1. Introduction
Potato
Solanum tuberosum L. is the world’s fourth most important food crop after rice (
Oryza sativa L.), wheat (
Triticum spp.), and maize (
Zea mays L.) (Available online:
http://faostat.fao.org/; accessed on 1 July 2022) [
1]. The popularity of potatoes as a food product is based on the high content of starch, fiber, vitamins (C, B1, B3, and B6), essential amino acids (methionine and cysteine), and minerals [
1,
2].
Potatoes starch ranks third in global production after maize and wheat starches because of their unique molecular structure and organization [
3]. Starch is synthesized in chloroplasts of photosynthetic tissues during the daytime and is used as an energy source at night (transient starch) or stored in heterotrophic organs such as tubers (storage starch) [
4]. In potato tubers, starch constitutes 70–85% of the dry matter [
5], contributing to the breakage of tuber physiological dormancy during sprouting, when starch is degraded into low-molecular-weight sugars through the activity of several enzymes [
6]. Furthermore, in the roots, starch is involved in gravity sensing when sedimentation of starch-filled amyloplasts triggers a molecular pathway regulating root gravitropism and bending of the root tip at the elongation zone [
7].
Structurally, starch represents a mixture of linear amylose and branched amylopectin polysaccharides consisting of α-glucose monomers linked by alpha-1,4-glucosidic bonds [
8]. Changes in starch metabolism (mostly towards degradation) are one of the first biochemical reactions of plants to environmental signals and thus are involved in plant resistance to abiotic stresses [
9,
10]. Starch breakdown starts with amylopectin linearization by debranching enzymes; then, linear amylopectin and amylose are hydrolyzed by amylases [
8] and modified by α-glucan phosphorylases PHO1 and PHO2 belonging to glycosyltransferase family 35 (EC 2.4.1.1) [
11,
12].
PHO1 is a plastid-specific enzyme representing the main functional starch phosphorylase responsible for up to 96% of the total phosphorylase activity in plant cells [
12]. In addition to its role in starch breakdown, PHO1 is implicated in the initiation of starch biosynthesis, including that in potato tubers [
12]. As a key enzyme of starch metabolism, PHO1 is suggested to be involved in the regulation of the potato stress response. Thus, it has been reported that PHO1 activity is induced by low temperatures and is correlated with the increased accumulation of reducing sugars in tubers [
13,
14] and activated biosynthesis of starch with short glucan chains [
15]. Although PHO1′s protein structure, its regulatory and catalytic domains, and potential protein–protein interactions have been characterized [
12,
14], the importance of PHO1 in starch metabolism still needs clarification.
CRISPR-Cas9 technology enables the development of functional genomics [
16], including that of potatoes [
17,
18,
19], which could be relatively easily and efficiently transformed by agrobacteria and regenerate from protoplasts or tissue explants [
20]. In this study, we used the CRISPR-Cas9 system to edit the
PHO1a gene in four potato cultivars, which resulted in the generation of independent transgenic lines carrying a radical mutation in the PHO1a functional domain. The G261V mutation caused significant changes in the morphology and starch content of the transgenic plants and affected the expression of
PHO1a and other genes regulating starch turnover, including those of
S. tuberosum β-amylases (
StBAM1 and
StBAM9), amylase inhibitor (
StAI), and stress-responsive MADS (
MCM1,
AGAMOUS,
DEFICIENSE, and
SRF) domain transcription factors (
StFUL1 (
FRUITFUL),
StFUL2,
StMADS23, and
StSOC1 (
SUPPRESSOR OF CONSTANS OVEREXPRESSION 1)) both in normal and cold stress conditions. Our results suggest that the G261V substitution affects the functional activity of PHO1a, highlighting an important role of the enzyme in starch metabolism, shoot and root development, and abiotic stress response in potatoes.
2. Materials and Methods
2.1. Plant Material and Growth Conditions
Tetraploid S. tuberosum cultivars Mishka, Zhukovskii rannii, Lux, and Terra (the Lorch Potato Research Institute (Kraskovo, Russia)) used in the study had an average tuber starch content of 14.2–17.7%, 10–12%, 11–15%, and 15.5–16.5% of the fresh tissue, respectively. All cultivars were early maturing, cold-resistant, and had medium susceptibility to cold-induced tuber sweetening. Plants were maintained in vitro by sub-culturing the apical portion of 3–4 week-old stems on A2 medium: Murashige and Skoog (M&S) basal nutrient medium including vitamins (Sigma-Aldrich, Burlington, MA, USA; M5519), 10 g/L sucrose (Sigma-Aldrich, S9378), and 7.5 g/L agar (Sigma-Aldrich, A7921) (pH 6.0). Plants were cultured in a growth chamber under long-day conditions (16 h light/8 h dark) at 21 °C.
2.2. Plasmid Construction for CRISPR/Cas9 Editing of the PHO1a Gene
The
PHO1a coding sequence was analyzed for possible guide (g) RNA as a CRISPR target using CRISPRdirect (Available online:
http://crispr.dbcls.jp; accessed on 20 August 2021) [
21] and CRISPR MultiTargeter (Available online:
http://www.multicrispr.net/basic_input.html; accessed on 20 August 2021). Plasmid p201N Cas9 (AddGene, Watertown, MA, USA; Cat. #59175; Available online:
https://www.addgene.org/; accessed on 1 April 2020) was used for CRISPR/Cas9 editing. The vector included the
Cas9 gene driven by the double CaMV35S promoter, the
NPTII gene for plant selection, and the I-
PpoI site to accept the gRNA expression cassette (driven by the
Medicago truncatula U6.6 promoter) preliminary assembled in the pUC gRNA shuttle plasmid (AddGene Cat. #47024) [
22]. The construct was designed to introduce random mutations, including insertions/deletions (with or without frame shift) into the
PHO1a coding sequence corresponding to the selected gRNA.
