Identification and Expression Analysis of CKX Gene Family in Brassica juncea var. tumida and Their Functional Analysis in Stem Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of CKX Gene Family Members in B. juncea
2.2. Sequence Analysis and Phylogenetic Tree Construction
2.3. Chromosomal Localization and Collinearity Analysis
2.4. Analysis of Gene Structure, Conserved Motifs and Cis-Acting Elements in Promoter Region
2.5. Expression Pattern Analysis of CKX Genes in B. juncea var. tumida
2.6. Plant Materials, RNA Isolation and cDNA Synthesis, and Real-Time Quantitative PCR (RT-qPCR) Validation
2.7. Hormone Extraction and Purification
3. Results
3.1. Identification and Analysis of Physicochemical Properties of CKX Gene Family Members in B. juncea
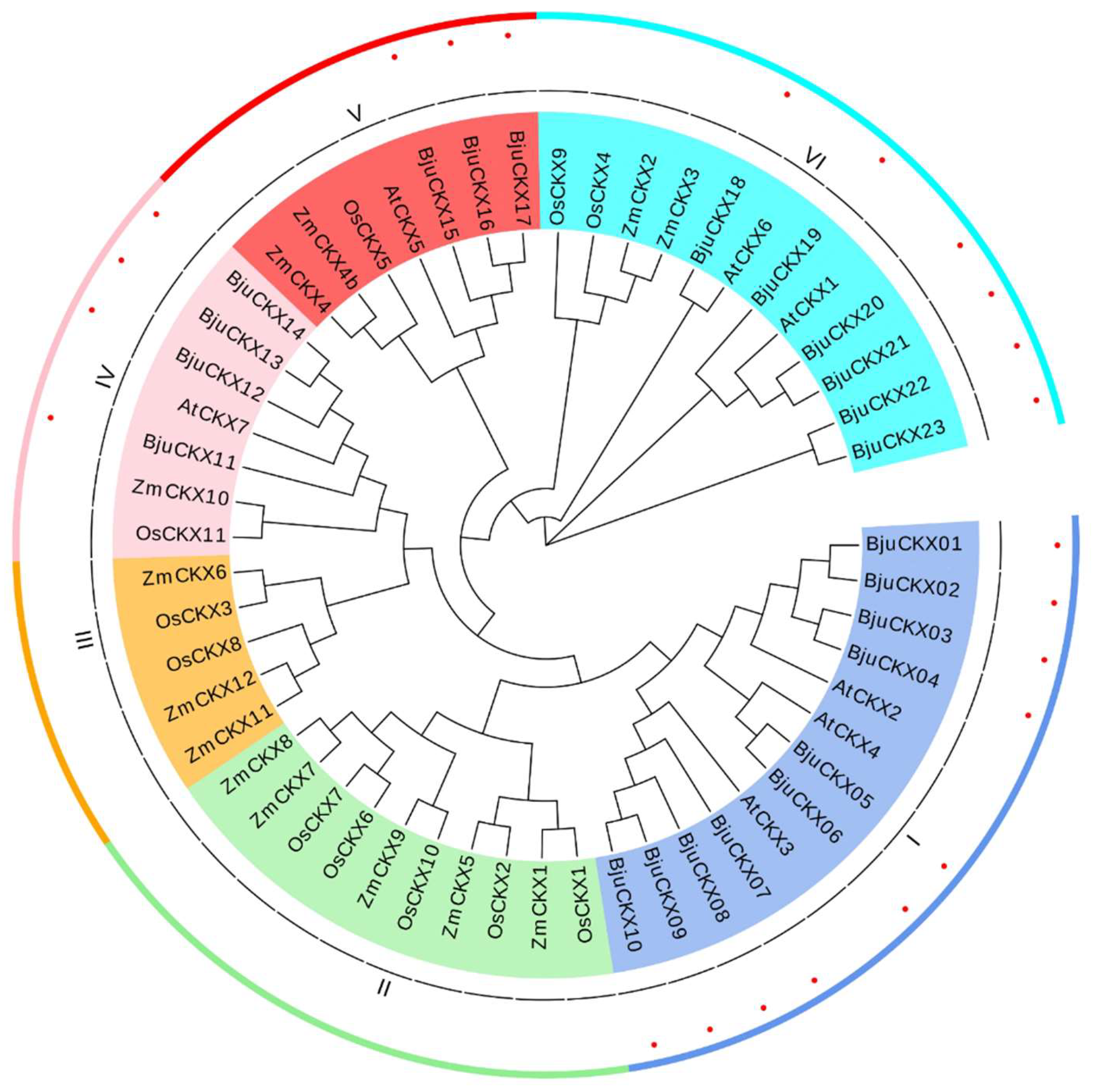
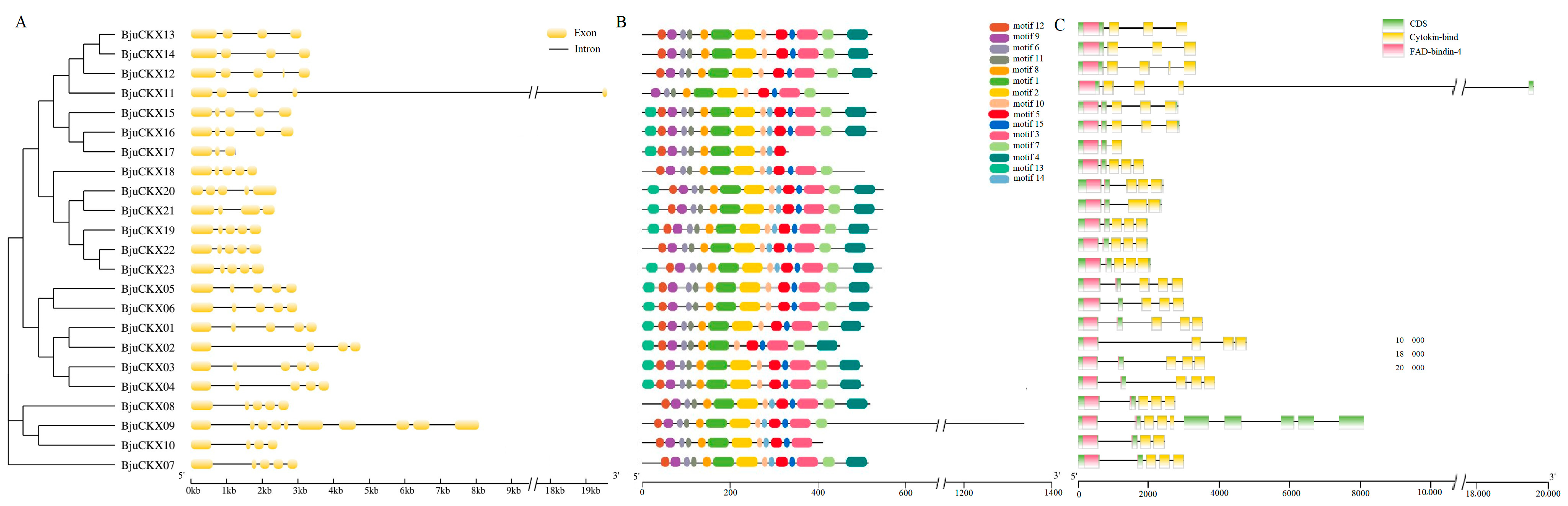
3.2. Phylogenetic Analysis of CKX Gene Family
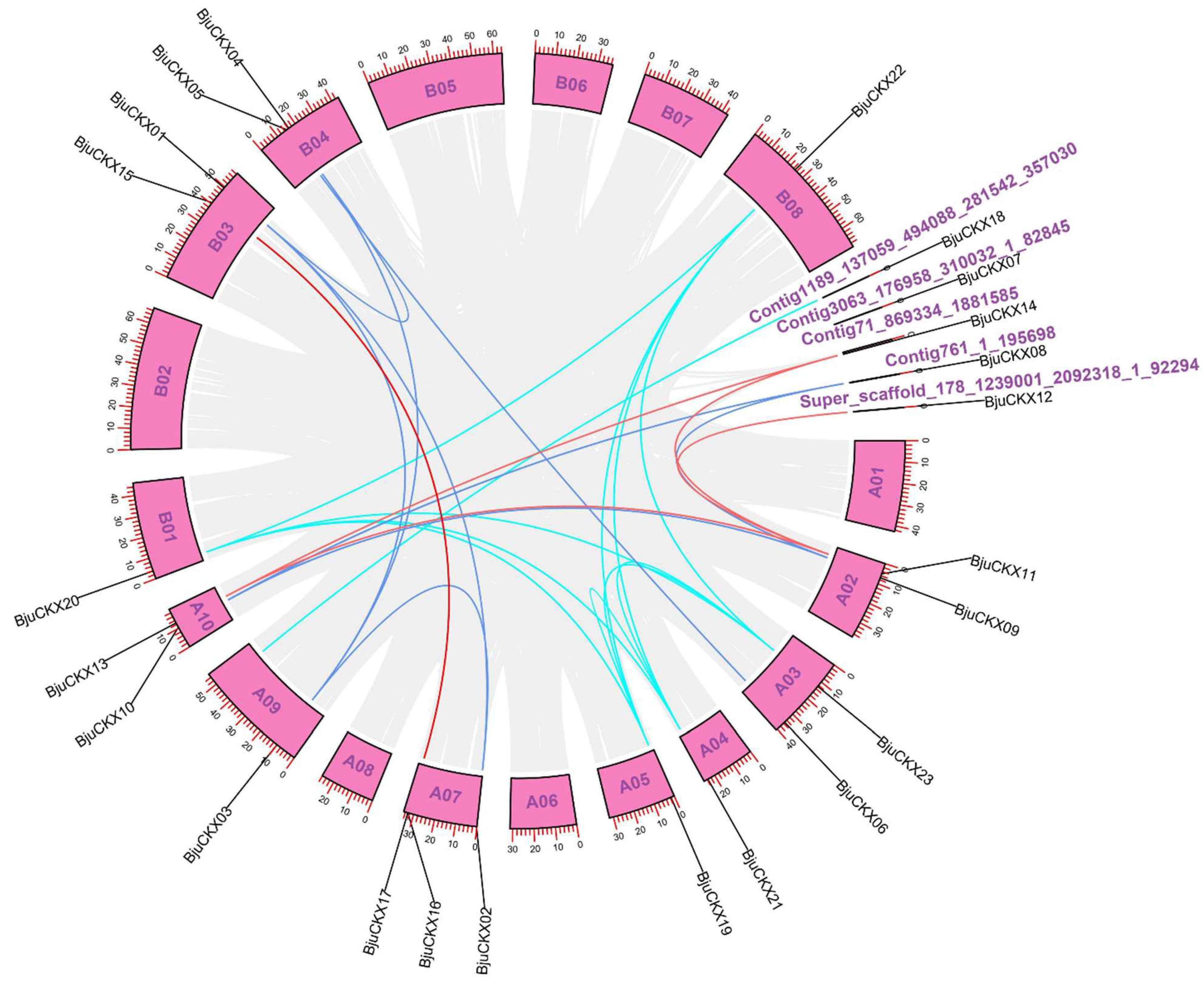
3.3. Analysis of Chromosome Localization and Gene Replication
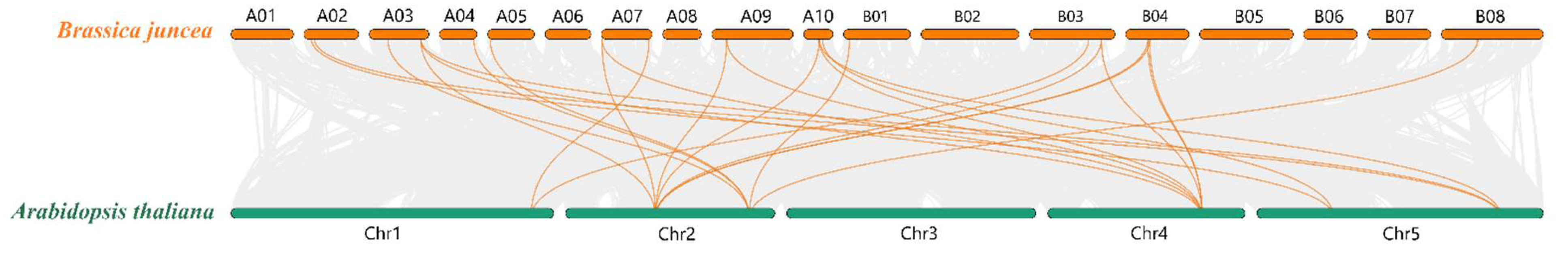
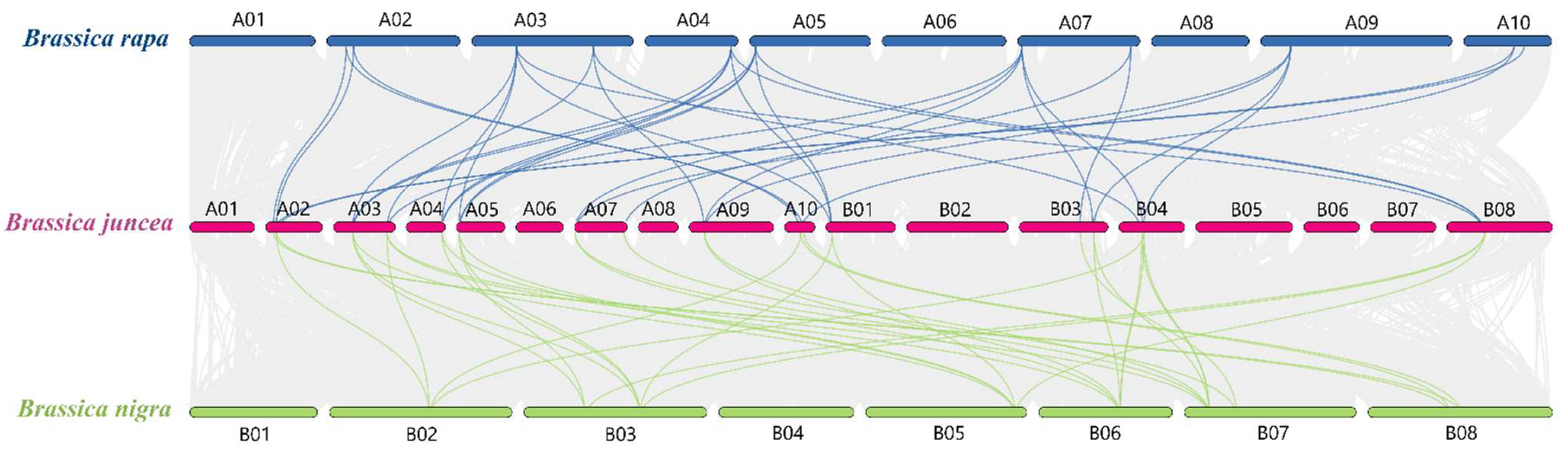
3.4. Collinearity Analysis
3.5. Analysis of Gene Structure and Conserved Motifs
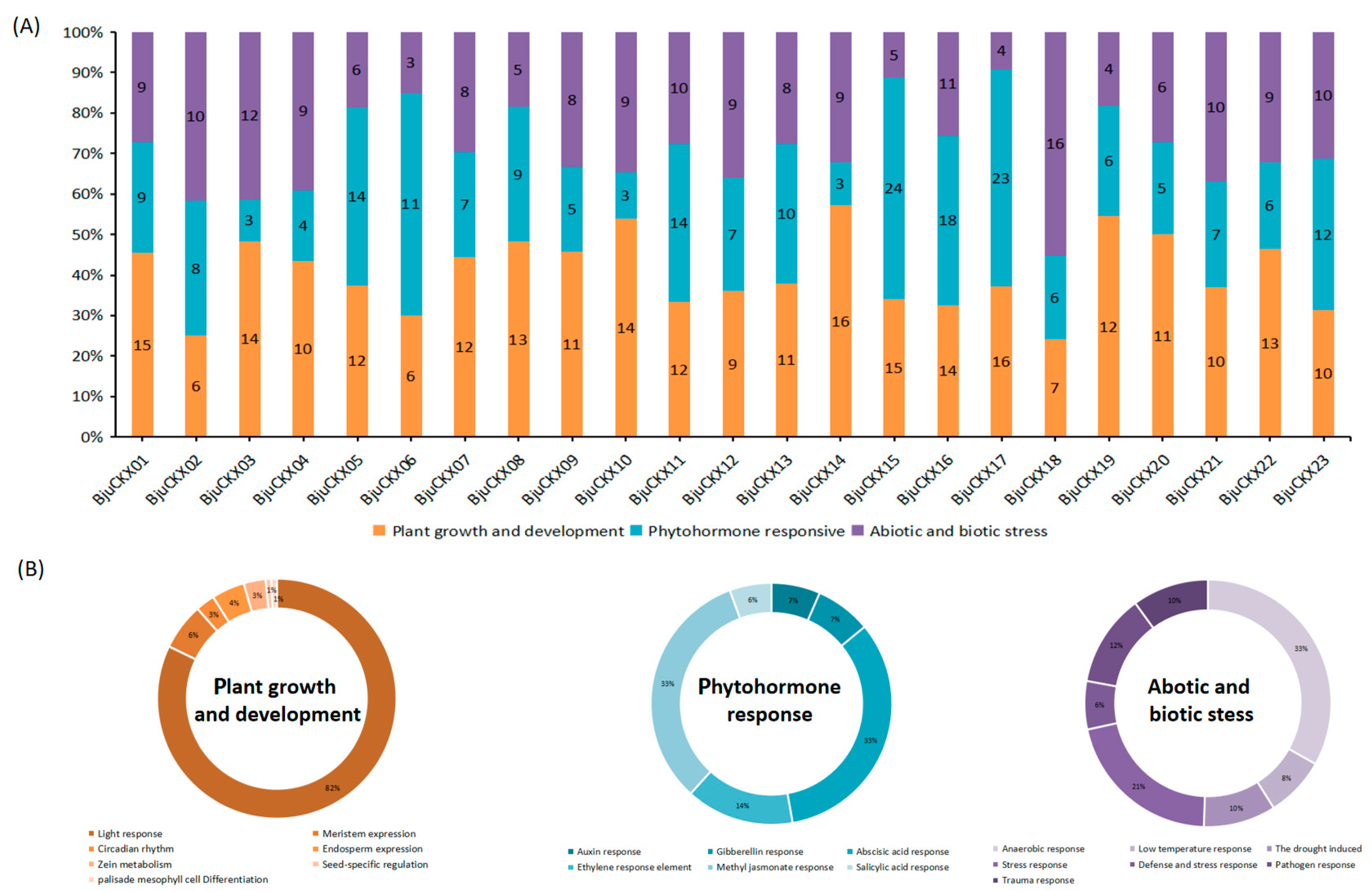
3.6. Analysis of Promoter Cis-Acting Elements
3.7. Transcriptional Expression Analysis of CKX Gene during Tumorous Stem Development
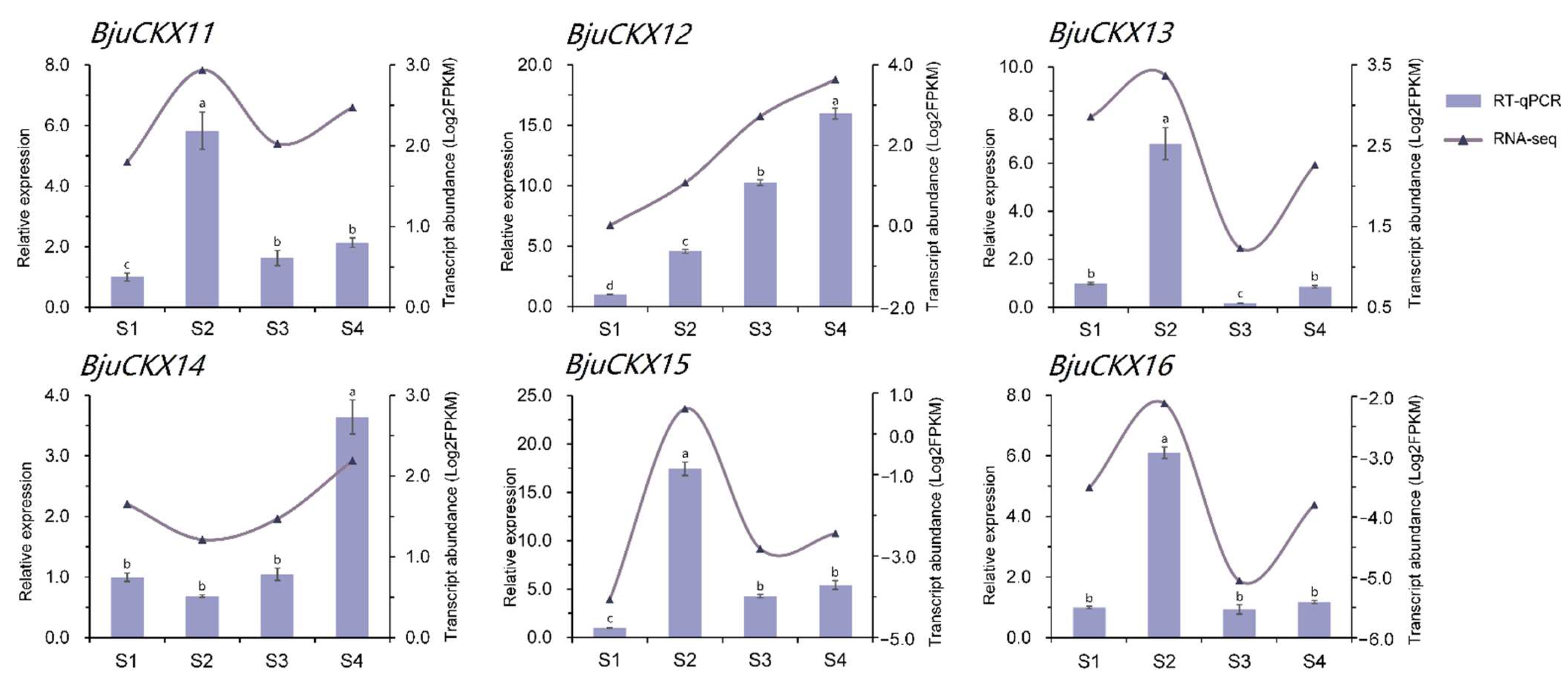
3.8. RT-qPCR Validation of CKX Gene in B. juncea var. tumida during Stem Formation
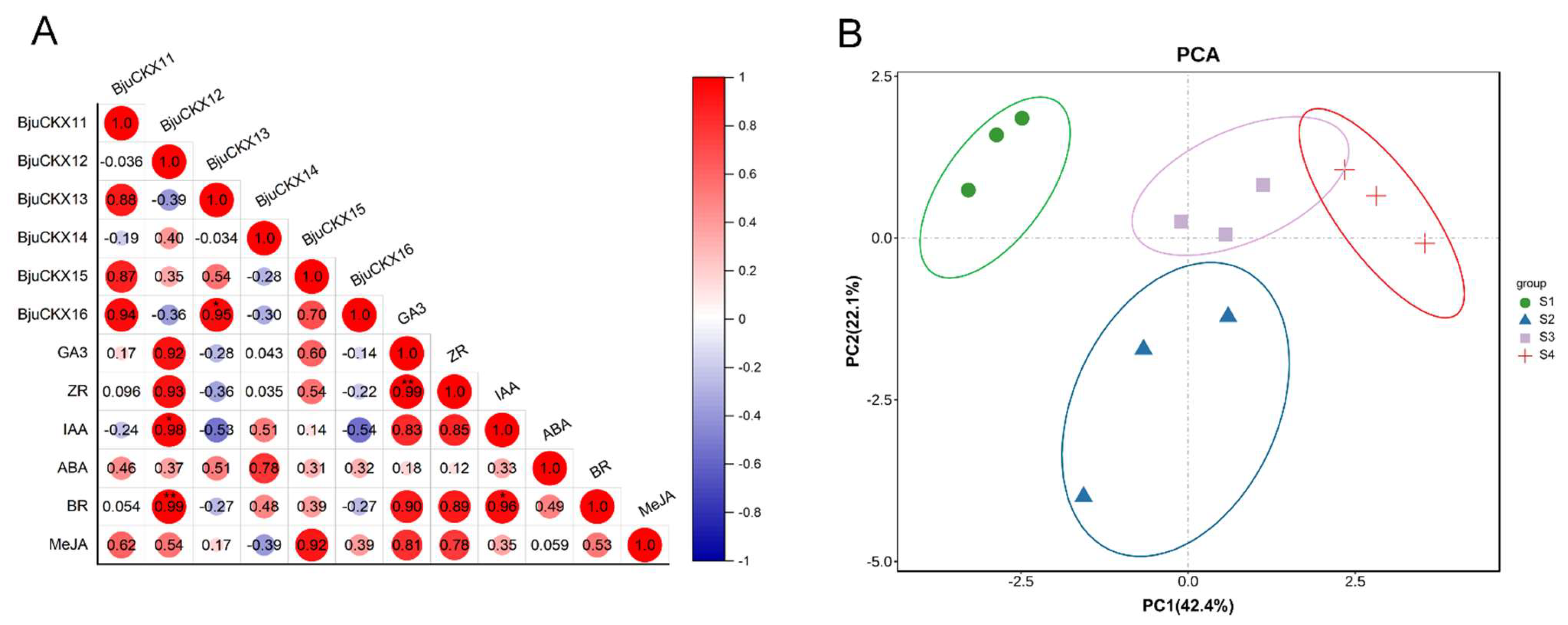
3.9. Dynamic Changes in Hormone Levels during Stem Development
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ha, S.; Vankova, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S.P. Cytokinins: Metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 2012, 17, 172–179. [Google Scholar]
