Abstract
SIMILAR TO RCD-ONE (SRO) is a plant-specific small protein family that controls many biological processes including physiological development and stress responses. The SRO gene family has been studied in several plant species, but no detailed characterization and expression profiles of this important gene family were performed in cucumber. In this study, we characterize the SRO genes in cucumber, and determined their transcript levels in various tissues and under exposure to diverse biotic and abiotic stressors. Four SRO genes (named as CsSRO1–CsSRO4) were identified and isolated, which were distributed on three different chromosomes. Gene duplication analysis showed that only one pair of segmental duplication event was identified, but no tandem duplication events were detected. All CsSROs consist of the PARP domain and a C-terminal RST domain, while the N-terminal WWE domain was only present in CsSRO2 and CsSRO4. SROs from 15 plant species are divided into two groups (I and II), and group I can be further divided into four subgroups (Ia to Id) according to the phylogenetic tree. The conserved motif and gene structure analyses showed that SROs within the same branch of the phylogenetic tree have analogous conserved motifs configuration and gene structures. However, SRO genes possessed variable numbers of introns in different subgroups, which may affect the evolution of new family members. RNA-Seq data and qRT-PCR results showed that the four CsSRO genes have distinct expression pattern in various tissues and under diverse stresses, suggesting their multiple functions in plant growth and stress responses. The findings provide a basis for further research aiming at functional characterization of the regulatory mechanism to reveal the roles of CsSRO genes in developmental and stress-related processes of cucumber.
1. Introduction
Plants have produced a series of regulatory networks through the transcriptional control of numerous stress-responsive genes to protect themselves from adverse environmental effects [1,2]. Transcription factors (TFs) are a class of proteins that can bind to the cis-regulatory elements in promoters of target genes and thus to activate or inhibit their expression, thereby regulating plant development and the response to various stresses [3,4]. SIMILAR TO RCD-ONE (SRO) is a unique type of small TF proteins in plants, which can act as key regulators involved in regulation of many biological pathways. Reports have found that SRO proteins consist of a conserved poly (ADP-ribose) polymerase catalytic center (PARP domain, PF00644) and a C-terminal RCD-SRO-TAF4 domain (RST domain, PF12174) that interacts with other TFs. Some SROs also contain a WWE domain (PF02825) at the N-terminus, which takes part in ADP ribosylation (ADPRylation) and ubiquitination of proteins [5,6,7].
The first identified SRO family member, RADICAL-INDUCED CELL DEATH1 (RCD1) in Arabidopsis, was able to complement the yeast yap1(-) mutant which has defects in oxidative stress sensitivity, suggesting that it might participate in oxidative stress [8]. AtRCD1 also has a role in salt tolerance through physically interacting with the plasma membrane localization Na+/H+ antiporter SALT OVERLY SENSITIVE 1 (SOS1) [9]. The rcd1 mutants exhibited pleiotropic phenotypes such as the aberrant leaf, root and rosette development, early flowering, and altered resistance to diverse abiotic stresses and hormones response [10,11,12,13,14,15]. Subsequently, five homologs (named as AtSRO1–5) of AtRCD1 were also identified in Arabidopsis. AtSRO1 is the closest homolog of AtRCD1, both of them contained WWE domain at the N-terminus and possessed similar expression patterns, but they play unequally redundant and independent roles in certain developmental processes and the response to various stresses [14,16,17,18]. For example, sro1-1 mutants displayed only unconspicuous morphological phenotypes, while rcd1-3 sro1-1 double mutants showed even more extreme phenotypes than rcd1-3 [13,14]. Additionally, rcd1 and sro1 mutants also showed different responses to oxidative stress [13], but they respond similarly to HgCl2 and H2O2 stress [19]. Another three SRO family members, AtSRO2, AtSRO3, and AtSRO5, displayed changed transcriptional levels under strong light, salt, HgCl2, H2O2, and ozone stress conditions. In addition, overexpression of AtSRO5 conferred salt tolerance of transgenic plants [18,19,20].
Besides Arabidopsis, SRO genes have also been extensively identified and functionally studied in other plant species. In rice, OsSRO1c functions as a core regulator to mediate abiotic stress responses via direct interaction with various TFs, such as SNAC1 (stress-responsive NAC 1) and DST (drought and salt tolerance) [21,22]. As an allele of OsSRO1c from the indica rice cultivar (Oryza sativa ssp. indica), BROWNING OF CALLUS1 (BOC1) can improve genetic transformation efficiency probably through inhibiting programmed cell death caused by oxidative stress, and thus reducing callus browning [23]. Another SRO member in rice, OsSRO1a, negatively regulates the rice-Xanthomonas oryzae interaction via the OsMYC2-mediated jasmonic acid (JA) signaling pathway [24]. A bread wheat (Triticum aestivum) SRO gene TaSRO1 showed promoted plant growth and multiple abiotic stress tolerance by controlling redox homoeostasis in transgenic wheat and Arabidopsis lines [25]. Overexpression of maize ZmSRO1b in Arabidopsis also elevated the tolerance of plants to salt, cadmium, and oxidative stress [26]. In contrast, ZmSRO1e-overexpressing maize and Arabidopsis plants exhibited increased sensitivity to oxidative and salt stress with reduced anthocyanin accumulation through disrupting the MYB–bHLH–WD40 (MBW) complex [27]. These findings demonstrated that SRO genes play vital roles in plant development, and response to multiple biotic and abiotic stresses.
Cucumber is an important vegetable crop with great economic value in the world, but it always encounters a variety of environmental stimulus (such as drought, salt, and pathogens) during its growth cycle [4,28]. In recent years, many stress-responsive TF gene families have been identified in cucumber, such as GARS [29,30], bHLH [31], WRKY [32], VQ [33], and TIFY [34], and some members of them were proven to function in stress responses [35,36]. However, no report has been carried out on the SRO family genes in cucumber until now. To elucidate whether the cucumber SRO family genes participate in stress response, we cloned and characterized four cucumber SRO genes, and the phylogenetic relationship, conserved motif, gene structure, and promoter region were analyzed by some routine bioinformatics tools. Additionally, the transcriptional levels of cucumber SRO genes under salt and drought stress conditions, as well as under the infection of Pseudoperonospora cubensis and Meloidogyne incognita, were also determined by qRT-PCR and RNA-seq analyses. Our findings provide a solid base for the classification and expression of the SRO family genes and help to facilitate further functional characterization of these genes in cucumber.