The final p201N-
PHO1a-gRNA plasmid for editing was assembled according to the published protocol (Available online:
https://media.addgene.org/data/plasmids/47/47024/47024-attachment_rNUYUYROwjgZ.pdf; accessed on 1 April 2020) [
22], verified by PCR and restriction analyses, and used to transform the
Agrobacterium tumefaciens LBA4404 strain. The empty vector p201N was used for plant transformation as a negative control.
2.3. Plant Transformation
Potato plants were transformed as follows. Stem internodes (pieces about 1 cm long) were taken from 3-week-old germ-free potato plants, put on sterile filter paper placed on the surface of A2 medium, and kept for 24 h in the dark at room temperature. The next day, 5 mL of overnight liquid
A.
tumefaciens culture grown at 28 °C with shaking at 200 rpm was centrifuged at 5000 rpm for 10 min, and the pellet was re-suspended in 0.5 mL of liquid A2 medium (OD
600 1.9–2.0) and spread over the filter paper with explants. After 4 h in the dark at room temperature, the infected internodes were slightly dried on fresh sterile filter paper and placed on callus induction medium (CIM: A2 supplemented with 1 mg/L 6-benzylaminopurine, 1 mg/L zeatin, and 0.02 mg/L 1-naphthaleneacetic acid) for 24 h. Then, the explants were transferred to CIM containing 50 µg/L kanamycin (selective agent) and 250 µg/L carbenicillin (
Agrobacterium growth repressor) for callus induction and plant regeneration. The explants were sub-cultured every 7–10 days using fresh CIM to maintain selection pressure. Shoots that emerged after 5–7 weeks were cut and transferred to A2 medium with 50 µg/L kanamycin and 250 µg/L carbenicillin. Shoots that produced roots in the selective medium were screened by PCR for the presence of the transgene using
NPTII-specific primers and for residual
Agrobacterium infection using
VirB-specific primers (
Table 1). Transgenic lines were cultivated in a growth chamber under the 16 h light/8 h dark cycle at 21 °C.
2.4. DNA Sequencing
Total genomic DNA was extracted from the whole plant of each transgenic line using the ZR-96 Plant/Seed DNA kit (Zymo Research, Irvine, CA, USA), and the ~0.5-kb region of the
PHO1a gene comprising the gRNA target site was amplified by PCR using primers stp4f and stp5exR (
Table 1) at the following conditions: initial denaturation for 5 min at 95 °C, 35 cycles of denaturation (30 s at 95 °C), annealing (30 s at 59 °C), extension (1 min at 72 °C), and final extension for 10 min at 72 °C. PCR products of the expected size (449 bp) were isolated from agarose gels using the QIAEX
® II Gel Extraction kit (QIAGEN, Hilden, Germany), cloned in the pGEM
®-T Easy vector (pGEM
®-T Easy Vector System I, Promega, Madison, WI, USA), and sequenced on an ABI Prism 377 DNA Sequencer (Applied Biosystems, Waltham, MA, USA). The obtained DNA sequences were analyzed for possible polymorphisms using the MEGA 7.0 software (
https://www.megasoftware.net/; downloaded on 1 December 2020).
2.5. Starch Content Determination
To measure the total starch content, ~500 mg of leaf or root tissues was homogenized in 4.5 mL of 33% (v/v) dimethyl sulfoxide containing 0.44 M hydrochloric acid, incubated at 60 °C for 30 min in a water bath, cooled to 25 °C, and diluted 1:5 with milliQ water; pH was adjusted to 4.5 with 5 M sodium hydroxide, and the suspension was filtered through Miracloth (Merck, Kenilworth, NJ, USA). Starch content (mg/g of fresh weight) was measured in 100 μL of the filtrate using the “Starch” kit (BoehringerMannheim/R-Biopharm, Rotkreuz, Switzerland) according to the manufacturer’s protocol. The analysis was performed in three technical replicates of two biological replicates.
2.6. Plant Growth Phenotype and Cold Stress
Wild type (WT) and PHO1a mutant plants were grown in plant culture tubes (Sigma-Aldrich, C1048, St. Louis, MO, USA) containing A2 medium in a growth chamber with a 16 h light/8 h dark cycle at 21 °C for 5–6 weeks. The phenotype of the accessions was analyzed based on plant height, root weight and length, weight of the aerial part, and other characteristics (such as tendency to form multiple shoots and/or roots) using 10 biological replicates of each line.
To test plant response to cold stress, WT and PHO1a-edited lines were grown in 1 L plastic pots containing A2 medium in a growth chamber at normal conditions (16 h light/8 h dark, 21 °C) for 3 weeks. Next, half of the plants of each line were transferred to 4 °C, and half remained at 21 °C (control) for another 3 days and then analyzed for total starch content in the roots and leaves and gene expression.
2.7. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
The transcription of
PHO1a,
StBAM1,
StBAM9,
StAI,
StFUL1,
StFUL2,
StMADS23, and
StSOC1 genes was analyzed in the leaves and roots. Tissues (~0.2–0.5 g) were ground to powder in liquid nitrogen and used for total RNA extraction (RNeasy Plant Mini Kit, QIAGEN). After removal of genomic DNA using RNase-free DNase (QIAGEN) and analysis by gel electrophoresis, RNA samples were used for first-strand cDNA synthesis (GoScript Reverse Transcription System; Promega) with an oligo-dT primer. RNA and cDNA concentrations were determined using Qubit
® Fluorometer and Qubit RNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). Then, qRT-PCR was performed in a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA) with 3.0 ng of cDNA, gene-specific primers (
Table 1), and SYBR Green RT-PCR mixture (Syntol, Moscow, Russia) at the following cycling conditions: initial denaturation at 95 °C for 5 min, 40 cycles of denaturation at 95 °C for 15 s, and annealing/extension at 60 °C for 40 s.
The expression of the target genes was normalized to that of two potato reference genes,
ELF1 (elongation factor 1-alpha; LOC102600998) and
SEC3A (LOC102599118), and statistically analyzed using Graph Pad Prism version 8 (GraphPad Software Inc., San Diego, CA, USA; Available online:
https://www.graphpad.com/scientific-software/prism/; accessed on 1 May 2022). The results were expressed as the mean ± standard deviation (SD) based on three technical replicates of three biological replicates for each combination of cDNA and primer pairs. The unequal variance (Welch’s)
t-test was applied to assess differences in gene expression, and
p < 0.01 was considered to indicate statistical significance.