- Raspor, M.; Motyka, V.; Ninkovic, S.; Malbeck, J.; Dobrev, P.I.; Zdravkovic-Korac, S.; Simonovic, A.; Cosic, T.; Cingel, A.; Savic, J.; et al. Overexpressing AtCKX1 in potato plants grown in vitro: The effects on cytokinin composition and tuberization. J. Plant Growth Regul. 2020, 40, 37–47. [Google Scholar]
- Wu, C.; Cui, K.H.; Wang, W.C.; Li, Q.; Fahad, S.; Hu, Q.Q.; Huang, J.L.; Nie, L.X.; Mohapatra, P.K.; Peng, S.B. Heat-induced cytokinin transportation and degradation are associated with reduced panicle cytokinin expression and fewer spikelets per panicle in rice. Front. Plant Sci. 2017, 8, 371. [Google Scholar]
- Zhang, W.; Peng, K.X.; Cui, F.B.; Wang, D.L.; Zhao, J.Z.; Zhang, Y.J.; Yu, N.N.; Wang, Y.Y.; Zeng, D.L.; Wang, Y.H.; et al. Cytokinin oxidase/dehydrogenase OsCKX11 coordinates source and sink relationship in rice by simultaneous regulation of leaf senescence and grain number. Plant Biotechnol. J. 2021, 19, 335–350. [Google Scholar]
- Sakakibara, H. Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar]
- Frébortová, J. Function of plant defense secondary metabolite in cytokinin degradation. Plant Signal. Behav. 2010, 5, 523–525. [Google Scholar]
- Kopecny, D.; Koncitikova, R.; Popelka, H.; Briozzo, P.; Vigouroux, A.; Kopecna, M.; Zalabak, D.; Sebela, M.; Skopalova, J.; Frebort, I.; et al. Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site. FEBS J. 2016, 283, 361–377. [Google Scholar]
- Nisler, J.; Kopecny, D.; Pekna, Z.; Koncitikova, R.; Koprna, R.; Murvanidze, N.; Werbrouck, S.P.O.; Havlicek, L.; De Diego, N.; Kopecna, M.; et al. Diphenylurea-derived cytokinin oxidase/dehydrogenase inhibitors for biotechnology and agriculture. J. Exp. Bot. 2021, 72, 355–370. [Google Scholar]