2. Materials and Methods
2.1. Plant Materials, Growth Conditions and Stress Treatments
The Chinese Long cucumber inbred line 9930 plants were employed for stress treatments in this work. The seeds were soaked in water at 55 °C for 15 min, then placed on damp filter paper overnight in an artificial climate incubator at 28 °C to promote germination. After the cotyledons were fully opened, the seedlings were moved to Hoagland’s nutrition for cultivation. Cucumber seedlings were soaked in Hoagland nutrient solution for hydroponics with a 16:8 h light: dark photoperiod. We transferred the two-leaf stage seedlings to a nutrition solution containing NaCl (200 mM) and PEG6000 (10%), for salt stress and drought stress, respectively, as described previously [4]. The leaf tissues were harvested for each stress condition with three biological replicates at 0, 3, 6, and 12 h after treatment. Subsequently, these samples were stored in a −80 °C freezer for RNA extraction until further experiments.
2.2. Identification and Cloning of CsSRO Genes of Cucumber
The Hidden Markov Model (HMM) profiles of the RST domain (PF12174) and PARP domain (PF00644) were downloaded from the Pfam database (http://pfam.xfam.org/, accessed on 19 June 2022) [37], and these HMM profiles were employed to search against the cucumber protein database downloaded from Cucurbit Genomics Database (CuGenDB) (http://cucurbitgenomics.org/, accessed on 19 June 2022) using HMMER software. We also searched the cucumber SRO family members using the BLASTP with the published Arabidopsis and rice SRO protein sequences as query sequences. After removing the redundant sequences obtained from the above methods and checking through Pfam and InterProScan (http://www.ebi.ac.uk/Tools/InterProScan/, accessed on 19 June 2022), we get four candidate SRO protein sequences, and their coding sequences (CDS) and corresponding genomic DNA (gDNA) sequences were also obtained from CuGenDB.
Total RNA was isolated from the leaf samples of normally growing and stress-treated seedlings using RNA-easyTM Isolation Reagent Vazyme cat (Vazyme Biology Co., Ltd., Nanjing, China), and the first strand cDNAs were synthesized using the HiScript®Ⅱ Q RT SuperMix for qPCR (+gDNA wipe) (Vazyme Biology Co., Ltd., Nanjing, China) on the basis of to the manufacturer’s instructions. The cDNA samples were diluted 10 times, and used as the template for PCR amplification. The PCR amplification was carried out at: denaturation at 95 °C for 3 min; 95 °C for 1 min, 60 °C for 15 s, and 72 °C for 3 min (35 cycles); followed by 72 °C for 5 min. The PCR products were detected by 1.0% agarose gel electrophoresis and subsequently sequencing. Primer sequences used for PCR amplification of each CsSRO gene are listed in Table S1.
2.3. In Silico Sequence Analyses of Cucumber SRO Family Members
The physicochemical characteristics of cucumber SRO proteins, including molecular weight (MW), isoelectric point (pI), and grand average of hydropathicity (GRAVY), were calculated using the ProtParam tool on the ExPASy server (http://web.expasy.org/protparam, accessed on 19 June 2022) [38]. The subcellular location of cucumber SRO proteins was predicted using CELLO v.2.5 (http://cello.life.nctu.edu.tw/, accessed on 19 June 2022) [39]. The conserved domains including PARP, RST, and WWE were examined through Pfam and InterProScan. The physical locations of CsSRO genes were illustrated with the MapInspect software according to the initial position information provided in CuGenDB. Moreover, these genes involved in the tandem or segmental duplications were analyzed with MCScanX on the basis of the criterion in our previous report [4]. The 2000 bp promoter sequences located upstream of the initiation codon (ATG) of the cucumber SRO genes were retrieved from CuGenDB, and various cis-regulatory elements related to stresses and hormones were determined using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 19 June 2022) [40].
2.4. Phylogenetic, Conserved Motif and Gene Structure Analyses of SRO Family Members of Cucumber and Other Plant Species
The full-length protein sequence alignments of all SRO family members from cucumber and different plant species were performed using the MAFFT tool under default settings, and then MEGA version 7.0 was used to construct phylogenetic trees by the Neighbor-Joining (NJ) methods and the bootstrap analysis conducted with 1000 replications [41]. MEME (http://meme-suite.org/tools/meme, accessed on 19 June 2022) was employed to examine the conserved motifs of SRO proteins of cucumber and other plant species with the default settings except the number of diverse motifs was chosen as 10 [42]. The gene structures of the SRO genes of cucumber, Arabidopsis, maize, rice, and grape were determined with the GSDS 2.0 tool (Gene Structure Display Server, http://gsds.gao-lab.org/, accessed on 19 June 2022) according to their CDS and the correspondent full-length gDNA sequences [43].
2.5. qRT-PCR Analysis of CsSRO Genes in Response to Abiotic Stress
The quantitative RT-PCR was conducted using the Roche Lightcyler 480II PCR System based on the manufacturer’s instructions of the ChamQ Universal SYBR qPCR Master Mix Kit (Vazyme Biology Co., Ltd., Nanjing, China). The total qRT-PCR mixture was 20 μL including 10 μL 2 × ChamQ Universal SYBR qPCR Master Mix, 0.4 μL forward primer, 0.4 μL reverse primer, 7.2 μL ddH2O, and 2 μL template cDNA. The reaction conditions were as follows: 95 °C for 30 s, followed by 40 cycles of 95 °C for 10 s, and 60 °C for 30 s. All reactions were performed in triplicate, and CsAct3 was used as the reference gene to determine the relative expression with the 2−ΔΔCt method [4]. qRT-PCR-specific primers used for expression analysis of each CsSRO gene were listed in Table S1.
2.6. Expression Analysis of CsSRO Genes Based on RNA-seq
The expression data of CsSRO genes in various tissues, including root, leaf, stem, female and male flowers, ovaries, tendril, and the base of tendril, were retrieved from the RNA-seq in NCBI under the accession number of PRJNA80169. The expression data of CsSRO genes in different biotic stresses, including the infection of P. cubensis (downy mildew treatment) and M. incognita (root-knot nematode treatment), were available in the NCBI under the accession numbers of PRJNA388584 and PRJNA419665, respectively. Based on all expression values, TBtools was used to generate expression heatmaps using the log2 transformed transcripts per million reads (TPM) values, based on the methods in our previous studies [4,44].
