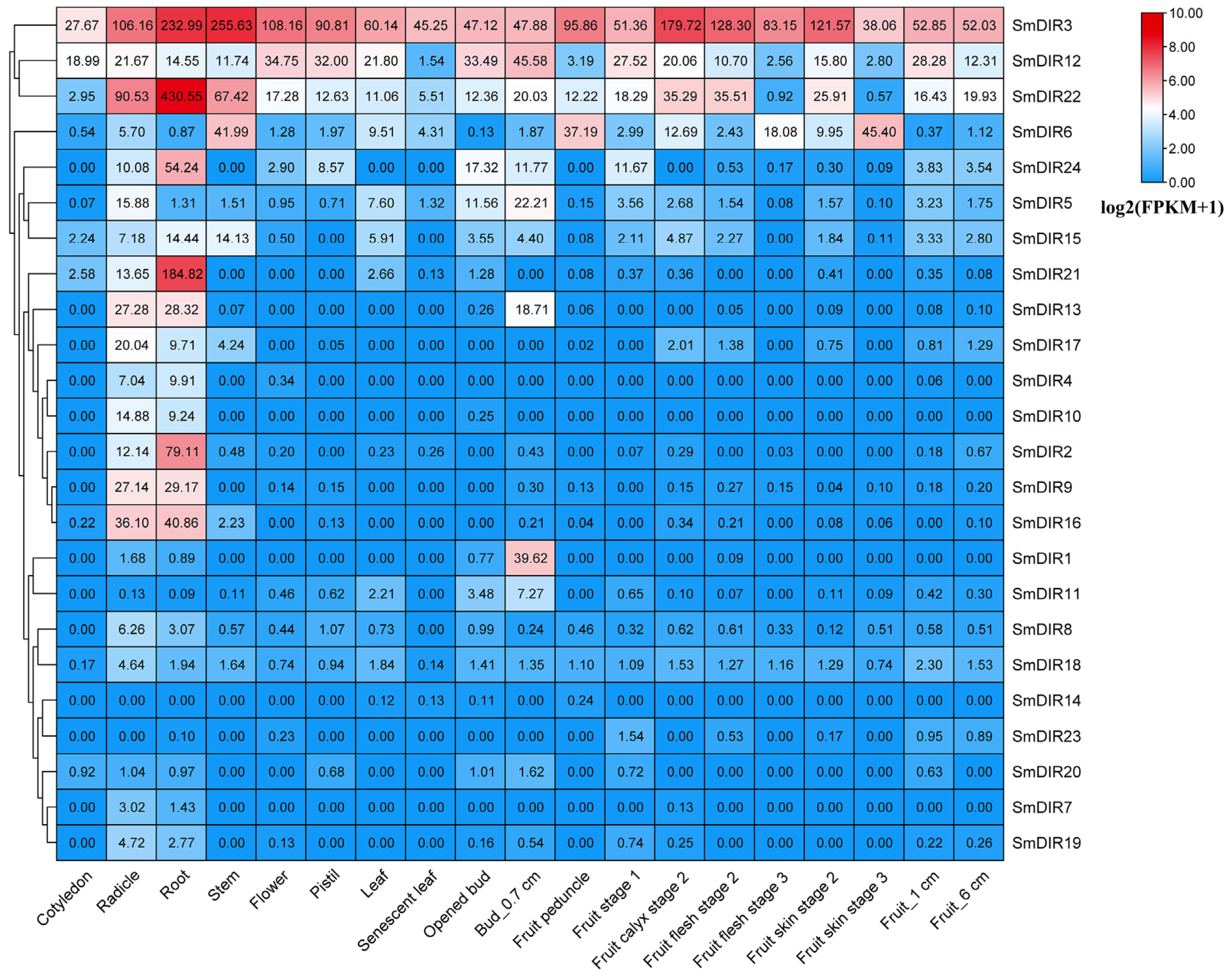
3.6. Organ-Specific Expression Analysis of Eggplant DIR Genes
3.7. The Expression Profiles of Eggplant DIR Genes under Abiotic Stresses
3.8. The Expression Profiles of Eggplant DIR Genes under Biotic Stresses
3.9. The Regulation Patterns Analysis of Eggplant DIR Family Genes under Stresses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Madison, I.; Long, T.A.; Williams, C.M. Computational solutions for modeling and controlling plant response to abiotic stresses: A review with focus on iron deficiency. Curr. Opin. Plant Biol. 2020, 57, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Ralph, S.; Park, J.-Y.; Bohlmann, J.; Mansfield, S.D. Dirigent proteins in conifer defense: Gene discovery, phylogeny, and differential wound-and insect-induced expression of a family of DIR and DIR-like genes in spruce (Picea spp.). Plant Mol. Biol. 2006, 60, 21–40. [Google Scholar] [CrossRef] [PubMed]
- Arasan, S.K.T.; Park, J.-I.; Ahmed, N.U.; Jung, H.-J.; Hur, Y.; Kang, K.-K.; Lim, Y.-P.; Nou, I.-S. Characterization and expression analysis of dirigent family genes related to stresses in Brassica. Plant Physiol. Biochem. 2013, 67, 144–153. [Google Scholar] [CrossRef]
- Cheng, X.; Su, X.; Muhammad, A.; Li, M.; Zhang, J.; Sun, Y.; Li, G.; Jin, Q.; Cai, Y.; Lin, Y. Molecular characterization, evolution, and expression profiling of the dirigent (DIR) family genes in Chinese white pear (Pyrus bretschneideri). Front. Genet. 2018, 9, 136. [Google Scholar] [CrossRef]
- Bonello, P.; Storer, A.J.; Gordon, T.R.; Wood, D.L.; Heller, W. Systemic effects of Heterobasidion annosum on ferulic acid glucoside and lignin of presymptomatic ponderosa pine phloem, and potential effects on bark-beetle-associated fungi. J. Chem. Ecol. 2003, 29, 1167–1182. [Google Scholar] [CrossRef]
- Miedes, E.; Vanholme, R.; Boerjan, W.; Molina, A. The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 2014, 5, 358. [Google Scholar] [CrossRef]
- Davin, L.B.; Jourdes, M.; Patten, A.M.; Kim, K.-W.; Vassao, D.G.; Lewis, N.G. Dissection of lignin macromolecular configuration and assembly: Comparison to related biochemical processes in allyl/propenyl phenol and lignan biosynthesis. Nat. Prod. Rep. 2008, 25, 1015–1090. [Google Scholar] [CrossRef]