The obtained gene transcription data were analyzed for correlation with starch content in the roots using Graph Pad Prism version 8. Correlations were considered strongly positive, putatively positive, or negative if the square of the multiple correlation coefficient (R2) was >0.7, 0.4–0.7, or <0.4, respectively (p-value < 0.001).
3. Results
3.1. Generation and Phenotyping of Potato Plants with the Edited PHO1a Gene
To edit the
PHO1a gene, region 5′-GATCAGATGGAAAGAGGTATTGG-3′ (position 782–804 nt in
PHO1a cDNA) was chosen as a gRNA, and the p201N-
PHO1a-gRNA binary vector was assembled, introduced into the
A. tumefaciens LBA4404 strain, and used to transform stem internodes of four potato cultivars. As a result, 39 transgenic (kanamycin-resistant) lines (6 of cv. Mishka, 11 of cv. Terra, 5 of cv. Zhukovskii rannii, and 17 of cv. Lux) were obtained and confirmed by PCR for transgene presence in the genome. All T
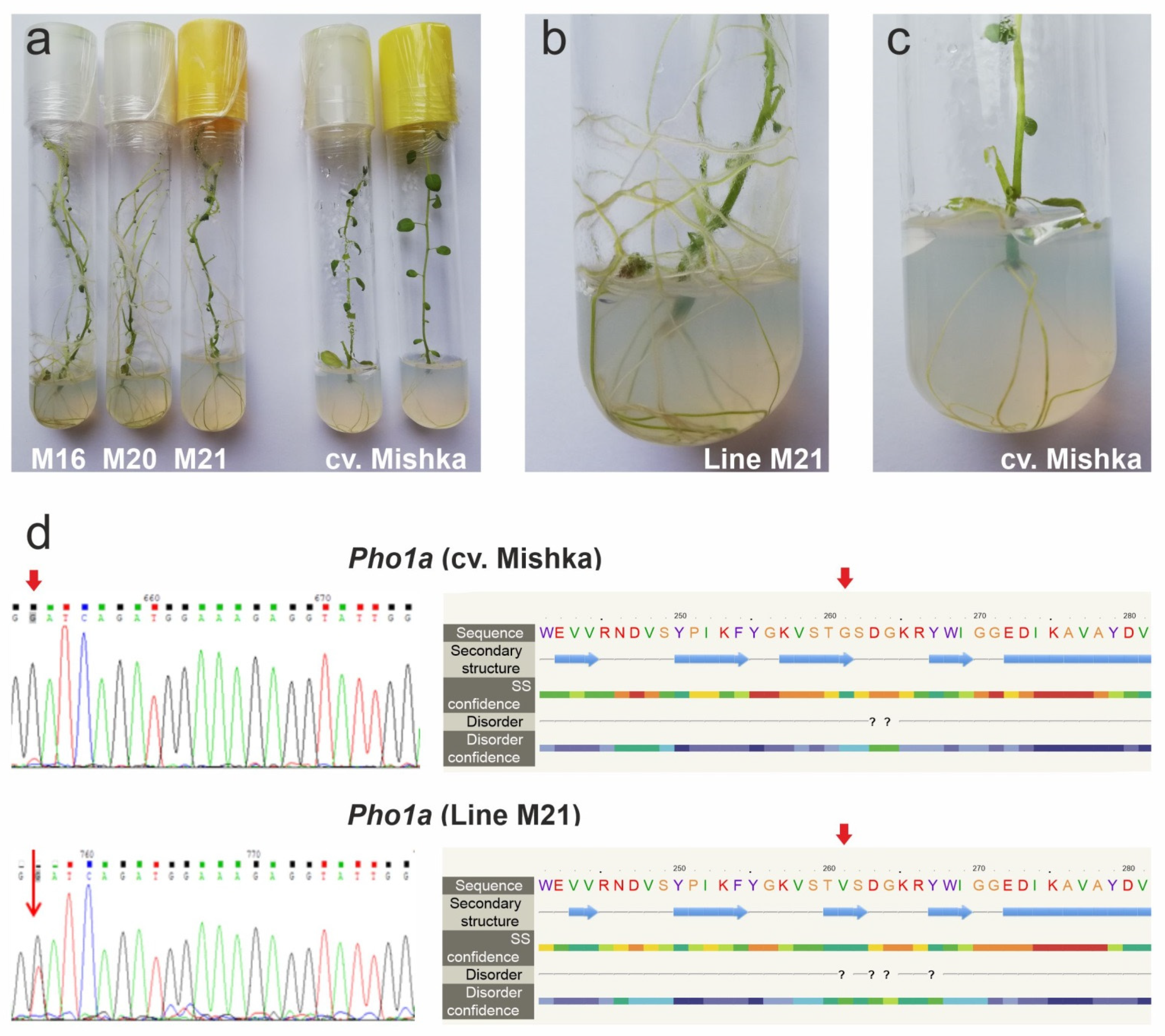
0 regenerants had altered morphology of the aerial part, which had a tendency to bushiness due to shoot initiation in axillary leaf meristems and enhanced root formation (
Figure 1a–c). Transgenic plants transformed with the empty p201N vector as a negative control did not show any morphological differences from the non-transgenic control.
Sequencing of the
PHO1a target site indicated that it was identical for all
PHO1a alleles in the four examined tetraploid potato cultivars. In all the 39 transgenic lines, 2 allelic
PHO1a variants, WT and mutant (782G→T, the first nucleotide of the gRNA sequence), were detected (
Figure 1d). Further cloning and sequencing of 10 clones for each line showed that 20–60% of them contained the 782G→T mutation; no other genetic alterations (including frameshift mutations) were detected.
These results indicated that the 782G→T single nucleotide polymorphism (SNP) in a PHO1a gene allele of 39 transgenic potato lines changed the potato phenotype, resulting in morphological differences.