3. Results
3.1. Cloning and Identification of SRO Gene Sequences of Cucumber
A total of four SRO genes were identified in cucumber, and they were numbered from CsSRO1 to CsSRO4, on the basis of their chromosomal locations (Table 1). To verify the accuracy of sequences acquired from the cucumber genome, we designed the specific primers based on the CDS sequences of CsSRO genes. The 1167 bp, 1398 bp, 1806 bp, and 1863 bp CDSs of the CsSRO genes were amplified, by using cDNA from cucumber leaf tissues as a template (Figure S1). Sequencing of PCR products confirmed the coding sequences of CsSRO genes, suggesting the reliability of the sequence information of the above identification (Table 1). The physicochemical characteristics including sequence length, MW, pI, and GRAVY values of CsSROs are provided in Table 1.

Table 1.
Identification and characterization of SRO family genes in cucumber.
The basic characteristic analysis showed that the gDNA and CDS lengths of four CsSRO genes varied in size, with gDNA lengths varied from 2586 bp (CsSRO1) to 9767 bp (CsSRO4) and CDS lengths ranged from 1167 bp (CsSRO1) to 1863 bp (CsSRO4) (Table 1). Their gene products had molecular weight (MW) and isoelectric points (pI) with MW ranging from 43,102.56 Da (CsSRO1) to 70,266.98 Da (CsSRO4) and pI ranging from 5.79 (CsSRO3) to 9.33 (CsSRO2). The GRAVY values of CsSROs ranged from −0.511 (CsSRO1) to −0.324 (CsSRO2), which were less than zero, suggesting that the CsSRO proteins were hydrophilic proteins. The subcellular localization prediction results showed that CsSRO1, CsSRO3, and CsSRO4 were predicted to be localized in the nucleus, while CsSRO2 might be located in mitochondria and nucleus (Table 1).
3.2. Phylogenetic Analysis of SRO Family Genes from Cucumber and Different Plant Species
To investigate the phylogenetic relationship and divergence of the SRO family in cucumber, a phylogenetic tree was created on the basis of multiple sequence alignment of protein sequences of SRO gene family from cucumber and other 14 different plant species. As shown in the phylogenetic tree, these SRO proteins were irregularly divided into two groups, namely group I and group II, and group I can be further divided into four subgroups (Ia to Id) (Figure 1). Consistent with previous reports [45,46], the SRO proteins of these species were clustered in dicot- or monocot-specific patterns. For example, group II had only dicotyledonous SRO proteins, while group I contained both monocotyledonous and dicotyledonous SRO proteins. For CsSRO proteins, CsSRO3 and CsSRO4 were clustered in group Ic, while CsSRO3 and CsSRO4 were clustered in group Id and group II, respectively (Figure 1).
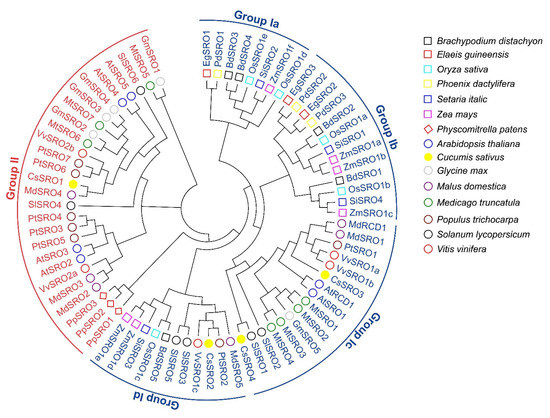
Figure 1.
Phylogenetic relationships of SRO family genes from cucumber and 14 other plant species. The phylogenetic tree was created with MEGA 7.0 using the Neighbor-Joining (NJ) method with bootstrap analysis performed using 1000 replicates. Two groups, namely group I and group II, are colored with blue and red, respectively, and the colored circles, rhombuses and squares represent different plant species.
3.3. Conserved Motifs of SRO Proteins from Cucumber and Other Plant Species
Domain analysis by Pfam and InterProScan tools showed that all CsSROs consist of the PARP domain and a C-terminal RST domain, while the N-terminal WWE domain was only present in CsSRO2 and CsSRO4 (Figure S2). To further investigate the diversification of SRO proteins, the conserved motif analysis of SRO proteins from cucumber and other plant species was performed based on the results of MEME, and a phylogenetic tree was also created. The results showed that 10 conversed motifs were predicted and their details are shown, and the SROs in the same group had similar motif composition (Figure 2). Motifs 9, 4, 8, 2, and 1 made up the PARP domain, which were widely distributed in a large proportion of SRO proteins, with the exception of several SROs, such as OsSRO1d and VvSRO1c (lack of motif 2), ZmSRO1d, ZmSRO1e, and AtSRO3 (lack of motif 9). Motifs 7 and 5 were annotated as the RST domain, which were present in most SRO proteins, except for CsSRO2, OsSRO1d, VvSRO1c, ZmSRO1d, and ZmSRO1e. In addition, motif 3 and motif 6 were present in the N-terminal region of all SRO proteins of group I, while they were absent in group II SRO proteins. Motif 10 was specific to SRO proteins of group Ia and Ib, as well as present in most members of group II, with the exception of AtSRO4 and AtSRO5 (Figure 2).
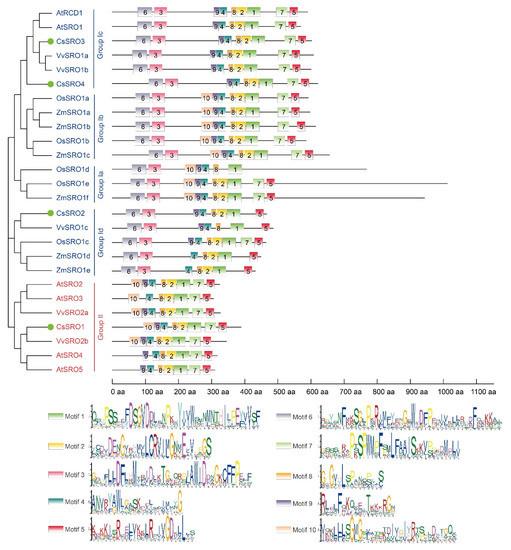
Figure 2.
Conserved motif configurations of the SRO family members from cucumber, Arabidopsis, maize, rice, and grape on the basis of phylogenetic relationship. The clustering was shown according to the results of phylogenetic analysis. The colorful boxes delineate different motifs (numbered 1–10) and their details were shown at the bottom of the figure.