- Davin, L.B.; Lewis, N.G. Lignin primary structures and dirigent sites. Curr. Opin. Biotech 2005, 16, 407–415. [Google Scholar] [CrossRef]
- MacRae, W.D.; Towers, G.N. Biological activities of lignans. Phytochemistry 1984, 23, 1207–1220. [Google Scholar] [CrossRef]
- Harmatha, J.; Dinan, L. Biological activities of lignans and stilbenoids associated with plant-insect chemical interactions. Phytochem. Rev. 2003, 2, 321–330. [Google Scholar] [CrossRef]
- Ralph, S.G.; Jancsik, S.; Bohlmann, J. Dirigent proteins in conifer defense II: Extended gene discovery, phylogeny, and constitutive and stress-induced gene expression in spruce (Picea spp.). Phytochemistry 2007, 68, 1975–1991. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Wang, L.; Wang, Z.; Shang, H.; Liu, X.; Zhu, Y.; Qi, D.; Deng, X. Cloning and expression analysis of a dirigent protein gene from the resurrection plant Boea hygrometrica. Prog. Nat. Sci. 2009, 19, 347–352. [Google Scholar] [CrossRef]
- Li, Q.; Chen, J.; Xiao, Y.; Di, P.; Zhang, L.; Chen, W. The dirigent multigene family in Isatis indigotica: Gene discovery and differential transcript abundance. BMC Genom. 2014, 15, 388. [Google Scholar] [CrossRef] [PubMed]
- Davin, L.B.; Wang, H.-B.; Crowell, A.L.; Bedgar, D.L.; Martin, D.M.; Sarkanen, S.; Lewis, N.G. Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science 1997, 275, 362–367. [Google Scholar] [CrossRef]
- Paniagua, C.; Bilkova, A.; Jackson, P.; Dabravolski, S.; Riber, W.; Didi, V.; Houser, J.; Gigli-Bisceglia, N.; Wimmerova, M.; Budínská, E. Dirigent proteins in plants: Modulating cell wall metabolism during abiotic and biotic stress exposure. J. Exp. Bot. 2017, 68, 3287–3301. [Google Scholar] [CrossRef]
- Liao, Y.; Liu, S.; Jiang, Y.; Hu, C.; Zhang, X.; Cao, X.; Xu, Z.; Gao, X.; Li, L.; Zhu, J. Genome-wide analysis and environmental response profiling of dirigent family genes in rice (Oryza sativa). Genes Genom. 2017, 39, 47–62. [Google Scholar] [CrossRef]
- Singh, D.K.; Mehra, S.; Chatterjee, S.; Purty, R.S. In silico identification and validation of miRNA and their DIR specific targets in Oryza sativa Indica under abiotic stress. Non-Coding RNA Res. 2020, 5, 167–177. [Google Scholar] [CrossRef]