3.2. Structural Comparison of the WT and Mutant PHO1a Proteins
Structural analysis of the edited PHO1a sequence showed that the non-synonymous 782G→T SNP resulted in amino acid substitution G261V in the N-terminal functional GT35_Glycogen_Phosphorylase domain located before the large L78 cleft comprising the catalytic center. According to PROVEAN and Phyre2 prediction, the substitution was radical and slightly affected the size of one of PHO1a β-shifts (
Figure 1d); however, positionally, it did not match any catalytic or binding sites, as indicated by a NCBI-CDD search.
3.3. Starch Content in WT and Transgenic Potato Lines
Considering that the G261V mutation in PHO1a caused significant morphological changes in the transgenic plants, it can be hypothesized that they may be a result of disturbances in starch metabolism. To analyze the possible correlation of the introduced PHO1a polymorphism with starch metabolism, we compared starch content in WT and transgenic plants at normal conditions (+21 °C) and in response to cold stress (+4 °C).
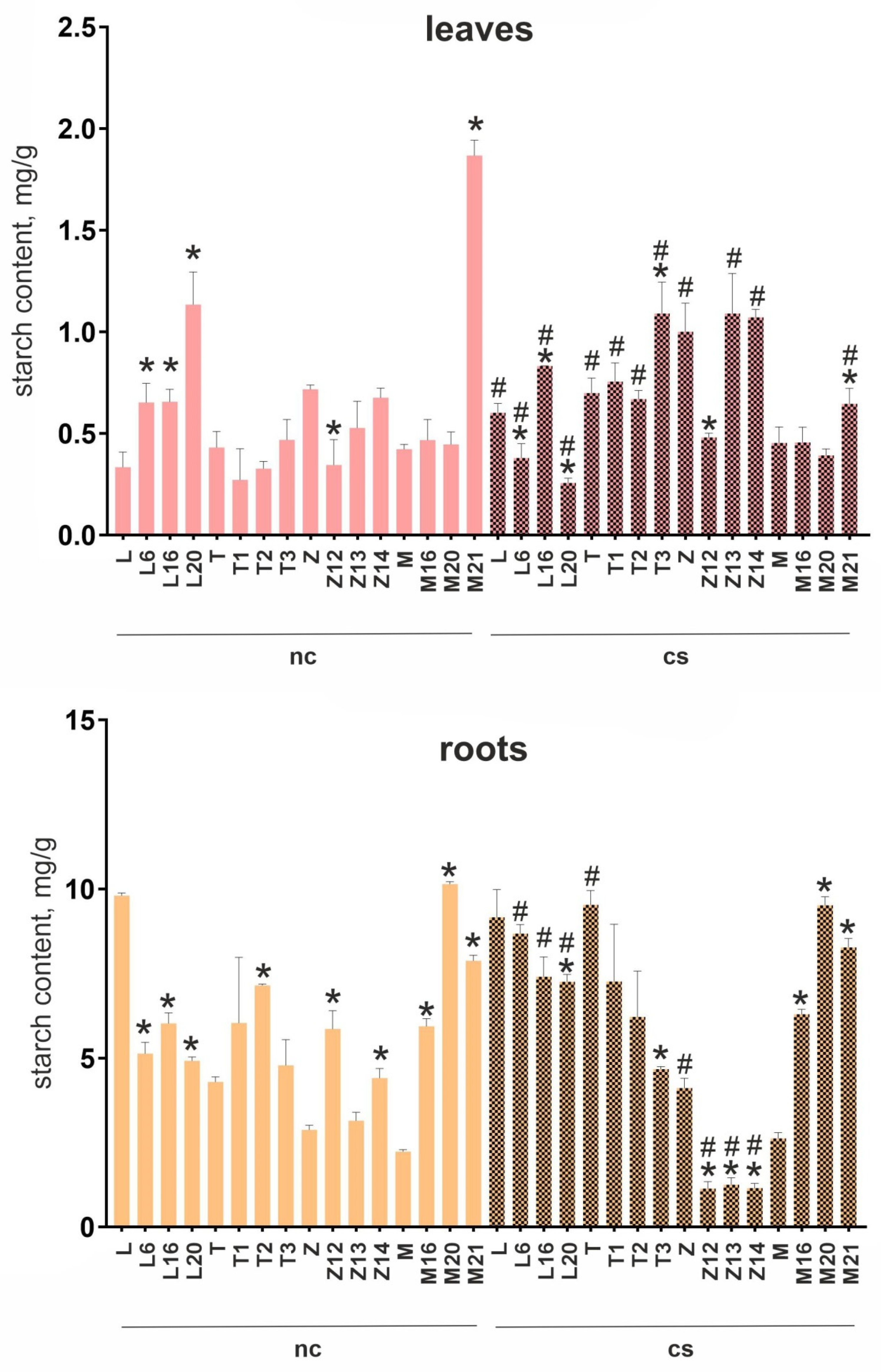
Several lines with the most pronounced morphological deviations from the WT control were selected for analysis: L6, L16, and L20 (cv. Lux); T1–T3 (cv. Terra), Z12–Z14 (cv. Zhukovskii rannii); and M16, M20, and M21 (cv. Mishka). At normal conditions, starch content in the leaves was decreased in T2 and Z12, increased in M21 and all L-lines, and was unchanged in the remaining lines compared to WT controls. In the roots, starch content was significantly decreased in all analyzed L-lines; increased in T2, Z12, Z14, and all M-lines; and remained unchanged in T1, T3, and Z13 (
Figure 2). These data indicated that despite a similar phenotype, plants with the edited PHO1a gene differed in starch content in the roots and leaves. At the same time, all analyzed lines of cv. Lux showed similar changes in starch content in the roots (decrease) and leaves (increase), whereas those of cv. Mishka had increased starch content in the roots.