- Gupta, M.K.; Gouda, G.; Donde, R.; Vadde, R.; Behera, L. Novel cytokinin oxidase/dehydrogenase inhibitors for enhancing grain yield in crop plants and potential applications in the biotechnology industry. J. Exp. Bot. 2021, 72, 153–156. [Google Scholar]
- Wang, C.N.; Wang, H.; Zhu, H.; Ji, W.K.; Hou, Y.L.; Meng, Y.Y.; Wen, J.Q.; Mysore, K.S.; Li, X.S.; Lin, H. Genome-wide identification and characterization of cytokinin oxidase/dehydrogenase family genes in Medicago truncatula. J. Plant Physiol. 2021, 256, 153308. [Google Scholar]
- Ma, Y.Y.; Zheng, L.; Xie, R.J.; He, S.L.; Deng, L. Genome-wide identification and analysis of CKX genes in Poncirus trifoliata. J. Hortic. Sci. Biotechnol. 2016, 91, 592–602. [Google Scholar]
- Schmülling, T.; Werner, T.; Riefler, M.; Krupková, E.; Manns, I.B. Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J. Plant Res. 2003, 116, 241–252. [Google Scholar]
- Gu, R.L.; Fu, J.J.; Guo, S.; Duan, F.Y.; Wang, Z.K.; Mi, G.H.; Yuan, L.X. Comparative expression and phylogenetic analysis of maize cytokinin dehydrogenase/oxidase (CKX) gene family. J. Plant Growth Regul. 2010, 29, 428–440. [Google Scholar]
- Liu, Z.N.; Lv, Y.X.; Zhang, M.; Liu, Y.P.; Kong, L.J.; Zou, M.H.; Lu, G.; Cao, J.S.; Yu, X.L. Identification, expression, and comparative genomic analysis of the IPT and CKX gene families in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom. 2013, 14, 594. [Google Scholar]
- Liu, P.; Zhang, C.; Ma, J.Q.; Zhang, L.Y.; Yang, B.; Tang, X.Y.; Huang, L.; Zhou, X.T.; Lu, K.; Li, J.N. Genome-wide identification and expression profiling of cytokinin oxidase/dehydrogenase (CKX) genes reveal likely roles in pod development and stress responses in oilseed rape (Brassica napus L.). Genes 2018, 9, 168. [Google Scholar]