- Li, L.; Sun, W.; Zhou, P.; Wei, H.; Wang, P.; Li, H.; Rehman, S.; Li, D.; Zhuge, Q. Genome-wide characterization of dirigent proteins in Populus: Gene expression variation and expression pattern in response to Marssonina brunnea and phytohormones. Forests 2021, 12, 507. [Google Scholar] [CrossRef]
- Khan, A.; Li, R.-J.; Sun, J.-T.; Ma, F.; Zhang, H.-X.; Jin, J.-H.; Ali, M.; Wang, J.-E.; Gong, Z.-H. Genome-wide analysis of dirigent gene family in pepper (Capsicum annuum L.) and characterization of CaDIR7 in biotic and abiotic stresses. Sci. Rep. 2018, 8, 5500. [Google Scholar] [CrossRef]
- Shi, Y.; Shen, Y.; Ahmad, B.; Yao, L.; He, T.; Fan, J.; Liu, Y.; Chen, Q.; Wen, Z. Genome-wide identification and expression analysis of dirigent gene family in strawberry (Fragaria vesca) and functional characterization of FvDIR13. Sci. Hortic. 2022, 297, 110913. [Google Scholar] [CrossRef]
- Yadav, V.; Wang, Z.; Yang, X.; Wei, C.; Changqing, X.; Zhang, X. Comparative analysis, characterization and evolutionary study of dirigent gene family in cucurbitaceae and expression of novel dirigent peptide against powdery mildew stress. Genes 2021, 12, 326. [Google Scholar] [CrossRef] [PubMed]
- Jin-Long, G.; Li-Ping, X.; Jing-Ping, F.; Ya-Chun, S.; Hua-Ying, F.; You-Xiong, Q.; Jing-Sheng, X. A novel dirigent protein gene with highly stem-specific expression from sugarcane, response to drought, salt and oxidative stresses. Plant Cell Rep. 2012, 31, 1801–1812. [Google Scholar] [CrossRef] [PubMed]
- Andrade, L.M.; Peixoto-Junior, R.F.; Ribeiro, R.V.; Nóbile, P.M.; Brito, M.S.; Marchiori, P.E.R.; Carlin, S.D.; Martins, A.P.B.; Goldman, M.H.S.; Llerena, J.P.P. Biomass accumulation and cell wall structure of rice plants overexpressing a dirigent-jacalin of sugarcane (ShDJ) under varying conditions of water availability. Front. Plant Sci. 2019, 10, 65. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Qin, Z.; Zhou, X.; Xin, M.; Wang, C.; Liu, D.; Li, S. Expression and functional analysis of the Propamocarb-related gene CsDIR16 in cucumbers. BMC Plant Biol. 2018, 18, 16. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cao, Y.; Liang, X.; Zhuang, J.; Wang, X.; Qin, F.; Jiang, C. A dirigent family protein confers variation of Casparian strip thickness and salt tolerance in maize. Nat. Commun. 2022, 13, 2222. [Google Scholar] [CrossRef]