3.4. Gene Structure of SRO Family Genes
A detailed SRO structure analysis was performed by comparing the CDS sequences with the corresponding gDNA sequences of SRO family genes from cucumber, Arabidopsis, maize, rice, and grape. As a result, the number of introns in these SRO genes ranged from 3 to 7, and the structural characteristics of SRO genes clustered together were similar but varied among different groups (Figure 3). Notably, SRO genes in group Ia were found to have 4 or 7 introns, group Ib SRO genes had 5 or 6 introns, group Ic SRO genes harbored 4–7 introns, group Id SRO genes contained 3 introns each, while group II SRO genes were found to have 3 or 4 introns (Figure 3). The numbers of introns in four CsSRO genes were 4, 3, 5, 7, respectively.
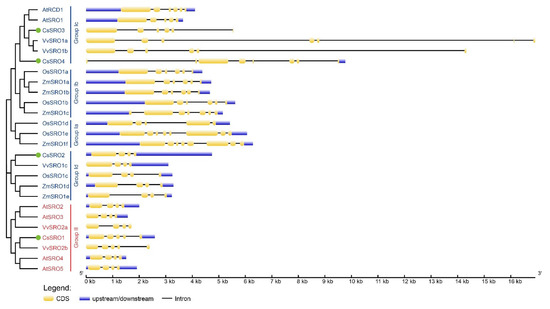
Figure 3.
The gene structures of SRO family genes from cucumber, Arabidopsis, maize, rice, and grape on the basis of phylogenetic relationship. The clustering was shown based on the results of phylogenetic analysis. The boxes colored blue and yellow indicate upstream/downstream and CDS, respectively, the black line indicates intron.
3.5. The Distribution and Gene Duplication of CsSRO Genes
Chromosomal mapping of CsSRO genes showed that these genes were located on 3 out of 7 chromosomes of the cucumber genome. Chromosome 3 had the most CsSRO genes (two), whereas chromosomes 1 and 2 each possessed only one gene (Figure 4). The analysis of the duplication event indicated that a segmental duplication event was identified (CsSRO3 and CsSRO4), while no tandem duplication event was detected (Figure 4).
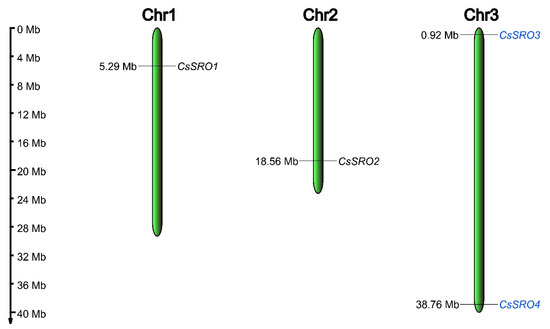
Figure 4.
Distribution of CsSRO genes on cucumber chromosomes. The segmental duplication genes are colored with blue. The chromosome numbers are labeled on the top of the bars, and the chromosome length represents the size of chromosomes. Positions are in Mb.
3.6. Bioinformatics Analysis of Putative Promoters of CsSRO Genes
The cis-regulatory elements within the promoter regions of genes would provide valuable information for their expression and possible regulatory patterns. Therefore, the PlantCARE tool was employed to analyze the stress- and hormone-related cis-regulatory elements within the promoter regions of the CsSRO genes, and 16 kinds of cis-regulatory elements were identified (Figure 5). Eight kinds of cis-regulatory elements were found to be responsive to various hormones, such as ABRE for abscisic acid (ABA), TGACG-motif and CGTCA-motif for methyl jasmonate (MeJA), TGA-element for auxin, ERE for ethylene, TCA-element for salicylic acid (SA), P-box and TATC-box for gibberellin. It is worth noting that ABRE was most abundant in the promoter sequences of CsSRO2, implying that it may play a critical role in responsive to ABA. In addition, eight kinds of cis-regulatory elements are stress-responsive, including ARE (anaerobic induction), GC-motif (anoxia), LTR (low-temperature responsive), STRE (stress-responsive), W-box (fungal elicitors), TC-rich repeats (defense and stress response), WRE3 and WUN-motif (wounding responses) (Figure 5).
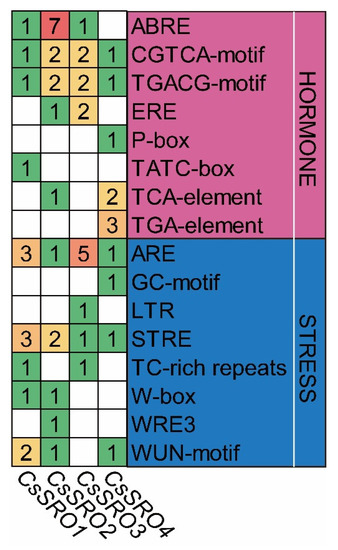
Figure 5.
Stress- and hormone-responsive cis-regulatory elements in the promoter regions of CsSRO genes. The number in box represents the copy of the cis-element of individual CsSRO genes. ABRE, cis-element involved in the abscisic acid (ABA) responsiveness; CGTCA-motif and TGACG-motif, cis-element involved in the MeJA responsiveness; ERE, ethylene-responsive element; P-box and TATC-box, cis-elements involved in the gibberellin responsiveness; TCA-element, salicylic acid (SA)-responsive element; TGA-element, auxin-responsive element; ARE, cis-acting regulatory element essential for the anaerobic induction; GC-motif, anoxia-specific inducibility element; LTR, low-temperature responsive element; STRE, stress responsive element; W-box, WRKY binding site involved in fungal elicitors; TC-rich repeats, cis-acting element involved in defense and stress responsiveness, WRE3 and WUN-motif, wound-responsive elements.
3.7. Tissue Expression Profiles Analysis of CsSRO Genes
RNA-Seq data from different tissues (root, stem, leaf, flowers, and tendrils) and three stages of developing ovaries were used to examine the tissue-specific expression profiles of CsSRO genes, and an expression heat map was generated. As a result, CsSRO genes were universally expressed in all tested tissues, and CsSRO3 and CsSRO4 exhibited relatively higher expression levels than the other two CsSRO genes (Figure 6). CsSRO1 displayed a much higher expression in root than in other tissues, and CsSRO2 showed the most abundant gene expression levels in root, unfertilized ovary, male and female flowers (Figure 6).