- Tsago, Y.; Chen, Z.Y.; Cao, H.; Sunusi, M.; Khan, A.U.; Shi, C.H.; Jin, X.L. Rice gene, OsCKX2-2, regulates inflorescence and grain size by increasing endogenous cytokinin content. Plant Growth Regul. 2020, 92, 283–294. [Google Scholar]
- Reid, D.E.; Heckmann, A.B.; Novak, O.; Kelly, S.; Stougaard, J. Cytokinin oxidase/dehydrogenase3 maintains cytokinin homeostasis during root and nodule development in Lotus japonicus. Plant Physiol. 2016, 170, 1060–1074. [Google Scholar]
- Khandal, H.; Gupta, S.K.; Dwivedi, V.; Mandal, D.; Sharma, N.K.; Vishwakarma, N.K.; Pal, L.; Choudhary, M.; Francis, A.; Malakar, P.; et al. Root-specific expression of chickpea cytokinin oxidase/dehydrogenase 6 leads to enhanced root growth, drought tolerance and yield without compromising nodulation. Plant Biotechnol. J. 2020, 18, 2225–2240. [Google Scholar]
- Mackova, H.; Hronkova, M.; Dobra, J.; Tureckova, V.; Novak, O.; Lubovska, Z.; Motyka, V.; Haisel, D.; Hajek, T.; Prasil, I.T.; et al. Enhanced drought and heat stress tolerance of tobacco plants with ectopically enhanced cytokinin oxidase/dehydrogenase gene expression. Plant Biotechnol. J. 2013, 64, 2805–2815. [Google Scholar]
- Nagaharu, U. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 1935, 7, 389–452. [Google Scholar]
- Xu, Z.; Wang, Q.M.; Guo, Y.P.; Guo, D.P.; Shah, G.A.; Liu, H.L.; Mao, A.N. Stem-swelling and photosynthate partitioning in stem mustard are regulated by photoperiod and plant hormones. Environ. Exp. Bot. 2008, 62, 160–167. [Google Scholar]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 384–3788. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Tae-ho, L.; Jin, H.; Barry, M.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar]
- Lescost, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar]