The starch content was different between cultivars, as well as between mutants of the same cultivar. In the first case, it can be explained by a variety of specificity of the starch metabolism. In the second case, individual mutants may differ in genome localization of transgene insertion and, accordingly, in its effect. Additionally, the difference may be due to the different number of edited alleles. Besides, the introduction of an editing tool into the plant genome is not a guarantee of the editing event in every cell; thus, the mutants may have some mosaic editing and, as a result, different effects.
Exposure of unedited plants to cold stress for 3 days decreased starch content in the roots of cv. Terra and Zhukovskii rannii and increased it in the leaves of cv. Lux, Terra, and Zhukovskii rannii (
Figure 2). Among the transgenic lines, cold exposure increased starch content in the roots of all L-lines and decreased it in those of all Z-lines; in the leaves, starch content was decreased in L6, L20, and M21 and increased in L16, T3, and M21 compared to normal conditions (
Figure 2). Thus, the reaction of starch metabolism to cold stress in WT plants appeared to be cultivar-specific, whereas, in the transgenic lines, the response pattern was similar to that of WT (roots of all M-lines), opposite to that (roots of Z-lines), or of a mixed type (leaves of L-lines).
3.4. Expression of Starch-Degradation-Related Genes in WT and Transgenic Plants
Considering that the amount of a substrate and/or product of metabolic pathways affects the expression of relevant genes [
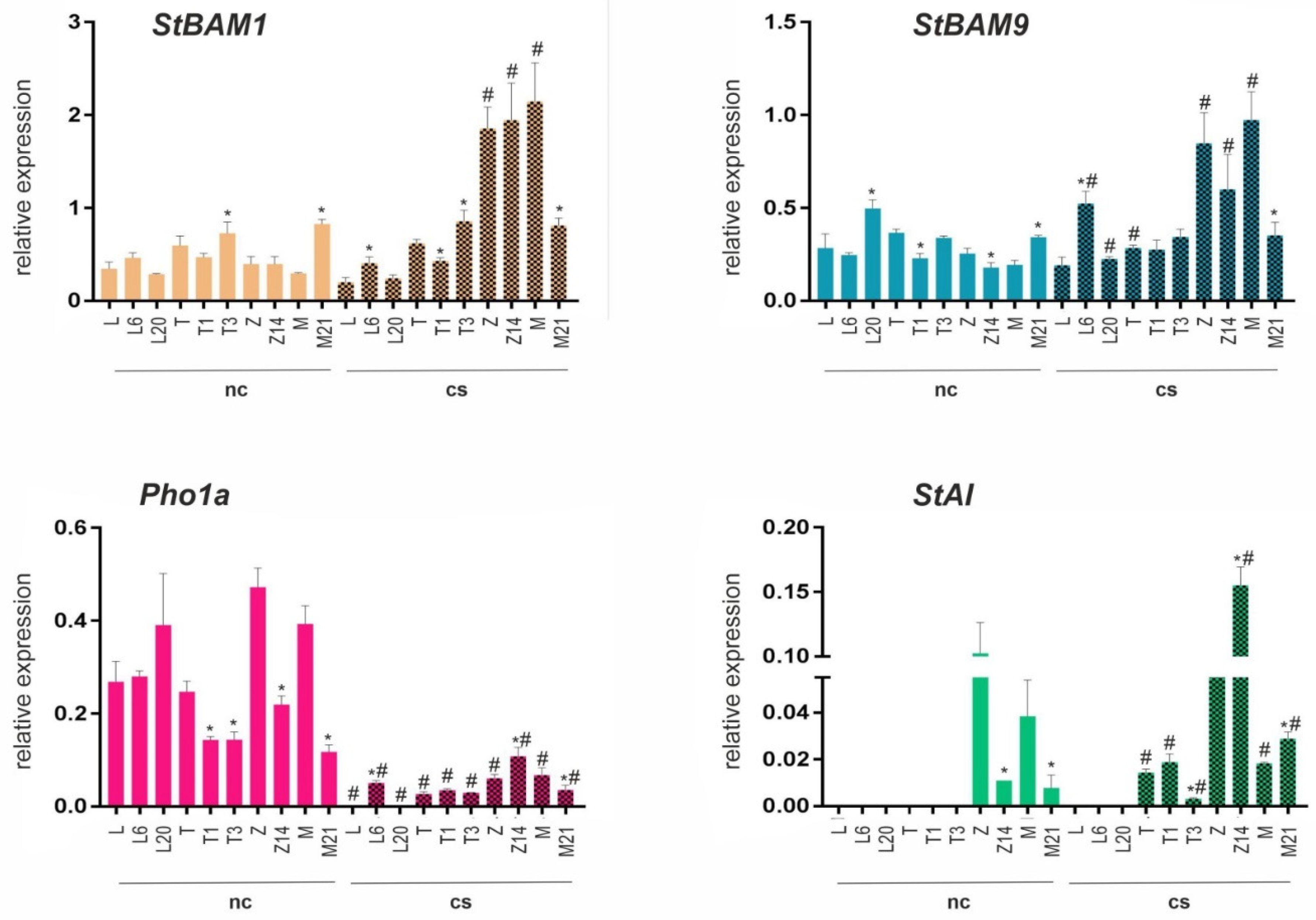
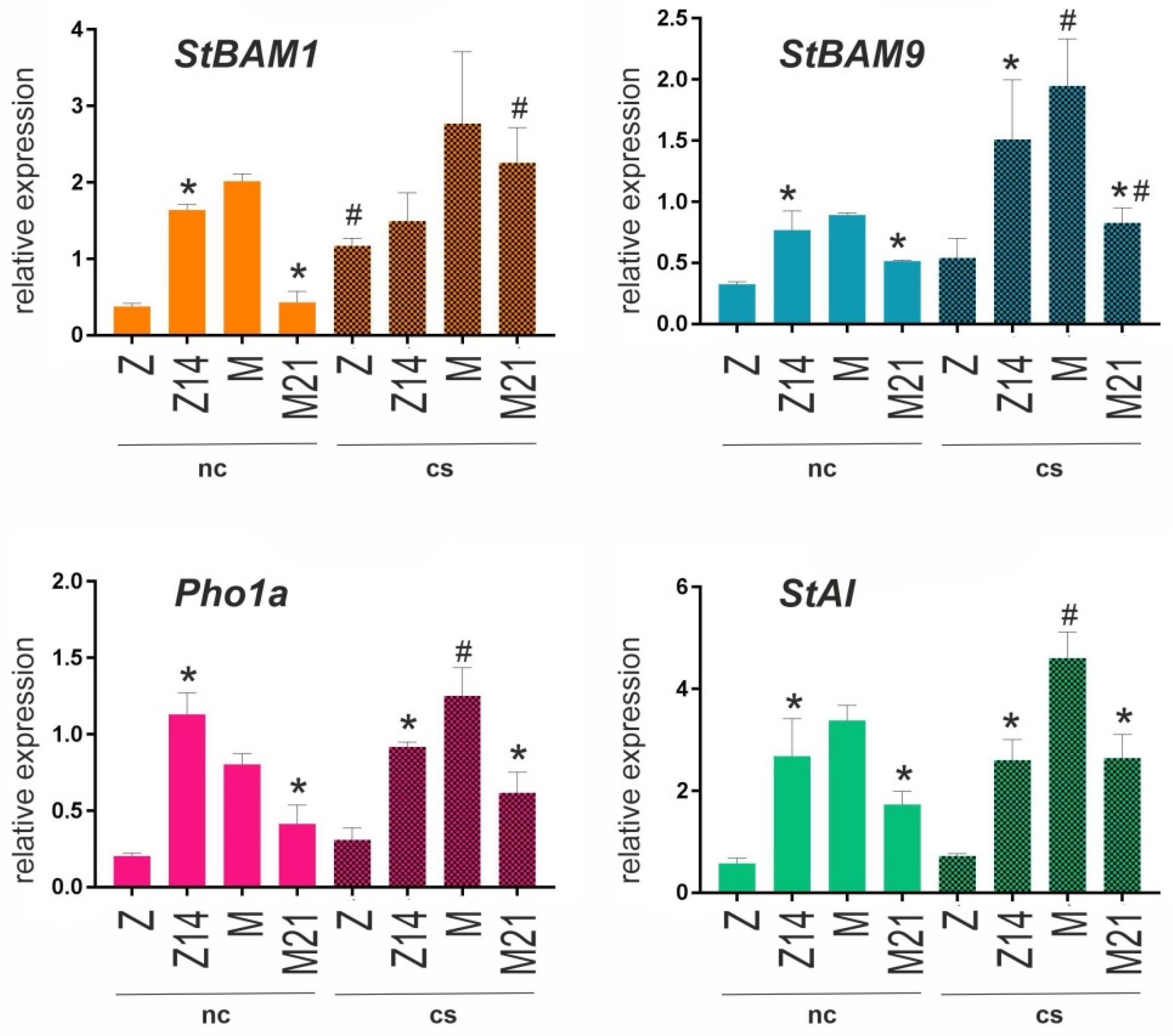
26], we tested whether changes in starch content influenced the transcription of PHO1a, StBAM1, StBAM9, and StAI genes in the leaves and roots of WT and transgenic plants grown under normal conditions and cold stress. The results indicated that PHO1a expression in the leaves of transgenic lines T1, T3, Z14, and M21 was decreased compared to respective WT controls and that cold treatment significantly downregulated PHO1a transcription in both WT and transgenic plants (
Figure 3). In the roots, PHO1a expression was increased in Z14 and decreased in M21 compared to WT controls and was not affected by cold exposure (
Figure 4).
The expression of key β-amylase genes was increased in the leaves of M21 (StBAM1 and StBAM9), T3 (StBAM1), and L20 (StBAM9) and decreased in T1 and Z14 (StBAM9) compared to WT. Cold treatment upregulated StBAM1 and StBAM9 expression in cv. Mishka, Zhukovskii rannii, and Z14; StBAM9 expression was also upregulated in L6 and downregulated in L20 (