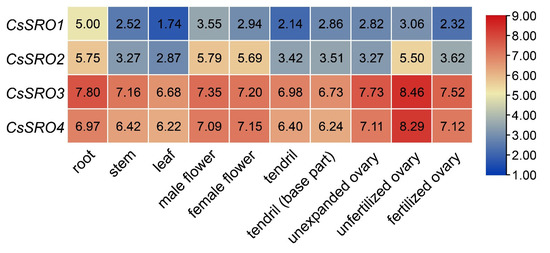
Figure 6.
Expression profiles of the CsSRO genes in different tissues of cucumber based on RNA-Seq data. Data were normalized based on the log2-transformed TPM values of each gene in all tissues analyzed, and genes highly or weakly expressed are colored red or blue, respectively.
3.8. Expression Patterns of CsSRO Genes under Salt and Drought Stress
Since the promoter analysis revealed several stress-related cis-elements, we examined the expression patterns of CsSRO genes under salt and drought stress. Under salt stress, the expression of CsSRO1, CsSRO3, and CsSRO4 was induced and peaked at 3 h, followed by an obvious decrease at 6 h and 12 h (Figure 7A). The expression level of CsSRO2 was increased rapidly and then maintained at 6 h, and then dropped at 12 h. Under drought treatment, the CsSRO genes exhibited similar expression profiles as under salt treatment (Figure 7B). These results indicated that CsSRO genes may play a role in response to various abiotic stresses.
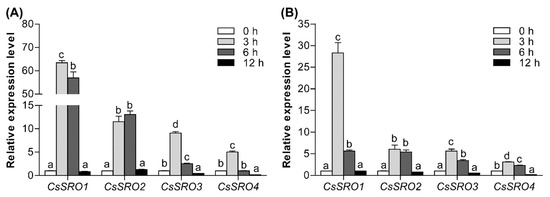
Figure 7.
qRT-PCR analysis of CsSRO genes expression patterns under salt (A) and drought (B) stress. Cucumber seedlings were treated with salt (200 mM NaCl) and drought (10% PEG6000) conditions at different time points (0, 3, 6, and 12 h), and leaf samples were collected from three biological replicates of each treatment. Relative gene expression levels were normalized to actin expression levels, and the expression levels of the control (0 h) were set to 1.0. Error bars represent the standard deviations. The small letters above the bars indicate the significant differences between means (p < 0.05).
3.9. Expression Patterns of CsSRO Genes in Responses to Various Pathogen Infections
To analyze whether the CsSRO genes responded to biotic stress, their expression patterns under the infection of P. cubensis (DM treatment) and M. incognita (RKN treatment) were determined on the basis of the published RNA-Seq data [4,47]. In the Vlaspik (DM-susceptible) cucumber plants, CsSRO1 displayed increased expression after P. cubensis infection at 2, 4, and 6 dpi compared to the control, while CsSRO3 and CsSRO4 were down-regulated at certain time points. In the PI 197088 (DM-resistant cucumber plant), CsSRO1 displayed increased expression after P. cubensis infection at the earlier time point (1 dpi), while the other three CsSRO genes displayed decreased expression at certain time points, especially at 2 dpi (Figure 8A). Under RKN inoculation, the transcription levels of CsSRO1 and CsSRO2 were remarkably increased in susceptible CC3 lines, but no changed regulation of CsSRO genes was found during resistant IL10-1 lines response to RKN inoculation (Figure 8B).
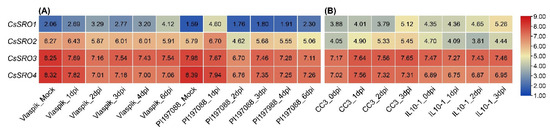
Figure 8.
Expression profiles of CsSRO genes in response to DM (A) and RKN (B) based on RNA-Seq data. (A) Expression profiles of CsSRO genes in Vlaspik (DM-susceptible cucumber plant) and PI 197088 (DM-resistant cucumber plant) leaves that were mock inoculated or collected 1, 2, 3, 4, and 6 dpi with P. cubensis infection. (B) Expression profiles of CsSRO genes in root tips from CC3 (RKN susceptible cucumber plant) and IL10–1 (RKN resistant cucumber plant) at 0, 1, 2 and 3 dpi with M. incognita inoculation. Genes highly or weakly expressed are colored red or blue, respectively, based on the log2 transformed TPM values.
4. Discussion
In recent decades, SRO gene family has been identified in diverse plant species whose genomes were sequenced [7,21,48]. However, no comprehensive identification and characterization has been performed for SRO genes in cucumber. In this study, the cucumber SRO family genes were identified and cloned, and these genes were named as CsSRO1–CsSRO4, making it a small gene family. The number of SRO family genes was compared to those of previously identified plant species, such as sesame, rice, Arabidopsis, maize, Brachypodium distachyon, tomato, banana, and apple, which generally contained 4–6 SRO genes in their genomes [46,48,49,50], but was much less than that in Chinese cabbage (12 genes) [51], and wheat (30 genes) [45]. These findings indicated that the expansion of the SRO gene family was relatively conservative, and the number of SRO family genes may not possess an absolute correlation with genome size during plant evolution. In addition, a segmental duplication event was found in this study (Figure 4), and similar results were also found in other plants, including wheat [45], and tomato [50].
Phylogenetic tree analysis showed that CsSRO proteins along with SRO proteins from 14 other different plant species were grouped into two groups (group I and group II) based on their sequence relationships, and group I can be further divided into four subgroups (Ia to Id) (Figure 1). The SRO proteins of these tested species were clustered in dicot- or monocot-specific patterns, and monocot species contained only group I SROs. In group I, monocotyledonous and dicotyledonous SRO proteins were also had individual distribution in special subgroups, of which group Ia and group Ib SROs were monocotyledonous, while SROs in group Ic and group Id were coming from mono- and dicot plants (Figure 1), suggesting that the evolution of SROs in these groups followed the monocot and dicot divergence. Notably, SROs including the WWE domain were distributed in group I, while all SROs in group II lack the WWE domain, which is consistent with the previous studies [7,48]. It can be speculated that the variation of the WWE domain is a divergent force for the expansion of the SRO gene family. We also identified 10 diverse conserved motifs, and there is an obvious difference in the arrangements of these conserved motifs in the two groups, while the SROs in the same group showed conservative conserved motif arrangements (Figure 2). Gene structure analysis provides extra clues into the procedure of evolution in different gene families [3]. In the current study, most SRO genes in the same subgroup displayed conservative gene structure, but they have variable numbers of introns in different subgroups (Figure 3), indicating that gain or loss of introns may present in SRO genes during the evolution, and the functions of phylogenetically related SRO genes may be conserved.