- Li, M.Y.; Xie, F.J.; He, Q.; Li, J.; Liu, J.L.; Sun, B.; Luo, Y.; Zhang, Y.; Chen, Q.; Zhang, F.; et al. Expression analysis of XTH in stem swelling of stem mustard and selection of reference genes. Genes 2020, 11, 113. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e4. [Google Scholar]
- Wang, G.L.; Xiong, F.; Que, F.; Xu, Z.S.; Wang, F.; Xiong, A.S. Morphological characteristics, anatomical structure, and gene expression: Novel insights into gibberellin biosynthesis and perception during carrot growth and development. Hortic. Res. 2015, 2, 15028. [Google Scholar]
- Xu, J.X.; Li, Q.Z.; Yang, L.Y.; Li, X.; Wang, Z.; Yang, Z.; Zhang, Y.C. The effect and the physiological mechanism of gibberellin and cytokinin on the regulation of Lycoris radiata bulb development. J. Plant Physiol. 2019, 55, 1029–1037. [Google Scholar]
- Nehnevajova, E.; Ramireddy, E.; Stolz, A.; Gerdemann-Knorck, M.; Novak, O.; Strnad, M.; Schmulling, T. Root enhancement in cytokinin-deficient oilseed rape causes leaf mineral enrichment, increases the chlorophyll concentration under nutrient limitation and enhances the phytoremediation capacity. BMC Plant Biol. 2019, 19, 83. [Google Scholar]
- Lysak, M.A.; Koch, M.A.; Pecinka, A.; Schubert, I. Chromosome triplication found across the tribe Brassiceae. Genome Res. 2015, 15, 516–525. [Google Scholar]
- Maere, S.; De Bodt, S.; Raes, J.; Casneuf, T.; Van Montagu, M.; Kuiper, M.; Van de Peer, Y. Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 5454–5459. [Google Scholar]
- Li, M.; Sun, B.; Xie, F.; Gong, R.; Luo, Y.; Zhang, F.; Yan, Z.; Tang, H. Identification of the GRAS gene family in the Brassica juncea genome provides insight into its role in stem swelling in stem mustard. PeerJ 2019, 7, e6682. [Google Scholar]