- Burlat, V.; Kwon, M.; Davin, L.B.; Lewis, N.G. Dirigent proteins and dirigent sites in lignifying tissues. Phytochemistry 2001, 57, 883–897. [Google Scholar] [CrossRef]
- Li, N.; Zhao, M.; Liu, T.; Dong, L.; Cheng, Q.; Wu, J.; Wang, L.; Chen, X.; Zhang, C.; Lu, W. A novel soybean dirigent gene GmDIR22 contributes to promotion of lignan biosynthesis and enhances resistance to Phytophthora sojae. Front. Plant Sci. 2017, 8, 1185. [Google Scholar] [CrossRef]
- Seneviratne, H.K.; Dalisay, D.S.; Kim, K.-W.; Moinuddin, S.G.; Yang, H.; Hartshorn, C.M.; Davin, L.B.; Lewis, N.G. Non-host disease resistance response in pea (Pisum sativum) pods: Biochemical function of DRR206 and phytoalexin pathway localization. Phytochemistry 2015, 113, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Reboledo, G.; Del Campo, R.; Alvarez, A.; Montesano, M.; Mara, H.; Ponce de León, I. Physcomitrella patens activates defense responses against the pathogen Colletotrichum gloeosporioides. Int. J. Mol. Sci. 2015, 16, 22280–22298. [Google Scholar] [CrossRef]
- Borges, A.F.; Ferreira, R.B.; Monteiro, S. Transcriptomic changes following the compatible interaction Vitis vinifera-Erysiphe necator. Paving the way towards an enantioselective role in plant defence modulation. Plant Physiol. Biochem. 2013, 68, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Rotino, G.L.; Sala, T.; Toppino, L. Eggplant. In Alien Gene Transfer in Crop Plants, Volume 2; Springer: New York, NY, USA, 2014; pp. 381–409. [Google Scholar]
- Chapman, M.A. Introduction: The importance of eggplant. In The Eggplant Genome; Springer: Cham, Switzerland, 2019; pp. 1–10. [Google Scholar]
- Hirakawa, H.; Shirasawa, K.; Miyatake, K.; Nunome, T.; Negoro, S.; Ohyama, A.; Yamaguchi, H.; Sato, S.; Isobe, S.; Tabata, S. Draft genome sequence of eggplant (Solanum melongena L.): The representative solanum species indigenous to the old world. DNA Res. 2014, 21, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Barchi, L.; Pietrella, M.; Venturini, L.; Minio, A.; Toppino, L.; Acquadro, A.; Andolfo, G.; Aprea, G.; Avanzato, C.; Bassolino, L. A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution. Sci. Rep. 2019, 9, 11769. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Wang, J.; Wang, W.; Hu, T.; Hu, H.; Bao, C. A high-quality chromosome-level genome assembly reveals genetics for important traits in eggplant. Hortic. Res. 2020, 7, 153. [Google Scholar] [CrossRef]