Previous reports revealed that the transcription of SRO genes exhibited constitutive expression or tissue-specific expression patterns. For example, Arabidopsis AtSRO1 and AtRCD1 [16], rice OsSRO1d and OsSRO1e [21], tomato SolySRO5 and SolySRO6 [50] were found to have a broad expression in various tissues. In sesame, SiSRO1a and SiSRO1b were highly transcribed in various tissues, while SiSRO2a and SiSRO2b exhibited the highest transcript levels in root [46]. We thus investigated the spatio-temporal expression patterns of CsSRO genes during the development of various tissues based on the published RNA-Seq data. The results showed that CsSRO1 was expressed at a much higher level in root than other tested tissues, CsSRO2 was highly expressed in root, unfertilized ovary, female and male flowers, while CsSRO3 and CsSRO4 were ubiquitously expressed in the tested tissues (Figure 6). In apple, the expression of MdSRO1 and MdSRO4 was highest in roots, whereas MdSRO5 displayed the highest expression in flowers [52]. In tomato, the highest transcript levels of SolySRO2, SolySRO3, and SolySRO5 were observed in flowers, roots, and 3 cm fruits, respectively [50]. In addition, CsSRO2 and CsSRO4 displayed obviously higher transcription levels in unfertilized ovary than unexpanded and fertilized ovaries (Figure 6), thus fueling our speculation that they may participate in ovary development of cucumber. Similarly, SolySRO4 was highly expressed in mature tomato fruits (breaker + 10 fruits), and its values were much higher than that in mature green fruits and breaker fruits, suggesting that this gene may participate in fruit development and ripening [50]. Moreover, the identified segmental duplication gene pairs, CsSRO3 and CsSRO4, showed very similar spatio-temporal expression patterns (Figure 6), suggesting their possible similar functions.
It was reported that genes in SRO family play vital roles in the resistance to various abiotic stresses. In this work, many cis-elements responding to stresses (especially ARE and STRE) were distributed in the promoters of CsSRO genes (Figure 5), and similar results were reported in the promoter regions of SolySRO and SiSRO genes [46,50]. The qRT-PCR analysis also revealed that the expression of four CsSRO genes were significantly induced under salt and drought stress (Figure 7), suggesting their involvement in stress tolerance. In previous reports, rice SRO genes (OsSRO1a–OsSRO1e) were induced by a series of abiotic stresses, including heat, cold, drought, UV, salt, wound, and submergence, and OsSRO1c was found to play positive roles in cold, drought, and oxidative stress [21,22]. Transgenic Arabidopsis plants overexpressing IcSRO1 from Ipomoea cairica also exhibited enhanced tolerance to salt and drought stress [53]. Overexpression of MdRCD1 in Arabidopsis promotes plant adaptation to drought, salt, and H2O2 stresses [52]. Overexpression of SiSRO2b also conferred the tolerance of transgenic yeast cells to osmotic, salt, and oxidative stress [46]. SRO family genes are also known as regulators function in plant defense response against pathogens, and the regulatory mechanisms depend on the hormone signaling pathways, such as JA, SA, and so on. For example, OsSRO1a was found to regulate JA-dependent disease susceptibility to the Xanthomonas oryzae pv oryzae (Xoo) [24]. The presence of cis-elements associated with hormones and defense responses in the promoters of CsSRO genes implied the potential roles of CsSRO genes in response to biotic stress through the hormone signal transduction pathway (Figure 5). Additionally, all CsSRO genes showed changed expression levels in both sensitive and resistant cucumber plants inoculated with DM, except for CsSRO2, whose expression was unchanged in DM-susceptible Vlaspik plants (Figure 8). CsSRO1 and CsSRO2 displayed significantly increased expression at certain time points in susceptible CC3 plants inoculated with RKN (Figure 8). In banana, a total of four out of six MaSRO genes showed increased expression levels during Fusarium oxysporum f. sp. Cubense tropical race 4 (Foc TR4) infection [48]. Therefore, CsSRO genes may also play important roles in the response of plants to adverse environmental conditions.
5. Conclusions
In this work, four SRO family genes were isolated from cucumber, and their evolutionary relationship, conserved motif composition, gene structure, chromosomal location, cis-regulatory elements within the promoter region, followed by expression patterns in various tissues and differential expression in response to abiotic and biotic stresses were systematically analyzed. Expression analysis showed that the four CsSRO genes were differentially expressed in various tissues and under diverse stresses, suggesting their roles in plant physiological processes and defense response. Our findings facilitate further study to the functional characterization of CsSRO genes in response to abiotic and biotic stresses in cucumber, and these CsSRO genes can be considered as potential candidates for breeding of cucumber stress-tolerant varieties in the future.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/horticulturae8070634/s1, Figure S1: PCR amplification of CsSRO genes. M, DL ladder 2000 DNA Marker; 1, CsSRO1; 2, CsSRO2; 3, CsSRO3; 4, CsSRO4, Figure S2: Schematic diagram of domain distributions in CsSRO proteins; Table S1: The gene-specific primers used in this study.