- Li, M.; Xie, F.; Li, Y.; Gong, L.; Luo, Y.; Zhang, Y.; Chen, Q.; Wang, Y.; Lin, Y.; Zhang, Y.; et al. Genome-wide analysis of the heat shock transcription factor gene family in Brassica juncea: Structure, evolution, and expression profiles. DNA Cell Biol. 2020, 39, 1990–2004. [Google Scholar]
- Kollmer, I.; Novak, O.; Strnad, M.; Schmulling, T.; Werner, T. Overexpression of the cytosolic cytokinin oxidase/dehydrogenase (CKX7) from Arabidopsis causes specific changes in root growth and xylem differentiation. Plant J. 2014, 78, 359–371. [Google Scholar]
- Zalabak, D.; Johnova, P.; Plihal, O.; Senkova, K.; Samajova, O.; Jiskrova, E.; Novak, O.; Jackson, D.; Mohanty, A.; Galuszka, P. Maize cytokinin dehydrogenase isozymes are localized predominantly to the vacuoles. Plant Physiol. Biochem. 2016, 104, 114–124. [Google Scholar]
- Todorova, D.; Vaseva, I.; Malbeck, J.; Trávníčková, A.; Macháčková, I.; Karanov, E. Cytokinin oxidase/dehydrogenase activity as a tool in gibberellic acid/cytokinin cross talk. Plant Biol. 2007, 51, 579–583. [Google Scholar]
- Jiang, Y.; Mi, X.N.; Lin, Y.; Wu, H.; Gu, T.T.; Ding, J.; Li, Y. Evolution and expression patterns of cytokinin oxidase genes in Fragaria vesca. Sci. Hortic. 2016, 212, 115–125. [Google Scholar]
- Li, S.X.; An, Y.R.; Hailati, S.; Zhang, J.; Cao, Y.M.; Liu, Y.S.; Geng, J.C.; Hu, T.M.; Yang, P.Z. Overexpression of the cytokinin oxidase/dehydrogenase (CKX) from medicago sativa enhanced salt stress tolerance of Arabidopsis. J. Integr. Plant Biol. 2019, 62, 374–386. [Google Scholar]
- Nemhauser, J.L.; Hong, F.; Chory, J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 2006, 126, 467–475. [Google Scholar]

| Gene Name | Accession Number | Amino Acids | Molecular Weight/Da | Isoelectric Point | Protein Hydrophobicity | Subgroup | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| BjuCKX01 | BjuB029649 | 505 | 56,097.41 | 5.75 | −0.019 | I | chlo/- |