Figure 3). Under normal conditions, the expression patterns of StBAM1 and StBAM9 in the roots were similar: both genes were downregulated in M21 and upregulated in Z14 compared to corresponding WT plants. After cold treatment, StBAM1 was upregulated in cv. Zhukovskii rannii and M21 and StBAM9 in cv. Mishka and M21 (
Figure 4).
Transcripts of the β-amylase inhibitor gene StAI were detected only in cv. Zhukovskii rannii and Mishka, and their levels were decreased in the leaves of the corresponding transgenic lines Z14 and M21 compared to WT (
Figure 3). Cold stress activated StAI transcription in cv. Terra and the corresponding transgenic lines T1 (at a similar level) and T3 (at a lower level) and significantly upregulated it in M21 and Z14 (
Figure 3). In the roots, StAI expression was stronger in cv. Mishka than in cv. Zhukovskii rannii and was downregulated in M21 and upregulated in Z14 compared to WT plants. Cold exposure significantly induced StAI expression in cv. Mishka and slightly in M21 (
Figure 4).
Overall, these data indicated that the expression patterns of the PHO1a, StBAM1, StBAM9, and StAI genes were cultivar-specific and regulated differentially in the leaves and roots. Cv. Mishka and Zhukovskii rannii and the corresponding transgenic lines were the most responsive to cold stress in terms of starch-metabolism-related gene expression, which was mostly upregulated by cold exposure, except PHO1a expression, which was downregulated in the leaves but unchanged in the roots.
3.5. Correlation between Starch Content and Gene Expression in WT and Mutant Potato Plants
Next, we analyzed the association of starch content in the leaves with
PHO1a,
StBAM1, and
StBAM9 expression levels (
Figure 5). The results revealed a putative positive correlation (R
2 = 0.6170) between
PHO1a expression and starch content in the leaves of WT plants in normal conditions; however, no such correlation was observed for transgenic lines in normal conditions or for any plants after cold stress.
There was no correlation between StBAM1 expression and starch content.
StBAM9 expression was strongly associated with starch content in the leaves of transgenic M21 and Z14 lines (R2 = 0.8116) and putatively associated with that of WT plants (R2 = 0.4081) after cold treatment, whereas no such correlation was observed in normal growth conditions.