- Li, D.; Qian, J.; Li, W.; Yu, N.; Gan, G.; Jiang, Y.; Li, W.; Liang, X.; Chen, R.; Mo, Y. A high-quality genome assembly of the eggplant provides insights into the molecular basis of disease resistance and chlorogenic acid synthesis. Mol. Ecol. Resour. 2021, 21, 1274–1286. [Google Scholar] [CrossRef]
- Barchi, L.; Rabanus-Wallace, M.T.; Prohens, J.; Toppino, L.; Padmarasu, S.; Portis, E.; Rotino, G.L.; Stein, N.; Lanteri, S.; Giuliano, G. Improved genome assembly and pan-genome provide key insights into eggplant domestication and breeding. Plant J. 2021, 107, 579–596. [Google Scholar] [CrossRef]
- Wang, S.; Chen, Z.; Ji, T.; Di, Q.; Li, L.; Wang, X.; Wei, M.; Shi, Q.; Li, Y.; Gong, B. Genome-wide identification and characterization of the R2R3MYB transcription factor superfamily in eggplant (Solanum melongena L.). Agri Gene 2016, 2, 38–52. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, J.; Zhou, X.; Liu, S.; Zhuang, Y. Identification of WRKY gene family and characterization of cold stress-responsive WRKY genes in eggplant. PeerJ 2020, 8, e8777. [Google Scholar] [CrossRef]
- Wan, F.-x.; Gao, J.; Wang, G.-l.; Niu, Y.; Wang, L.-z.; Zhang, X.-g.; Wang, Y.-q.; Pan, Y. Genome-wide identification of NAC transcription factor family and expression analysis of ATAF subfamily members under abiotic stress in eggplant. Sci. Hortic. 2021, 289, 110424. [Google Scholar] [CrossRef]
- Chen, J.; Wang, S.; Wu, F.; Wei, M.; Li, J.; Yang, F. Genome-wide identification and functional characterization of auxin response factor (ARF) genes in eggplant. Int. J. Mol. Sci. 2022, 23, 6219. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Yu, C.S.; Lin, C.J.; Hwang, J.K. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004, 13, 1402–1406. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Lv, L.L.; Feng, X.F.; Li, W.; Li, K. High temperature reduces peel color in eggplant (Solanum melongena) as revealed by RNA-seq analysis. Genome 2019, 62, 503–512. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, J.; Zhou, X.; Liu, S.; Zhuang, Y. Transcriptomics analysis unravels the response to low temperature in sensitive and tolerant eggplants. Sci. Hortic. 2020, 271, 109468. [Google Scholar] [CrossRef]