| BjuCKX02 | BjuA046407 | 450 | 49,748.58 | 5.17 | −0.055 | I | chlo/- |
| BjuCKX03 | BjuA017172 | 502 | 55,439.79 | 5.63 | 0.019 | I | extr/- |
| BjuCKX04 | BjuB028610 | 504 | 55,806.39 | 6.23 | 0.024 | I | plas/- |
| BjuCKX05 | BjuB046928 | 524 | 58,189.70 | 5.78 | 0.016 | I | chlo/- |
| BjuCKX06 | BjuA013849 | 524 | 57,978.45 | 5.54 | 0.030 | I | chlo/- |
| BjuCKX07 | BjuO004995 | 515 | 58,506.51 | 5.62 | −0.220 | I | vacu/- |
| BjuCKX08 | BjuO009289 | 519 | 59,054.18 | 5.88 | −0.215 | I | vacu/- |
| BjuCKX09 | BjuA006752 | 1337 | 148,490.79 | 6.00 | −0.214 | I | nucl/- |
| BjuCKX10 | BjuA016599 | 411 | 46,156.67 | 6.81 | −0.201 | I | cyto = vacu/- |
| BjuCKX11 | BjuA006478 | 470 | 51,673.56 | 5.44 | −0.183 | IV | cyto/- |
| BjuCKX12 | BjuO011542 | 534 | 59,220.25 | 5.21 | −0.126 | IV | cyto/- |
| BjuCKX13 | BjuA038850 | 523 | 57,967.61 | 4.94 | −0.136 | IV | cyto/- |
| BjuCKX14 | BjuO008975 | 525 | 57,914.52 | 4.97 | −0.127 | IV | cyto/- |
| BjuCKX15 | BjuB029847 | 533 | 59,905.92 | 5.69 | −0.249 | V | vacu/- |
| BjuCKX16 | BjuA029462 | 535 | 60,082.13 | 5.56 | −0.241 | V | E.R./- |
| BjuCKX17 | BjuA029463 | 333 | 36,584.35 | 5.15 | −0.074 | V | E.R./- |
| BjuCKX18 | BjuO000929 | 507 | 57,217.29 | 6.69 | −0.257 | VI | vacu/- |
| BjuCKX19 | BjuA017827 | 535 | 60,758.18 | 7.14 | −0.260 | VI | vacu = golo/- |
| BjuCKX20 | BjuB027127 | 549 | 62,108.95 | 8.80 | −0.259 | VI | mito/- |
| BjuCKX21 | BjuA017334 | 548 | 61,990.78 | 9.10 | −0.247 | VI | mito/- |
| BjuCKX22 | BjuB017009 | 526 | 59,226.41 | 7.02 | −0.237 | VI | chlo/- |
| BjuCKX23 | BjuA010583 | 545 | 61,374.01 | 8.72 | −0.191 | VI | mito/- |
| Period | GA3 (ng/g·FW) | ZR (ng/g·FW) | IAA (ng/g·FW) | ABA (ng/g·FW) | BR (ng/g·FW) | MeJA (ng/g·FW) |
|---|---|---|---|---|---|---|