Author Contributions
Data curation, L.X., Z.Z., C.Z. and Y.Z.; formal analysis, C.Z. and J.Z.; funding acquisition, S.L. and Y.Z.; investigation, L.X., Z.Z. and J.Z.; methodology, L.X., Z.Z., C.Z., J.Z., Z.H. and Y.Z.; resources, L.X., Z.Z., Z.H. and S.L.; software, C.Z. and Y.Z.; writing—original draft, L.X. and Y.Z.; writing—review and editing, S.L. and Y.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Academic and Technical Leader Plan of Jiangxi Provincial Main Disciplines (20204BCJ22023), the Natural Science Foundation of Jiangxi Province (20202BABL205002), and the National Natural Science Foundation of China (31860566).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available within the article and Supplementary Materials.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Dos Santos, T.B.; Ribas, A.F.; de Souza, S.G.H.; Budzinski, I.G.F.; Domingues, D.S. Physiological responses to drought, salinity, and heat stress in plants: A review. Stresses 2022, 2, 113–135. [Google Scholar] [CrossRef]
- Kalia, V.C.; Gong, C.; Patel, S.K.S.; Lee, J.K. Regulation of plant mineral nutrition by signal molecules. Microorganisms 2021, 9, 774. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Li, G.; Zhang, L.; Xu, J.; Hu, L.; Jiang, L.; Liu, S. Comprehensive genomic analysis and expression profiling of the BTB and TAZ (BT) genes in cucumber (Cucumis sativus L.). Czech J. Genet. Plant Breed. 2020, 56, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.; Xiao, L.; Hu, Y.; Liu, L.; Liu, H.; Hu, Z.; Liu, S.; Zhou, Y. Genome-wide survey and expression analysis of B-box family genes in cucumber reveal their potential roles in response to diverse abiotic and biotic stresses. Agriculture 2022, 12, 827. [Google Scholar] [CrossRef]
- Aravind, L. The WWE domain: A common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem. Sci. 2001, 26, 273–275. [Google Scholar] [CrossRef]
- Jaspers, P.; Brosché, M.; Overmyer, K.; Kangasjärvi, J. The transcription factor interacting protein RCD1 contains a novel conserved domain. Plant Signal. Behav. 2010, 5, 78–80. [Google Scholar] [CrossRef]
- Jaspers, P.; Overmyer, K.; Wrzaczek, M.; Vainonen, J.P.; Blomster, T.; Salojärvi, J.; Reddy, R.A.; Kangasjärvi, J. The RST and PARP-like domain containing SRO protein family: Analysis of protein structure, function and conservation in land plants. BMC Genom. 2010, 11, 170. [Google Scholar] [CrossRef] [Green Version]
- Belles-Boix, E.; Babiychuk, E.; Van Montagu, M.; Inzé, D.; Kushnir, S. CEO1, a new protein from Arabidopsis thaliana, protects yeast against oxidative damage. FEBS Lett. 2000, 482, 19–24. [Google Scholar] [CrossRef]
- Katiyar-Agarwal, S.; Zhu, J.; Kim, K.; Agarwal, M.; Fu, X.; Huang, A.; Zhu, J.K. The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 18816–18821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overmyer, K.; Tuominen, H.; Kettunen, R.; Betz, C.; Langebartels, C.; Sandermann, H., Jr.; Kangasjärvi, J. Ozone-sensitive arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell 2000, 12, 1849–1862. [Google Scholar] [CrossRef] [Green Version]
- Ahlfors, R.; Lång, S.; Overmyer, K.; Jaspers, P.; Brosché, M.; Tauriainen, A.; Kollist, H.; Tuominen, H.; Belles-Boix, E.; Piippo, M.; et al. Arabidopsis RADICAL-INDUCED CELL DEATH1 belongs to the WWE protein-protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. Plant Cell 2004, 16, 1925–1937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujibe, T.; Saji, H.; Arakawa, K.; Yabe, N.; Takeuchi, Y.; Yamamoto, K.T. A methyl viologen-resistant mutant of Arabidopsis, which is allelic to ozone-sensitive rcd1, is tolerant to supplemental ultraviolet-B irradiation. Plant Physiol. 2004, 134, 275–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teotia, S.; Lamb, R.S. The paralogous genes RADICAL-INDUCED CELL DEATH1 and SIMILAR TO RCD ONE1 have partially redundant functions during Arabidopsis development. Plant Physiol. 2009, 151, 180–198. [Google Scholar] [CrossRef] [Green Version]
- Teotia, S.; Lamb, R.S. RCD1 and SRO1 are necessary to maintain meristematic fate in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 1271–1284. [Google Scholar] [CrossRef] [Green Version]
- Vainonen, J.P.; Jaspers, P.; Wrzaczek, M.; Lamminmäki, A.; Reddy, R.A.; Vaahtera, L.; Brosché, M.; Kangasjärvi, J. RCD1-DREB2A interaction in leaf senescence and stress responses in Arabidopsis thaliana. Biochem. J. 2012, 442, 573–581. [Google Scholar] [CrossRef] [Green Version]
- Jaspers, P.; Blomster, T.; Brosché, M.; Salojärvi, J.; Ahlfors, R.; Vainonen, J.P.; Reddy, R.A.; Immink, R.; Angenent, G.; Turck, F.; et al. Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J. 2009, 60, 268–279. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, Y.; Björn, L.O.; Li, S. Arabidopsis radical-induced cell death1 is involved in UV-B signaling. Photochem. Photobiol. Sci. 2009, 8, 838–846. [Google Scholar] [CrossRef]
- Li, B.Z.; Zhao, X.; Zhao, X.L.; Peng, L. Structure and function analysis of Arabidopsis thaliana SRO protein family. Yi Chuan 2013, 35, 1189–1197. [Google Scholar] [CrossRef]
- Zhao, X.; Gao, L.; Ren, J.; Pan, F. Arabidopsis SIMILAR TO RCD-ONE genes are ubiquitous and respond to multiple abiotic stresses through diverse signaling pathways. J. Biosci. 2019, 44, 129. [Google Scholar] [CrossRef]
- Borsani, O.; Zhu, J.; Verslues, P.E.; Sunkar, R.; Zhu, J.K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 2005, 123, 1279–1291. [Google Scholar] [CrossRef] [Green Version]