- Li, J.; Gao, Z.; Zhou, L.; Li, L.; Zhang, J.; Liu, Y.; Chen, H. Comparative transcriptome analysis reveals K+ transporter gene contributing to salt tolerance in eggplant. BMC Plant Biol. 2019, 19, 67. [Google Scholar] [CrossRef]
- Peng, J.; Wang, P.; Fang, H.; Zheng, J.; Zhong, C.; Yang, Y.; Yu, W. Weighted gene co-expression analysis network-based analysis on the candidate pathways and hub genes in eggplant bacterial wilt-resistance: A plant research study. Int. J. Mol. Sci. 2021, 22, 13279. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Y.; Cheng, Y.; Chen, X. Transcriptome analysis reveals multiple signal network contributing to the Verticillium wilt resistance in eggplant. Sci. Hortic. 2019, 256, 108576. [Google Scholar] [CrossRef]
- Chen, L.-m.; Li, X.-w.; He, T.-j.; Li, P.-j.; Liu, Y.; Zhou, S.-x.; Wu, Q.-c.; Chen, T.-t.; Lu, Y.-b.; Hou, Y.-m. Comparative biochemical and transcriptome analyses in tomato and eggplant reveal their differential responses to Tuta absoluta infestation. Genomics 2021, 113, 2108–2121. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Shang, L.; Zhu, Q.-H.; Fan, L.; Guo, L. Twenty years of plant genome sequencing: Achievements and challenges. Trends Plant Sci. 2021, 27, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Davin, L.B.; Lewis, N.G. Dirigent proteins and dirigent sites explain the mystery of specificity of radical precursor coupling in lignan and lignin biosynthesis. Plant Physiol. 2000, 123, 453–462. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, N.; Song, W.; Yin, G.; Qin, Y.; Yan, Y.; Hu, Y. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef]
- Alvarez, A.; Montesano, M.; Schmelz, E.; Ponce de León, I. Activation of shikimate, phenylpropanoid, oxylipins, and auxin pathways in Pectobacterium carotovorum elicitors-treated moss. Front. Plant Sci. 2016, 7, 328. [Google Scholar] [CrossRef]
- Weidenbach, D.; Esch, L.; Möller, C.; Hensel, G.; Kumlehn, J.; Höfle, C.; Hückelhoven, R.; Schaffrath, U. Polarized defense against fungal pathogens is mediated by the Jacalin-related lectin domain of modular Poaceae-specific proteins. Mol. Plant 2016, 9, 514–527. [Google Scholar] [CrossRef]