| S1 | 6.601 ± 0.816 a | 12.191 ± 1.197 a | 62.815 ± 5.003 a | 55.976 ± 3.521 b | 8.654 ± 0.656 a | 16.922 ± 1.084 a |
| S2 | 4.718 ± 0.264 b | 4.870 ± 0.728 b | 58.301 ± 7.164 a | 53.812 ± 2.329 b | 7.364 ± 0.396 b | 12.235 ± 0.901 b |
| S3 | 4.471 ± 0.229 b | 2.915 ± 0.329 c | 47.196 ± 5.811 b | 84.153 ± 7.329 a | 6.904 ± 0.310 b | 13.975 ± 1.131 b |
| S4 | 4.053 ± 0.309 b | 1.817 ± 0.219 c | 38.476 ± 2.237 b | 25.348 ± 2.466 c | 5.272 ± 0.464 c | 13.871 ± 1.026 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Zhou, J.; Gong, L.; Zhang, R.; Wang, Y.; Wang, C.; Du, X.; Luo, Y.; Zhang, Y.; Wang, X.; et al. Identification and Expression Analysis of CKX Gene Family in Brassica juncea var. tumida and Their Functional Analysis in Stem Development. Horticulturae 2022, 8, 705. https://doi.org/10.3390/horticulturae8080705
Li M, Zhou J, Gong L, Zhang R, Wang Y, Wang C, Du X, Luo Y, Zhang Y, Wang X, et al. Identification and Expression Analysis of CKX Gene Family in Brassica juncea var. tumida and Their Functional Analysis in Stem Development. Horticulturae. 2022; 8(8):705. https://doi.org/10.3390/horticulturae8080705
Chicago/Turabian StyleLi, Mengyao, Jin Zhou, Li Gong, Ran Zhang, Yan Wang, Chao Wang, Xiaoming Du, Ya Luo, Yong Zhang, Xiaorong Wang, and et al. 2022. "Identification and Expression Analysis of CKX Gene Family in Brassica juncea var. tumida and Their Functional Analysis in Stem Development" Horticulturae 8, no. 8: 705. https://doi.org/10.3390/horticulturae8080705
APA StyleLi, M., Zhou, J., Gong, L., Zhang, R., Wang, Y., Wang, C., Du, X., Luo, Y., Zhang, Y., Wang, X., & Tang, H. (2022). Identification and Expression Analysis of CKX Gene Family in Brassica juncea var. tumida and Their Functional Analysis in Stem Development. Horticulturae, 8(8), 705. https://doi.org/10.3390/horticulturae8080705