Thus, starch content was associated with PHO1a expression at a normal temperature only in WT plants and with StBAM9 expression in both WT and transgenic plants only in response to cold stress.
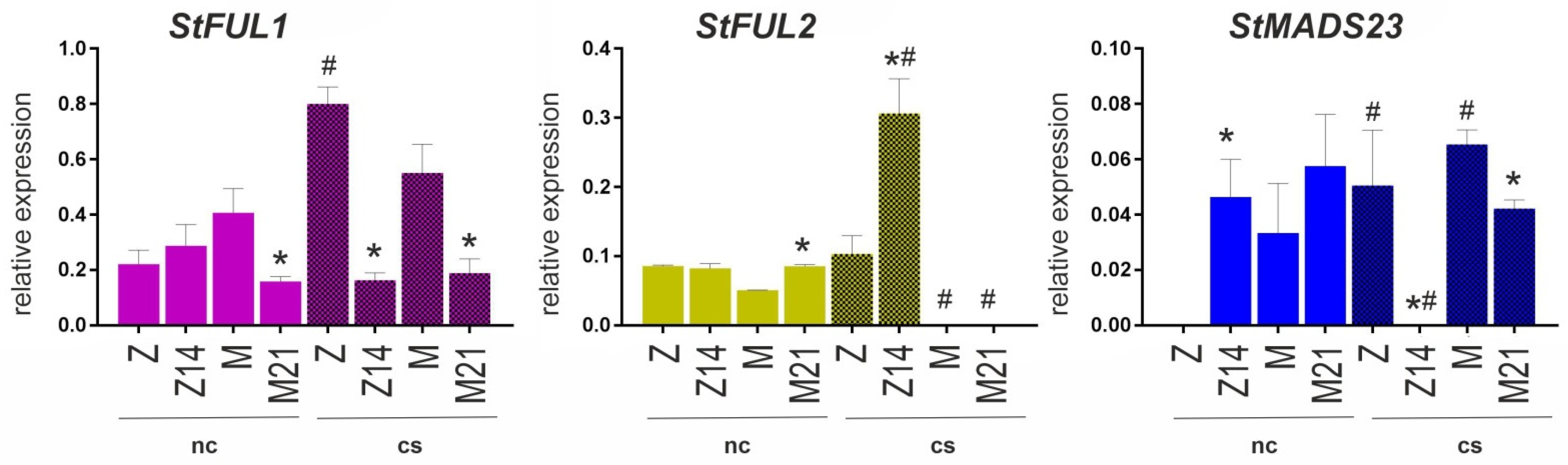
3.6. MADS-Box Gene Expression in WT and Mutant Potato Plants
Potato MADS-box genes StSOC1, StFUL2, StMADS23, and StFUL1 are homologous to those of Arabidopsis, where they are activated in the roots in response to stress [
27,
28,
29,
30]. However, the StSOC1 gene was not transcribed in the roots of two cultivars (Mishka and Zhukovskii rannii) and transgenic M21 and Z14 lines, whereas the other analyzed genes were expressed in all samples (except for StMADS23 in cv. Zhukovskii rannii) (
Figure 6). In normal conditions, StFUL1 expression was the highest in cv. Mishka and similar in the other accessions; it was upregulated by cold stress in cv. Zhukovskii rannii. StFUL2 showed the same expression level in all four accessions under normal conditions but was downregulated below the detection level in cv. Mishka and M21 and significantly upregulated in Z14 by cold stress. StMADS23 was not expressed in cv. Zhukovskii rannii under normal conditions but was strongly activated by cold stress; the opposite pattern was observed for the transgenic Z14 line, where StMADS23 was expressed under normal conditions but repressed by cold treatment. In cv. Mishka, StMADS23 was upregulated by cold stress.
Overall, these results revealed that PHO1a mutation affected MADS-box gene expression in a cultivar-specific manner and influenced transcriptional response to cold stress.
4. Discussion
Carbon is reserved in plants, mainly in the form of starch, whose synthesis and degradation are regulated by complex enzymatic and transcriptional mechanisms [
8,
31]. Plastid L-type starch phosphorylase PHO1 is a unique enzyme involved both in starch anabolism and catabolism [
12]. In potatoes, PHO1 is represented by two isoforms: PHO1a, which functions in all tissues and is mainly active in tuber amyloplasts, and PHO1b, which is specific for chloroplasts [
32,
33]. PHO1a may act differently in heterotrophic and photosynthetic potato tissues, as evidenced by its upregulation in tubers during long-term cold storage and downregulation in the leaves after short-term cold stress; however, both changes result in starch degradation [
14]. Studies on transgenic potatoes show that
PHO1a transcriptional downregulation does not affect plant morphology [
32] or the size and structure of starch granules [
34]. Thus, the functional dualism of this enzyme remains an intriguing research problem, especially given the prospective use of the
PHO1 gene for crop improvement [
12].
In this study, we edited the
PHO1a gene of four potato cultivars using CRISPR/Cas9 and obtained independent transgenic lines. In the tetraploid genome of
S. tuberosum, editing events may occur at each of the four alleles [
35]. In our case, all the regenerants had the same non-synonymous mutation 782G→T in 1–2 of the 4 alleles (
Figure 1d). It should be noted that there were no transgenic lines carrying this mutation in all four alleles or those carrying other mutations in the targeted
PHO1a region, including insertions or deletions, which could potentially repress the translation of the functional protein or dramatically modify the PHO1a protein structure. Moreover, the resultant G261V amino acid substitution, despite its radical character, did not affect any of the known catalytically important PHO1a regions. These observations do not rule out the possibility that our targeting strategy might have produced plants homozygous for the 782G→T mutation or carrying other
PHO1a mutations, which could have resulted in the disruption or knockdown of the PHO1a enzyme and the emergence of non-viable cells or transgenic plants that failed to develop. These considerations highlight the critical role of PHO1a in potatoes.