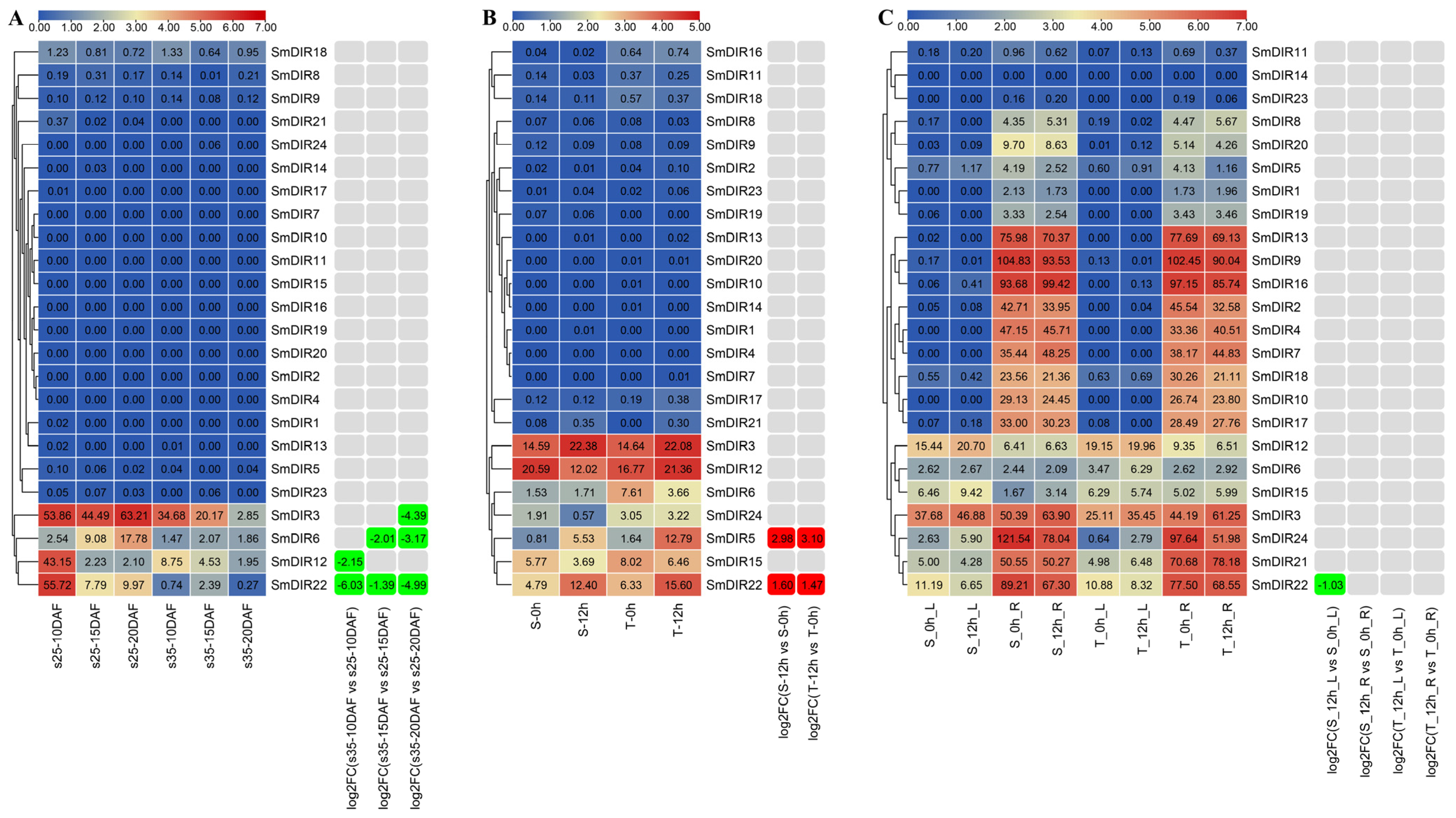
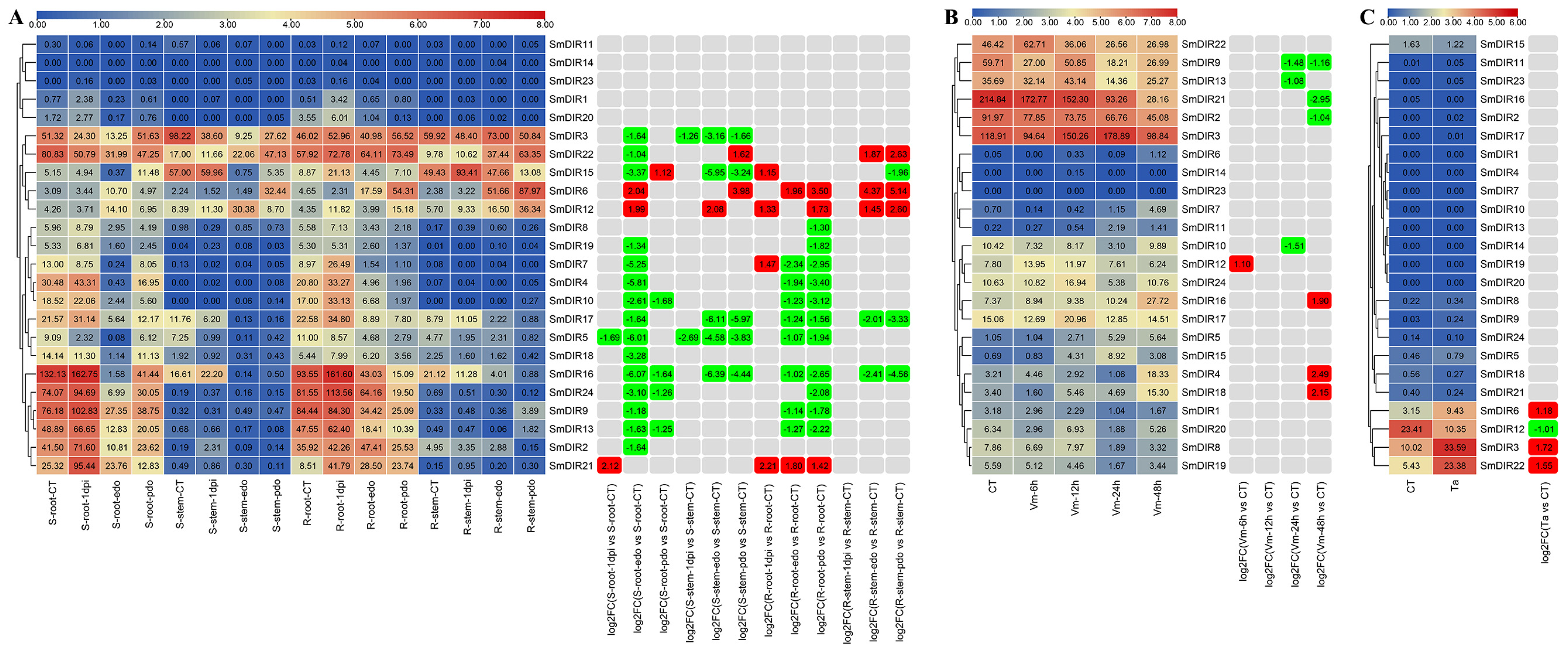
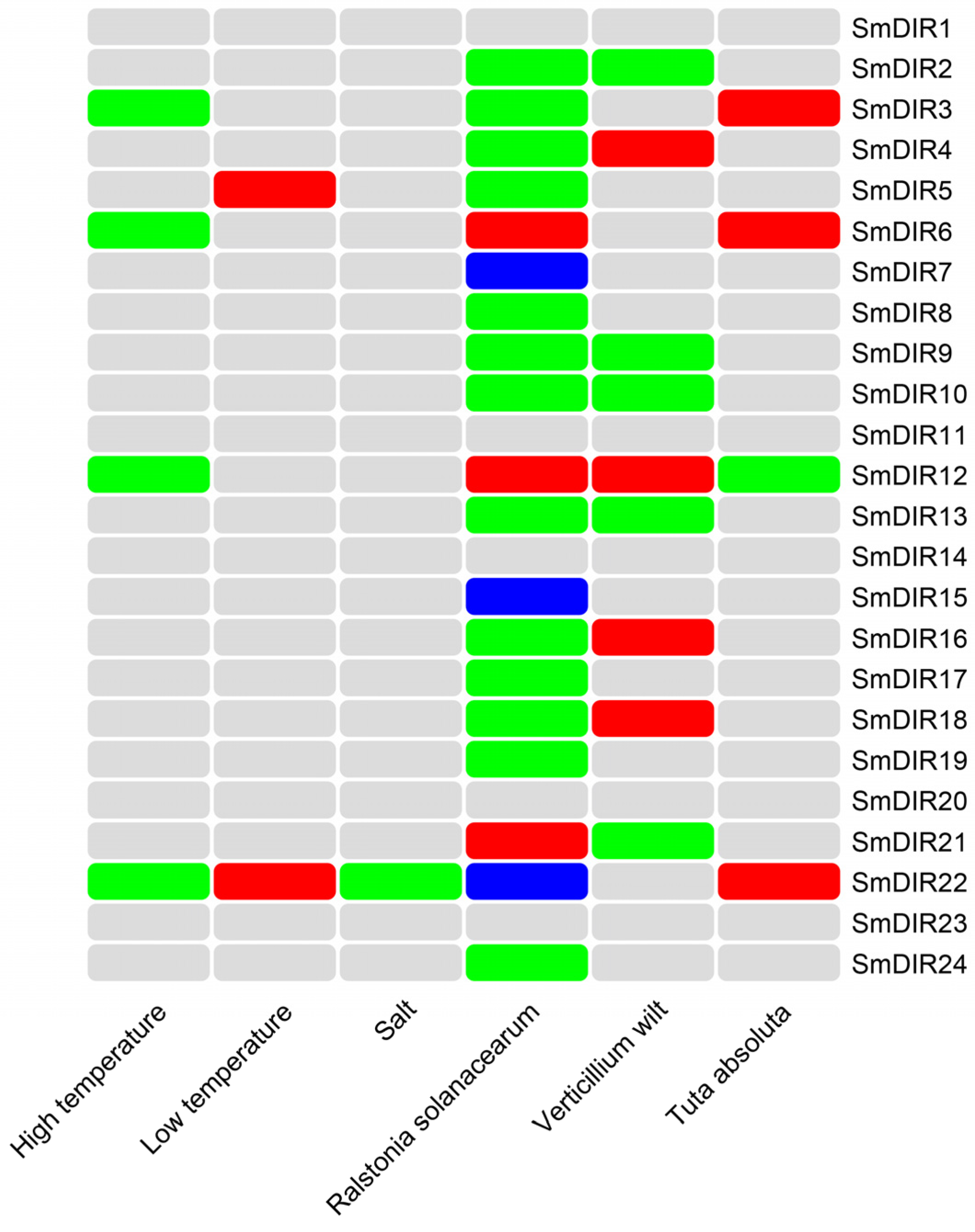
| Gene Name | Gene ID | Number of Amino Acid (aa) | Molecular Weight (kDa) | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Subcellular Location |
|---|---|---|---|---|---|---|---|---|
| SmDIR1 | SMEL4.1_01g003540.1 | 156 | 16.97 | 9.36 | 45.55 | 76.92 | −0.108 | Mitochondrion |
| SmDIR2 | SMEL4.1_01g003550.1 | 343 | 36.23 | 6.13 | 11.54 | 95.54 | 0.171 | Plasma Membrane |
| SmDIR3 | SMEL4.1_01g031140.1 | 191 | 21.33 | 9.57 | 28.14 | 79.69 | −0.017 | Plasma Membrane |
| SmDIR4 | SMEL4.1_02g003080.1 | 190 | 21.06 | 9.67 | 24.42 | 94.95 | 0.065 | Mitochondrion |
| SmDIR5 | SMEL4.1_02g009260.1 | 186 | 20.69 | 7.06 | 36.88 | 91.24 | 0.196 | Plasma Membrane |
| SmDIR6 | SMEL4.1_02g018810.1 | 194 | 21.19 | 9.08 | 28.43 | 93.87 | 0.111 | Extracellular Space |
| SmDIR7 | SMEL4.1_04g004510.1 | 190 | 21.37 | 9.42 | 29.75 | 91.79 | 0.022 | Plasma Membrane |
| SmDIR8 | SMEL4.1_05g003580.1 | 244 | 27.03 | 5.90 | 43.08 | 79.88 | −0.167 | Plasma Membrane |
| SmDIR9 | SMEL4.1_05g003590.1 | 438 | 44.86 | 4.34 | 40.80 | 87.99 | 0.075 | Extracellular Space |
| SmDIR10 | SMEL4.1_05g019870.1 | 249 | 26.06 | 5.46 | 36.58 | 85.42 | 0.115 | Plasma Membrane |
| SmDIR11 | SMEL4.1_06g013950.1 | 146 | 16.14 | 8.82 | 27.09 | 91.51 | 0.135 | Plasma Membrane |
| SmDIR12 | SMEL4.1_06g020190.1 | 212 | 23.63 | 9.73 | 29.47 | 89.62 | 0.094 | Plasma Membrane |
| SmDIR13 | SMEL4.1_06g027190.1 | 372 | 38.41 | 4.75 | 26.34 | 96.02 | 0.230 | Plasma Membrane |
| SmDIR14 | SMEL4.1_07g016780.1 | 177 | 19.52 | 5.22 | 27.96 | 95.31 | 0.124 | Plasma Membrane |
| SmDIR15 | SMEL4.1_08g026590.1 | 190 | 21.28 | 7.79 | 13.38 | 88.84 | 0.088 | Plasma Membrane |
| SmDIR16 | SMEL4.1_08g026600.1 | 167 | 19.13 | 6.02 | 26.49 | 66.53 | −0.298 | Plasma Membrane |
| SmDIR17 | SMEL4.1_09g022870.1 | 546 | 59.69 | 8.52 | 38.73 | 97.33 | 0.354 | Plasma Membrane |
| SmDIR18 | SMEL4.1_11g000960.1 | 168 | 18.76 | 6.90 | 15.68 | 99.76 | 0.123 | Plasma Membrane |
| SmDIR19 | SMEL4.1_11g019830.1 | 178 | 19.93 | 9.85 | 35.35 | 100.34 | 0.069 | Plasma Membrane |
| SmDIR20 | SMEL4.1_11g026460.1 | 369 | 38.79 | 4.63 | 42.99 | 84.80 | −0.063 | Extracellular Space |