- You, J.; Zong, W.; Du, H.; Hu, H.; Xiong, L. A special member of the rice SRO family, OsSRO1c, mediates responses to multiple abiotic stresses through interaction with various transcription factors. Plant Mol. Biol. 2014, 84, 693–705. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Zong, W.; Li, X.; Ning, J.; Hu, H.; Li, X.; Xiao, J.; Xiong, L. The SNAC1-targeted gene OsSRO1c modulates stomatal closure and oxidative stress tolerance by regulating hydrogen peroxide in rice. J. Exp. Bot. 2013, 64, 569–583. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Su, J.; Xu, M.; Zhou, Z.; Zhu, X.; Ma, X.; Hou, J.; Tan, L.; Zhu, Z.; Cai, H.; et al. A common wild rice-derived BOC1 allele reduces callus browning in indica rice transformation. Nat. Commun. 2020, 11, 443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashihara, K.; Onohata, T.; Yariuchi, R.; Tanaka, S.; Akimitsu, K.; Gomi, K. The overexpression of OsSRO1a, which encodes an OsNINJA1- and OsMYC2-interacting protein, negatively affects OsMYC2-mediated jasmonate signaling in rice. Plant Cell Rep. 2020, 39, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, S.; Wang, M.; Wei, T.; Meng, C.; Wang, M.; Xia, G. A wheat SIMILAR TO RCD-ONE gene enhances seedling growth and abiotic stress resistance by modulating redox homeostasis and maintaining genomic integrity. Plant Cell 2014, 26, 164–180. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Xu, Y.; Liu, F.; Zhao, M.; Sun, Y.; Ma, Q. Maize similar to RCD1 gene induced by salt enhances Arabidopsis thaliana abiotic stress resistance. Biochem. Biophys. Res. Commun. 2018, 503, 2625–2632. [Google Scholar] [CrossRef]
- Qin, L.; Sun, L.; Wei, L.; Yuan, J.; Kong, F.; Zhang, Y.; Miao, X.; Xia, G.; Liu, S. Maize SRO1e represses anthocyanin synthesis through regulating the MBW complex in response to abiotic stress. Plant J. 2021, 105, 1010–1025. [Google Scholar] [CrossRef]
- Liao, L.; Hu, Z.; Liu, S.; Yang, Y.; Zhou, Y. Characterization of germin-like proteins (GLPs) and their expression in response to abiotic and biotic stresses in cucumber. Horticulturae 2021, 7, 412. [Google Scholar] [CrossRef]
- Lu, X.; Liu, W.; Xiang, C.; Li, X.; Wang, Q.; Wang, T.; Liu, Z.; Zhang, J.; Gao, L.; Zhang, W. Genome-wide characterization of GRAS family and their potential roles in cold tolerance of cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2020, 21, 3857. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Dong, S.; Liu, X.; Bo, K.; Miao, H.; Beckles, D.M.; Zhang, S.; Gu, X. Genome-wide characterization of cucumber (Cucumis sativus L.) GRAS genes and their response to various abiotic stresses. Horticulturae 2020, 6, 110. [Google Scholar] [CrossRef]
- Li, J.; Wang, T.; Han, J.; Ren, Z. Genome-wide identification and characterization of cucumber bHLH family genes and the functional characterization of CsbHLH041 in NaCl and ABA tolerance in Arabidopsis and cucumber. BMC Plant Biol. 2020, 20, 272. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, X.; Han, J.; Lu, W.; Ren, Z. Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC Plant Biol. 2020, 20, 443. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.; Zhu, C.; Yang, S.; Hu, Z.; Liu, S.; Zhou, Y. Comprehensive identification of the VQ family genes in cucumber and their roles in response to abiotic and biotic stresses. Sci. Hortic. 2022, 295, 110874. [Google Scholar] [CrossRef]
- Dai, Z.; Dong, S.; Miao, H.; Liu, X.; Han, J.; Li, C.; Gu, X.; Zhang, S. Genome-wide identification of TIFY genes and their response to various pathogen infections in cucumber (Cucumis sativus L.). Sci. Hortic. 2022, 295, 110814. [Google Scholar] [CrossRef]
- Luan, Q.; Chen, C.; Liu, M.; Li, Q.; Wang, L.; Ren, Z. CsWRKY50 mediates defense responses to Pseudoperonospora cubensis infection in Cucumis sativus. Plant Sci. 2019, 279, 59–69. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, H.; Yang, X.; Li, Q.; Ling, J.; Wang, H.; Gu, X.; Huang, S.; Jiang, W. CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner. Plant Physiol. Biochem. 2016, 108, 478–487. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic. Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, W.; Zhu, C.; Hu, Z.; Liu, S.; Wu, H.; Zhou, Y. Identification and transcriptional analysis of zinc finger-homeodomain (ZF-HD) family genes in cucumber. Biochem. Genet. 2021, 59, 884–901. [Google Scholar] [CrossRef]
- Jiang, W.; Geng, Y.; Liu, Y.; Chen, S.; Cao, S.; Li, W.; Chen, H.; Ma, D.; Yin, J. Genome-wide identification and characterization of SRO gene family in wheat: Molecular evolution and expression profiles during different stresses. Plant Physiol. Biochem. 2020, 154, 590–611. [Google Scholar] [CrossRef]
- Liu, A.; Wei, M.; Zhou, Y.; Li, D.; Zhou, R.; Zhang, Y.; Zhang, X.; Wang, L.; You, J. Comprehensive analysis of SRO gene family in Sesamum indicum (L.) reveals its association with abiotic stress responses. Int. J. Mol. Sci. 2021, 22, 13048. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cheng, C.; Zhang, K.; Tian, Z.; Xu, J.; Yang, S.; Lou, Q.; Li, J.; Chen, J.F. Comparative transcriptomics reveals suppressed expression of genes related to auxin and the cell cycle contributes to the resistance of cucumber against Meloidogyne incognita. BMC Genom. 2018, 19, 583. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhou, D.; Hu, H.; Li, W.; Hu, Y.; Xie, J.; Huang, S.; Wang, W. Genome-wide characterization of a SRO gene family involved in response to biotic and abiotic stresses in banana (Musa spp.). BMC Plant Biol. 2019, 19, 211. [Google Scholar] [CrossRef]
- Jiang, H.; Xiao, Y.; Zhu, S. Genome-wide identification, systematic analysis and characterization of SRO family genes in maize (Zea mays L.). Acta Physiol. Plant 2018, 40, 176. [Google Scholar] [CrossRef]
- Li, N.; Xu, R.; Wang, B.; Wang, J.; Huang, S.; Yu, Q.; Gao, J. Genome-wide identification and evolutionary analysis of the SRO gene family in tomato. Front. Genet. 2021, 12, 753638. [Google Scholar] [CrossRef]
- Qiao, Y.; Gao, X.; Liu, Z.; Wu, Y.; Hu, L.; Yu, J. Genome-wide identification and analysis of SRO gene family in Chinese cabbage (Brassica rapa L.). Plants 2020, 9, 1235. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, R.; Qu, F.; Yao, J.; Hao, Y.; Wang, X.; You, C. Identification of the SRO gene family in apples (Malus × domestica) with a functional characterization of MdRCD1. Tree Genet. Genomes 2017, 13, 94. [Google Scholar] [CrossRef]
- Yuan, B.; Chen, M.; Li, S. Isolation and Identification of Ipomoea cairica (L.) sweet gene IcSRO1 encoding a SIMILAR TO RCD-ONE protein, which improves salt and drought tolerance in transgenic Arabidopsis. Int. J. Mol. Sci. 2020, 21, 1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).