However, even a single radical G261V mutation significantly affected potato morphology by enhancing the development of shoots and roots (
Figure 1a–c) [
36]. Considering that all developmental processes in plants depend on energy supply and starch is the main energy source [
37], we hypothesized that the morphological changes observed in the obtained transgenic lines were associated with disturbances in starch metabolism due to the mutation in the
PHO1a gene. Indeed, the transgenic lines differed from the parental plants in starch content (
Figure 2) and the expression of
PHO1a and starch degradation-related genes under both normal and low-temperature growth conditions (
Figure 3 and
Figure 4). Although the structural analysis did not reveal interference of the G261V substitution with functionally important PHO1a sites, it is possible that it could still affect its catalytic activity and disrupt the coordinated operation of starch catabolism enzymes under both normal and cold stress conditions.
The correlation observed between the level of
PHO1a expression and starch content in the leaves of only WT but not transgenic plants at normal rather than cold temperatures (
Figure 5) indicated that the mutation interfered with the function of PHO1a in starch metabolism and points to the involvement of the enzyme in the potato’s response to cold stress. The downregulation of
PHO1a expression in almost all samples after cold treatment (
Figure 3 and
Figure 4) indicates that it was not associated with the mutation and is consistent with a previously shown cold-induced decrease in
PHO1 gene activity in rice endosperm [
38].
The observed changes in starch content may affect the activity of other starch-metabolism-related enzymes whose expression may be regulated by starch as a substrate or product [
26]. Starch is degraded by phosphorylases and amylases [
8,
31]. We analyzed two of the ten known potato β-amylases (BAM, EC 3.2.1.2), StBAM1 and StBAM9, which are known to participate in cold-induced starch degradation in tuber amyloplasts [
33,
39]. StBAM9 plays a dominant role in starch degradation: although it does not have catalytic activity, it directly binds to the surface of starch granules and, through the formation of a complex with enzymatically active StBAM1, attracts the latter to starch-degradation sites, which results in the release of soluble glucan molecules [
40,
41]. StBAM1 and StBAM9 are inhibited by StAI, which binds to the enzymes and blocks the active center or changes the molecular conformation, thus decreasing the catalytic activity or interaction with partner proteins [
39,
40]. Although no correlation has been found between
StAI expression and the cold resistance of potato species [
25], its transcription is strongly activated in response to short-term cold stress, with a simultaneous decrease in starch content in the leaves of cultivated and wild potato species [
42]. Our data showed that the expression of the
StBAM1,
StBAM9, and
StAI genes was affected by the
PHO1a mutation and was differentially regulated in the leaves and roots, although without any clear trend (
Figure 3 and
Figure 4), which may indicate a forced adaptation of starch hydrolytic enzymes to altered phosphorolysis. Still, some reactions remain stable; thus, there was a correlation between
StBAM9 expression and starch content in both WT and transgenic plants after cold stress (
Figure 5).
Starch breakdown is involved in plant response to low temperatures because the released sugars act as osmotic stabilizers and help plants resist cold [
43]. Given the dual role of PHO1a, which participates in both starch synthesis and degradation, it can be suggested that under cold stress, the expressed PHO1a is primarily used for polysaccharide phosphorolysis, whereas under normal conditions, it is shared between the two opposite processes.
MADS-domain transcription factors are important regulators of carbohydrate metabolism in plants and are known to exert control over starch content and composition in different conditions, including stress [
44,
45]. Consistent with previous data, we observed changes in the expression of MADS-box genes StMADS23, StFUL1, and StFUL2 in the transgenic plants carrying mutant PHO1a. The analyzed MADS genes were chosen because they are expressed in the roots [
27]. StFUL1 and StFUL2 are homologs of the A. thaliana AGL79 (AGAMOUS-LIKE 79) gene, which is transcribed in both vegetative and reproductive tissues and has been shown to modulate lateral root development and the response to nutritional stress [
28,
30]. In potatoes, StFUL1 (or POTM1 (Potato MADS-box 1)) has been reported to regulate axillary bud development [
46]. StMADS23 is a homolog of the A. thaliana ANR1 (ARABIDOPSIS NITRATE REGULATED 1) gene involved in root growth and response to nutritional changes [
29,
30]. The differential activity of the StMADS23, StFUL1, and StFUL2 genes in the roots of the WT and transgenic plants at both normal and cold temperatures (
Figure 6) suggests their role in the regulation of starch metabolism, especially since they are known to play such a role in sucrose metabolism [
47].
5. Conclusions
In this study, we edited the PHO1a gene in four potato cultivars using the CRISPR-Cas9 system and analyzed the generated transgenic lines for starch content and the expression of starch-degradation-related genes and MADS-box genes. The results suggest that the G261V radical mutation in the PHO1a functional domain due to 782G→T SNP, even in one allele, can affect starch metabolism in potato leaves and roots, leading to changes in plant morphology and response to cold stress.
In general, the data obtained cannot serve as a basis for unambiguous conclusions about the role of PHO1a in starch metabolism and plant response to cold stress, including due to the heterozygosity of the edited lines. Given the involvement of PHO1a in two opposite processes (starch synthesis and degradation), it can be assumed that PHO1a retains its dual role in mutant plants, most likely performing it at the expense of unedited alleles. At the same time, the disturbance introduced by editing into the PHO1a function causes a change in the starch content, including in response to cold stress. This may be the result of both a disturbance in the levels of starch synthesis/decomposition due to the mutation of PHO1a and a change in the regulation of other starch-metabolism genes in response to alteration in starch content. Considering the different shifts in starch content (compared to the WT control) in leaves and roots (including in response to cold), it can be assumed that PHO1a uses different ratios in these tissues for its roles in “starch synthesis” and “starch breakdown”. This duality of PHO1a, including variations in the ratio of the two roles, has been demonstrated by researchers. For example, low temperatures reduce PHO1a expression in rice grain endosperm (Hwang et al., 2016a, b) and increase it in cold-stored potato tubers (Schreiber et al., 2014), both reducing the starch content, which is associated with a dual function of the enzyme. We believe that our data can be used in further studies of the role of PHO1a.