| SmDIR21 | SMEL4.1_12g003420.1 | 184 | 20.19 | 9.52 | 26.98 | 90.11 | 0.123 | Mitochondrion |
| SmDIR22 | SMEL4.1_12g003430.1 | 194 | 20.87 | 9.24 | 22.53 | 101.55 | 0.198 | Extracellular Space |
| SmDIR23 | SMEL4.1_12g003440.1 | 193 | 21.21 | 6.96 | 23.74 | 93.37 | 0.130 | Extracellular Space |
| SmDIR24 | SMEL4.1_12g003620.1 | 194 | 21.36 | 7.87 | 19.27 | 79.85 | 0.055 | Extracellular Space |
| Motif | Sequence | Number of Amino Acid | Pfam Annotation |
|---|---|---|---|
| motif 1 | FGTLTVIDDPLTIGPEPNSKJIGRAQGIYGSASQNGVSLLMALNFVFTGGKY | 52 | Dirigent |
| motif 2 | YREMAIVGGTGKFRLARGYATAKTYWFB | 28 | Dirigent |
| motif 3 | HAKEKVTKLHFYFHDILSGKNPTAIQIAQ | 29 | - |
| motif 4 | TGDAIVEYNVVVLHY | 15 | - |
| motif 5 | GSTLSILGRNPVFHE | 15 | - |
| motif 6 | MPTAQGIDLGPKAVEKWFKKL | 21 | - |
| motif 7 | MAIIQNSNSFQLKVIISYLLILALTITFATAGRILDEEVATPTVPPNNPDPTDQPPVSGATAAGAAAGAT | 70 | - |
| motif 8 | GQHTTDGVETILHITVYJTY | 20 | - |
| motif 9 | DSLSFFGVHR | 10 | - |
| motif 10 | IYSGQIPFATPLGFQPPEDGVAIPNANGAMPTFNINGVPLGTGLAGTIFAGGNN | 54 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, K.; Xing, W.; Sheng, S.; Yang, D.; Zhen, F.; Jiang, H.; Yan, C.; Jia, L. Genome-Wide Identification and Expression Analysis of Eggplant DIR Gene Family in Response to Biotic and Abiotic Stresses. Horticulturae 2022, 8, 732. https://doi.org/10.3390/horticulturae8080732
Zhang K, Xing W, Sheng S, Yang D, Zhen F, Jiang H, Yan C, Jia L. Genome-Wide Identification and Expression Analysis of Eggplant DIR Gene Family in Response to Biotic and Abiotic Stresses. Horticulturae. 2022; 8(8):732. https://doi.org/10.3390/horticulturae8080732
Chicago/Turabian StyleZhang, Kaijing, Wujun Xing, Suao Sheng, Dekun Yang, Fengxian Zhen, Haikun Jiang, Congsheng Yan, and Li Jia. 2022. "Genome-Wide Identification and Expression Analysis of Eggplant DIR Gene Family in Response to Biotic and Abiotic Stresses" Horticulturae 8, no. 8: 732. https://doi.org/10.3390/horticulturae8080732
APA StyleZhang, K., Xing, W., Sheng, S., Yang, D., Zhen, F., Jiang, H., Yan, C., & Jia, L. (2022). Genome-Wide Identification and Expression Analysis of Eggplant DIR Gene Family in Response to Biotic and Abiotic Stresses. Horticulturae, 8(8), 732. https://doi.org/10.3390/horticulturae8080732







